Abstract
Human papillomavirus type 16 can integrate into the host genome, thereby rendering the viral coding genes susceptible to epigenetic modification. Using bisulfite genomic sequencing, we determined the methylation status of all 110 CpG sites within the viral epigenome in advanced stage III/IV HPV-16 associated head and neck cancers. We found that the viral genome was hypomethylated in the majority of head and neck cancers, in particular within the viral regulatory region, LCR, which controls transcription of the E6 and E7 oncogenes. The hypomethylation status of LCR correlated with detectable levels of E6 and E7 expression, which suggests the tumors may still be dependent on these viral oncogenes to maintain the malignant phenotype. In addition to the methylation status of LCR, we report other potential factors which may influence intratumoral E6 and E7 expression, including viral copy number and integration site. We were able to detect the viral epigenetic alterations in sampled body fluids, such as serum and saliva, which correlated with the changes observed in the primary tumors. Since viral epigenetic changes occurs in the setting of viral integration into the human genome, the detection of methylated HPV genes in the serum and/or saliva may have diagnostic potential for early detection strategies of viral integration and assessment of risk for cancer development in high risk individuals. Our findings also support continued targeting of the E6 and/or E7 antigens through various vaccine strategies against HPV-associated cancers.
Keywords: epigenetics, HPV-16, head and neck cancer, molecular markers
Introduction
Epigenetics describes the regulation of gene expression through heritable changes in DNA methylation and chromatin structure. DNA methylation can impact the transcription of genes by either physically impeding the binding of transcriptional proteins to the gene and/or by changing the chromatin structure to repress transcription. We now know that epigenetics plays an important role in tumorigenesis in mammals (1). DNA methylation occurs in cytosines (5-methylcytosine) that precede guanines in dinucleotide CpG sites. CpGs are asymmetrically distributed into CpG-poor regions and dense regions called “CpG islands,” which are located in the promoter regions. These CpG islands are usually unmethylated in normal cells, whereas the sporadic CpG sites in the rest of the genome are generally methylated (2). Methylation of CpG islands in promoter regions is often associated with gene silencing, and aberrant DNA methylation can occur in cancers, leading to the silencing of tumor suppressor genes (1).
Several oncogenic viruses can integrate within the host genome and become susceptible to modification by the host epigenetic machinery and, at times, can utilize the machinery to regulate its own viral gene expression. One such virus is the human papillomavirus. HPV type 16 (HPV-16) is the most common virus type associated with cervical and head and neck cancer and is present in greater than 90% of HPV-associated head and neck squamous cell carcinomas (HPV-HNSCC) (3). After viral entry into a cell, episomal HPV type 16 DNA can integrate into the host genome with resultant deletion of non-critical and regulatory viral genes. Late genes (L1 and L2) and some early genes (E1 and E2) are commonly deleted, and the viral oncogenes E6 and E7 are often the only open reading frames consistently expressed in cancer cell lines (4) and in primary HPV-associated cancers (5).
In order to evaluate the role of epigenetic alterations in HPV-induced carcinogenesis, we mapped all potential DNA methylation sites in the HPV-16 epigenome in patients with head and neck cancer. Since the long control region (LCR) is a key regulatory site for viral gene expression, we focused our analysis in this region and evaluated the methylation status of key inhibitory and activating transcriptional sites. Furthermore, we correlated the E6 and E7 expression levels to the methylation status of LCR, and also evaluated other potential factors which may influence E6 and E7 expression, such as viral copy number and integration status. We explored whether epigenetic alterations within the viral epigenome could be detected in body fluids, such as serum and saliva, which can have diagnostic significance for early detection strategies of virus integration as well as assessment of risk for cancer development in healthy individuals infected with the virus.
Materials and Methods
Cell lines, tissues, sera and saliva
Cervical cancer cell lines, CaSki (~600 copies of integrated HPV-16 DNA/genome) and SiHa (2 copies of integrated HPV-16 DNA/genome) were purchased from ATCC (Rockville, MD), and cultured as per manufacturer's instructions. Primary tumor tissue, serum, and saliva were collected from patients with HPV-positive, oropharyngeal squamous cell carcinoma (OPSCC) as determined by in-situ hybridization with an HPV-16 specific probe. Primary tumor was obtained from 22 patients with advanced stage III/IV HPV-associated OPSCC. Tissue was dissected to separate tumor from stromal elements, yielding at least 80% cancer cells. Tissue blocks were stained with hematoxylin/eosin, and tumor areas were subsequently outlined, cut by a head and neck pathologist, and processed for DNA and RNA extraction. RNA was available and extracted from nine patients. Matched saliva samples were available from 9 patients prior to any therapy. Oral rinsing was performed by gargling twice in 20 mls of saline. Matched serum samples were available from 10 patients prior to any therapy. This study was approved by the Institutional Review Board of the Johns Hopkins University and written informed consent was obtained from all patients.
DNA and RNA extraction
Tissue samples were centrifuged and digested in a solution of sodium dodecylsulfate and proteinase K at 50 °C overnight. Genomic DNA (gDNA) was isolated by phenol/chloroform extraction, and precipitated in ethanol. The DNA pellet was resuspended in TE buffer (EDTA, 2.5 mmol/l and Tris–HCl, 10 mmol/l) and stored at -20 °C. Total RNA was extracted from frozen tumor tissue using Trizol (Invitrogen, Carlsbad, CA). The RNA integrity was assessed by agarose gel electrophoresis.
Bisulfite treatment
Bisulfite-conversion of gDNA was performed using the EpiTect Bisulfite Kit (Qiagen, Valencia, CA) as per manufacturer's recommendations. This bisulfite-modified DNA was subsequently resuspended in 120 ul of Tris-EDTA buffer (EDTA, 2.5 mmol/l and Tris–HCl, 10 mmol/l) and stored at –80 °C until use.
Bisulfite-sequencing
The modified DNA was amplified in the form of 18 amplicons (Supplemental Table 1). Bisulfite-modified gDNA was amplified by PCR using 10X buffer (166 mM (NH4)2SO4, 670 mM Tris Buffer (pH 8.8), 67 mM MgCl2, 0.7% 2-mercaptoethanol, 1% DMSO) and primer sets that were designed to recognize DNA bases after bisulfite treatment. The conditions for PCR amplifications were as follows: A 5 minute incubation at 95°C was followed by 45 cycles of 1 minute at 95°C, 1 minute at 54°C, and 2 minutes at 72°C. A seven-minute elongation step at 72°C completed the PCR amplification. For amplification of some amplicons, touch-down PCR was performed as follows: a 5 minute 95°C incubation step was followed by 2 cycles of 1 min at 95°C for denaturation, 1 min at 66°C for annealing, and 1 min at 72°C for elongation. The annealing temperature was decreased by two degrees, and two PCR cycles were run each time until the annealing temperature was 56°C. The PCR was run for 35 cycles, with an additional 7 min at 72°C at the completion of the cycles for further elongation. PCR products were gel-extracted (Qiagen) and sequenced with forward and reverse primers using the ABI BigDye cycle sequencing kit (Applied Biosystems, Foster City, CA).
Methylation-specific PCR (MSP)
Bisulfite-treated DNA was amplified with methylation and unmethylation specific primer sets for each individual gene. Primer sequences are shown in Supplemental Table 2. PCR reactions were performed for 35 cycles of 95 °C for 30 sec, 58 °C for 30 sec, and 72 °C for 1 min.
Quantitative-PCR
To quantify the viral load of HPV-16, gDNA from patients was used for real-time PCR with primers and probes sets specific for the E6 and E7 region of HPV-16 DNA. PCR for β-actin was performed in parallel to normalize the input DNA. gDNA from the CaSki cell line was used to develop standard curves for the HPV-16 viral load, as it has been previously characterized to harbor 600 copies of HPV-16 DNA/genome equivalent. gDNA from CaSki cells was serially diluted to the following concentrations: 50ng, 5ng, 0.5ng, 0.05ng, and 0.005ng. A standard curve was developed for β-actin which has 2 copies/genome equivalent, using the same serial dilutions. All samples were run in triplicate. Taqman Fast Universal PCR Master Mix was used according to the manufacturer's instructions (Applied Biosystems, Foster City, CA, USA). Primer and probe sequences are shown in Supplemental Table 3.
To evaluate the viral integration status, we used a previously described real-time quantitative PCR assay (6). E2 and E6 primers and probes were synthesized using published sequences (6). The final primer and probe concentrations were 0.3 and 0.1 μM, respectively, in a total volume of 10 μl. A standard curve was obtained by amplification of a 10-fold dilution series of 0.3 to 0.0003 ng of a HPV16 plasmid clone (with a ratio of E2 to E6 1:1). At least three water controls were included in each run. All experiments were performed twice in duplicate with similar ratios. The integration status of HPV-16 DNA was assessed by comparing the levels of detected HPV-16 E2 and E6 genes and was expressed as an E2/E6 ratio. Ratios of E2 to E6 of less than 1 indicate the presence of both integrated and episomal forms. The ratio of E2 to E6 represents the amount of the episomal form in relation to the integrated form.
Quantitative RT-PCR
To examine the mRNA expression of E6 and E7, cDNA was synthesized from 1 μg of total RNA which was isolated from patients’ samples, treated with DNase I, and subsequently cleaned as recommended by the manufacturer (Qiagen). For cDNA preparation, 1μg of total RNA was transcribed with random hexamers and oligo dT using the Superscript II reverse transcriptase (Invitrogen). The same amount of RNA was processed without reverse transcriptase (RT) in the cDNA synthesis to assure that no gDNA was amplified in the reaction. cDNA from the CaSki cell line was used to develop standard curves for the E6 and E7 transcripts. β-actin was used as a loading control. All samples were run in triplicate. To examine the expression levels of E6 and E7, the mRNA levels of E6 and E7 were separately divided by the viral load within the tumor and multiplied by 10000.
Statistics
Pearson's product-moment correlation coefficients and Spearman's rank correlation coefficients were used to assess the linearity and the rank association between methylation and expression levels. P value of 0.05 was used to assess the significance of the association.
Results
The HPV-16 epigenome is hypomethylated in advanced stage HPV-related head and neck cancers
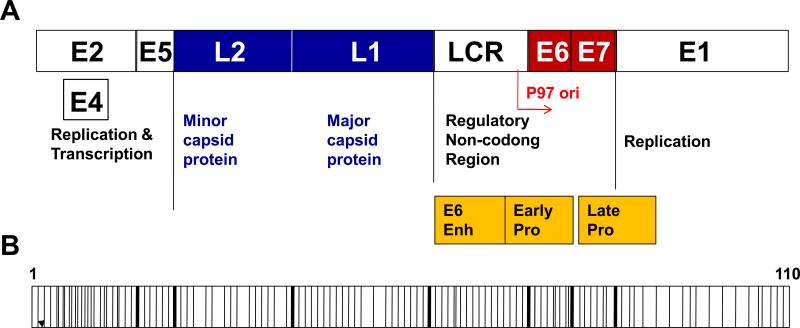
The genome and epigenome map of HPV-16 DNA is shown in Figure 1A and 1B. We identified 110 CpG sites within the HPV-16 viral methylome. We evaluated the methylation status of all 110 sites in two established invasive cervical cancer cell lines (CaSki and SiHa) and in 22 patients with advanced stage III/IV HPV-16 associated oropharyngeal squamous cell carcinoma (OPSCC) by performing bisulfite-sequencing analysis.
Figure 1. A topography of the HPV-16 genome structure.
A, The HPV-16 genome is a circular, double-stranded DNA, which is 8-kb in length. It consists of a long control region (LCR), six early genes encoding early proteins (E1, E2, E4–E7) and two late genes encoding L1 and L2. The E6 and E7 oncoproteins are essential for HPV-mediated cellular transformation. Enh, enhancer; Pro, promoter. B, A map of the CpG dinuleotides in the HPV-16 genome. Each vertical bar represents a CpG site. The thick bars indicate borders between each gene region.
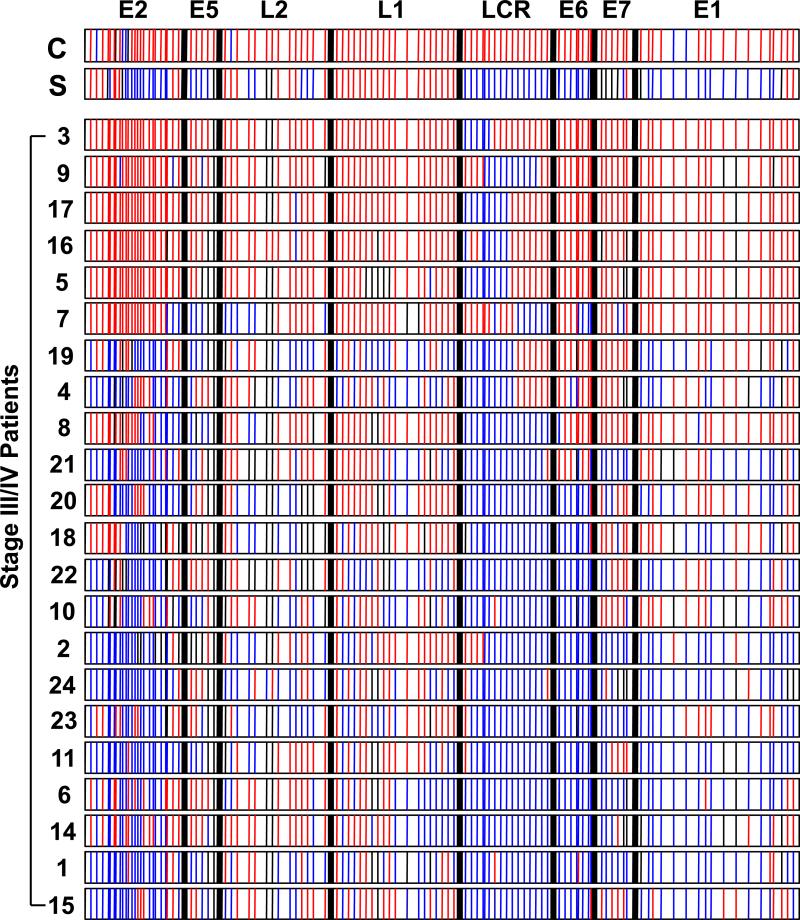
We found that the CaSki cell line harbored dense CpG methylation throughout the entire HPV-16 epigenome. Specifically, 94% of the genome was methylated and 5% of the genome was unmethylated in the E1 and E2 regions. In contrast, SiHa was methylated in 35% of the viral epigenome and 62% of the genome was unmethylated (Figure 2).
Figure 2. DNA methylation pattern of the HPV-16 genome.
Bisulfite-sequencing analysis of the HPV-16 genome was performed in two HPV-16 cell lines and 22 HPV-16 associated stage III/IV oropharyngeal squamous cell carcinomas (OPSCC). Each bar represents a CpG site, and the thick black bars indicate the borders of each gene region. A blue line indicates an unmethylated site. A red line indicates a methylated CpG site. A thin black indicates that site was unable to be assessed. C, CaSki cell line; S, SiHa cell line.
We evaluated the methylation status of the HPV-16 viral epigenome in 22 primary advanced stage head and neck cancers which consisted of one patient with stage III disease and 21 patients with stage IV disease. Interestingly, we found that the methylation pattern in primary head and neck cancers was comparable to SiHA. Specifically, 12 of the 22 (54.5%) advanced stage OPSCC were unmethylated in greater than 50% of the viral DNA CpG sites with areas lacking methylation clustered in the E2, LCR, and E6 regions. 5 of the 22 (22.7%) OPSCC were unmethylated in less than 10% of the HPV-16 epigenome with areas lacking methylation clustered within LCR (Figure 2).
The LCR region is preferentially hypomethylated in advanced stage head and neck cancers
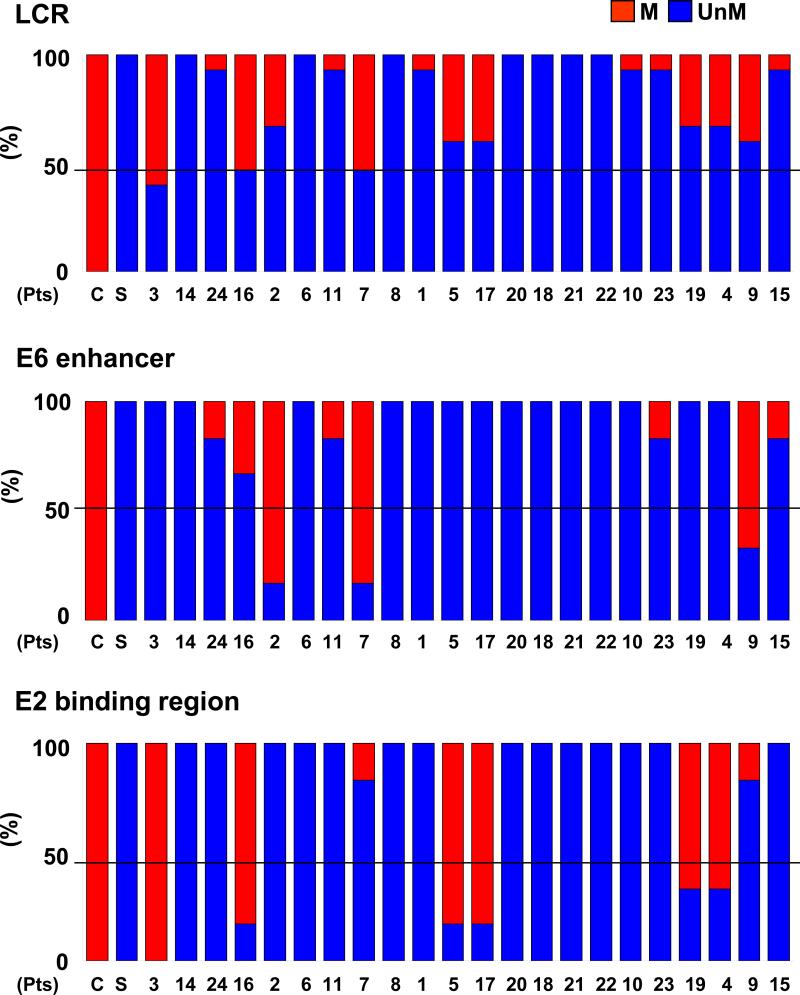
The long control region (LCR) contains the origin of replication (ori), the E6 enhancer as well as the E2 binding site (6-8). Since this region is critical in transcriptional regulation of the viral genome, we were interested in further evaluating the methylation status within this region. In the HPV cell lines, LCR was 100% unmethylated in SiHa and 100% methylated in CaSki. In advanced stage head and neck cancers, we found that 21/22 (95.5%) of the advanced stage tumors were unmethylated at or greater than 50% of the CpG sites within LCR. Interestingly, one tumor (sample #3) which was methylated in greater than 90% of the whole viral genome, was unmethylated in 40% of the CpG sites within LCR suggesting preferential hypomethylation within this viral gene regulatory region (Figure 3A).
Figure 3. Frequency of CpG methylation in the HPV-16 DNA LCR, E6 enhancer, and E2 binding site in primary head and neck cancer samples.
Methylation frequencies of CpG sites in the HPV-16 DNA LCR(A), E6 enhancer (B), and E2 binding site (C) are depicted. Blue bar represents the percent un-methylation (UnM) and the red bar represents the percent methylation (M) of CpGs sites within each respective region. C, CaSki cell line; S, SiHa cell line. Numbers represent the patient samples (Pts).
Both the E6 enhancer and the E2 binding site within LCR regulate transcription of the viral oncoproteins, E6 and E7. Therefore, we evaluated whether there was preferential methylation in either of these sites within LCR. The SiHa cell line demonstrated 100% unmethylation in both the E6 enhancer and the E2 binding site and the CaSki cell line was 100% methylated in both of these sites. We found that 19 of 22 (86%) of the advanced stage III/IV OPSCC were unmethylated in greater than 50% of the CpG sites within the E6 enhancer region (Figure 3B). Furthermore, 74% (14/19) of these tumors demonstrated 100% unmethylation in this region. For the E2 binding region, 16 of 22 (73%) of the tumors were unmethylated in greater than 50% of the CpG sites and 88% (14/16) of these tumors demonstrated 100% unmethylation within this region. Both the E6 enhancer and E2 binding region were unmethylated in greater than 50% of the CpG sites in 80% of the advanced stage cancers.
9 of 22 (41%) of the OPSCC demonstrated methylation in greater than 50% of the CpG sites at either the E6 enhancer region or the E2 binding site (Figure 3B and C). Interestingly, the tumors which were heavily methylated in the E2 binding site (samples #3, 16, 5, 17, 19 and 4) were unmethylated in the E6 enhancer region and those tumors which were heavily methylated in the E6 enhancer region (samples # 2, 7, and 9) were unmethylated in the E2 binding site. Our results demonstrate that the E6 enhancer region and/or the E2 binding site are hypomethylated in the majority of advanced stage OPSCC.
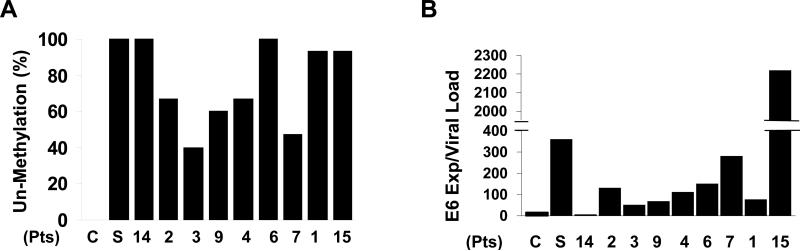
The methylation status within LCR correlates with E6 and E7 expression levels
LCR contains the E6 enhancer and the E2 binding site both of which regulate viral E6 and E7 expression. Therefore, we evaluated whether the methylation status of LCR correlated with the expression levels of E6 and E7 in the cell lines as well as nine primary tumor samples for which RNA was available. E6 and E7 expression levels were normalized to β-actin. The CaSki cell line, which was 100% methylated in LCR, expressed 16 copies of E6 per viral load (Figure 4A and B). SiHa, which was 100% unmethylated in LCR, expressed 361 copies of E6 per viral load. For the primary head and neck tumor samples, the majority of the cancers demonstrated a methylation pattern comparable to SiHa and were unmethylated in greater than 50% of the CpG sites in LCR. E6 expression levels in the primary samples ranged from 7 to 2220 copies of E6/viral load. In 78% (7 of 9) of these tumors, E6 expression ranged between 51-282 copies per viral load which is a higher level than CaSki but lower than SiHa (Figure 4B). One patient (#15) expressed 2220 copies of the E6/viral load and one sample (#14) expressed 7 copy of the E6/viral load. Interestingly, both of these patient samples were unmethylated in 93-100% of the CpG sites in LCR. Since the E2 binding site also regulates E7 expression, we performed the same analysis with E7 expression and found similar correlations between methylation status and expression (Figure 4A and Supplemental Figure 1).
Figure 4. Methylation frequency of HPV-16 LCR and expression of E6.
A, The unmethylation frequencies of CpG sites in the HPV-16 DNA LCR region is depicted. B, Real-time PCR was performed with cDNA from 9 OPSCC patients. Relative expression of E6/viral load was calculated by comparing the ratios of mRNA expression of E6 to viral load.
Although there was a trend between methylation status of LCR and E6 and E7 expression, there was no statistical significance using Pearson correlation coefficients or Spearman's rank correlation coefficients. The lack of statistical significance may be attributed to the low number of available samples (N=9) which could be evaluated. However, other factors may also be contributing to E6 and E7 expression and we explored these other potential confounding factors.
There is a variable viral load within head and neck cancers
It has been previously published that the CaSki cell line harbors between 500-600 copies of integrated HPV-16 DNA in more than 11 chromosomal sites of the hyperdiploid genome. However, the CaSki cell line has only one active papillomaviral transcriptional center per cell that maps to a low tandem copy integration site at chromosome 14. SiHa has 2 integrated copies of HPV-16 DNA at the 13q21 locus of the homologous chromosomes and both viral copies are transcriptionally active. Based on this observation, we evaluated the viral load in the primary cancers to determine if the viral copy number could influence the expression levels of E6 and E7 in the primary tumors. The CaSki cell line served as our reference for calculating the viral copy number in our tumors. In our study, we found that CaSki contained 613 copies of HPV-16 DNA per genome and SiHa 6 copies of HPV-16 DNA per genome. 78% (7 of 9) of the primary head and neck cancers contained integrated viral copy numbers which ranged between 24 to 283 copies of HPV-16 DNA per genome (Supplemental Figure 2). Outliers included patient sample #15 which harbored 3 copies of HPV-16 DNA and patient sample #14 which harbored 1866 integrated viral copies.
A mixture of episomal and integrated forms of the virus are present in OPSCC
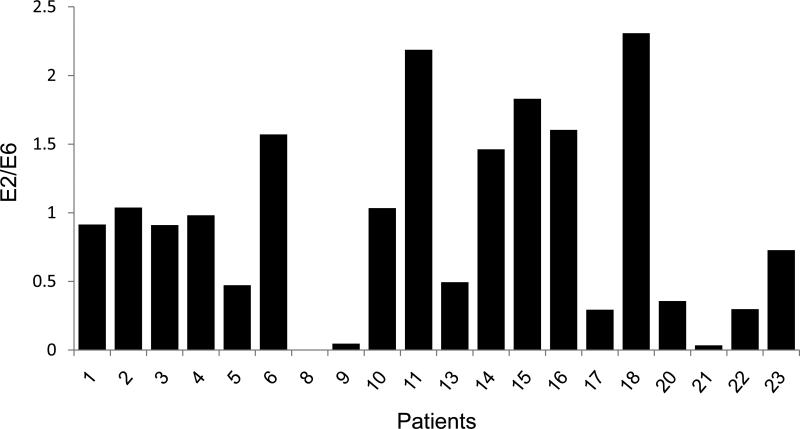
HPV DNA can exist in multiple forms within the genome, including integrated and episomal forms. Therefore, we assessed the ratio of integrated to episomal forms of the viral DNA to determine if this factor could affect E6 and E7 expression, independent of methylation status. Current assays measuring HPV-16 integration are based on quantification with real-time PCR of HPV-16 E6 relative to E2 DNA since the E2 gene is often lost during the viral integration process (6). Therefore, detection of a greater quantity of HPV-16 E6 compared to E2 suggests the presence of integrated HPV-16 DNA. We were able to assess the viral integration status in 20 of 22 of the OPSCC cases based on the availability of the tumor samples (Figure 6). We found there were 8/20 (40%) tumors which contained a E2/E6 ratio greater than 1.0, suggesting predominance of the episomal forms of the virus, within the tumor (Figure 6). There were 7/20 (35%) tumors with a E2/E6 <0.5, suggesting predominance of the integrated forms of the virus. Despite the lack of episomal forms of the virus within these tumors, there was significant hypomethylation detected within LCR (Figures 3 and 6), suggesting that there is preferential hypomethylation within this region in integrated forms of the viral DNA.
Figure 6. Assessment of Viral Integration.
The ratio of E2 to E6 was measured for 20 HPV-16 associated OPSCC using real-time quantitative PCR. An E2/E6 ratio of 0.5 indicates a mixture of integrated and episomal forms of detected HPV DNA. Values of greater than 1 indicate predominance of the episomal form. Numbers represent the patient samples.
Methylation status within LCR is detected in serum and saliva
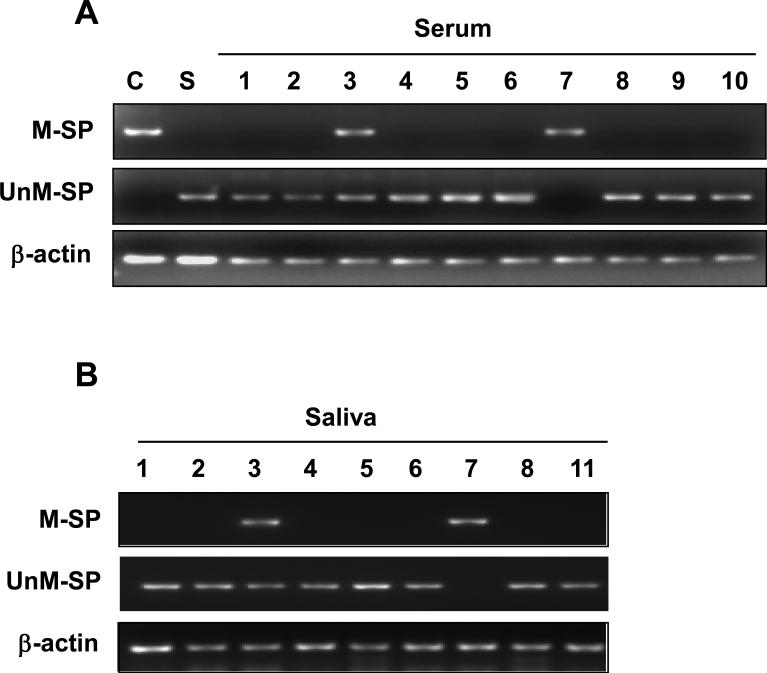
We evaluated whether the HPV-16 DNA methylation status of LCR was detectable in the serum and saliva of OPSCC patients. Therefore, we performed methylation-specific PCR (MSP) with primers designed in LCR of HPV-16 DNA. Using these primers, we were able to detect a methylated allele in CaSki and an unmethylated allele in SiHa. An unmethylated DNA allele was observed in all of the serum and saliva samples from OPSCC patients tested except for case #7 in which greater than 50% of the CpG sites was methylated in this region (Figures 5 and 3). Within the primary tumor #3, LCR was methylated in 60% of the CpG sites and both unmethylated and methylated alleles were detected in the serum and saliva using this assay (Figures 5 and 3). Our results demonstrate that the LCR methylation status within the primary tumor is detectable in the saliva and serum of advanced stage HPV-16 associated OPSCC and can serve as a biomarker for viral DNA integration.
Figure 5. Methylation-specific PCR analysis of the HPV-16 DNA.
The methylation status of LCR in serum (A) and saliva (B) was examined by MSP after PCR with methylation-specific (M-SP) and un-methylation-specific (UnM-SP) primers. Bisulfite-treated DNA from CaSki and SiHa were used as controls. β-actin was used to confirm the integrity of the bisulfite-treated DNA. Tumor sample #3 contained both methylated and unmethylated forms of viral DNA which was detected in this assay in both the serum and saliva. Tumor sample #7 contained greater than 70% methylation within LCR which was detected as a single methylated band in the serum and saliva. C, CaSki cell line; S, SiHa cell line. Numbers represent the patient samples.
Discussion
The HPV-16 genome consists of an 8-kb circular, double-stranded DNA, which encodes six early expressed regulatory genes (E1, E2, E4–E7) and two late expressed structural genes, L1 and L2. These two sets of viral coding regions are separated by a long control region (LCR) that contains the origin of replication (ori), P97, and non-coding cis-elements such as the E2 binding site (E2BS), E6 enhancer, and promoter regions (7-9). Binding of the E2 protein to the E2BS inhibits transcription factors from docking onto LCR and, thus, represses the transcription of the viral oncogenes, E6 and E7 (10, 11). Expression of the E6 and E7 proteins are essential to HPV-mediated transformation due to their binding and inhibition of the cellular gatekeepers, p53 and Rb proteins, which can result in dysregulation of a variety of cellular processes, including cell cycle control (12). The integration of the viral DNA into the chromosome of mammalian cells, as well as certain genetic or epigenetic alterations of the viral genome, can lead to loss of E2-mediated inhibition of E6 and E7 expression, which is a key event in papillomaviral carcinogenesis.
Integration of the viral DNA into the host genome, although requisite for carcinogenesis, makes it vulnerable to modification by the host's methylation machinery. This phenomenon is highlighted when cultured primary human foreskin keratinocytes are transfected with HPV-16 DNA (13, 14). In the pre-immortal keratinocytes, the episomal HPV-16 DNA remains in an unmethylated state. However, the immortal descendant cells with integrated HPV-16 DNA acquire a densely methylated viral genome. Several hypotheses exist to explain this observation both in vitro and in vivo. Some groups suggest that the host cell may actively be silencing the foreign genome as a cellular defense mechanism to inhibit the expression of non-self proteins which can disrupt normal cellular function. Other groups have suggested that the HPV-16 genome may not be a passive spectator in this process, but might actively participate by recruiting DNA methyltransferases (DNMTs) and/or histone deacetylases (HDACs) via the viral oncoprotein, E7, to strategically regulate viral gene expression during the viral life cycle in order to evade immune recognition by the host (15, 16).
In human cancers, methylation of oncogenic viral DNA may occur due to a combination of these factors. DNA methylation impacts the transcription of genes by either physically impeding the binding of transcriptional proteins to the gene and/or altering the chromatin structure to repress transcription. We know that alterations in methylation patterns play an important role in tumorigenesis in mammals (1). A hallmark of cancer is a paradoxical aberration of normal DNA methylation patterns, with a global loss of DNA methylation that coexists with regional hypermethylation of certain genes. For example, the hypermethylation of tumor suppressor genes has attracted significant attention, and DNA methylation inhibitors are being tested as potential anticancer agents. However, emerging data also suggests that hypomethylation can play a role in activating pro-oncogenic genes which may be required for metastasis and invasion. It has been proposed that hypermethylation and hypomethylation in cancer are independent processes, which target different cellular programs at different stages of cancer development. Therefore, evaluating the methylation status at specific sites, as opposed to a more global genomic assessment, may yield more valuable and predictive information regarding cancer progression.
This concept is exemplified in studies performed with the HPV-16 methylome in cervical lesions. Fernandez et al evaluated the DNA methylome of the HPV-16 virus in a collection of human cervical samples at different progressive stages of disease. They found that the DNA methylome evolved from an unmethylated to a highly methylated state in association with disease progression, from asymptomatic healthy carriers, through chronically infected tissues and pre-malignant lesions, to the development of invasive cervical cancer. Thus, the progression to cancer was associated with increasing numbers of methylated CpGs and increasing proportions of methylated HPV-16 genomes, which has been supported by other studies (17-19).
However, other groups have evaluated the methylation status of specific regions within the HPV-16 methylome, which has resulted in conflicting data. Badal et al evaluated 81 patients with HPV-16 associated cervical lesions and they found that the LCR and E6 genes of HPV-16 DNA were hypermethylated in 52% of asymptomatic smears, 21.7% of precursor lesions, and 6.1% of invasive carcinomas (17). Bisulfite modification and sequencing analysis revealed that in most of the HPV-16 genomes of the CaSki cell line and asymptomatic patients, all 11 CpG dinucleotides that overlap with the enhancer and the promoter regions in LCR were methylated, while in the SiHa cells and cervical lesions, the same subset of CpGs remained unmethylated (17). These results suggest that neoplastic transformation may be suppressed by CpG methylation, while hypomethylation seems to correlate with neoplastic progression. In contrast, Kalantari et al evaluated 115 cervical samples to establish the methylation patterns of 19 CpG dinucleotides within LCR and the L1 gene by bisulfite modification and sequencing and reported that methylation of most sites was highest in carcinomas and methylation was lowest in dysplasia (18).
The discrepancy between these studies may be attributed to the differing methylation sites evaluated with LCR. LCR has binding sites for both inhibitory and activating transcription factors. For example, transcription starts at the E6 promoter, P97, which is regulated by one binding site for Sp1 and two binding sites for the inhibitory HPV-encoded E2 protein (20, 21). The activity of P97 is stimulated by an enhancer with binding sites for several cellular factors, including AP1 (22), NF1 (23), and the progesterone receptor (24). Other factors which have been reported to be able to bind the LCR region are HIV tat-1 (25), YY1 (26), Octa (27), and TEF-2 (27). In addition, two specifically positioned nucleosomes can form over the enhancer and promoter regions (28) to repress transcription when they are modified by HDACs. Depending upon which of these sites one analyzes, correlations between methylation status and tumor progression might be directly or inversely related.
Due to the wide variance in the literature regarding HPV-associated cervical lesions, we were interested in characterizing the methylation status of the HPV-16 viral genome in HPV-associated head and neck cancers (HPV-HNSCC). We performed bisulfite sequencing analysis of all 110 CpG sites within the HPV-16 genome in 22 patients with advanced stage III/IV HPV-16 associated HNSCC. In contrast to the cervical lesions, we found that the majority of these advanced stage patients had significant hypomethylation of the viral epigenome, especially within LCR which is a regulatory region for viral oncogene expression.
Due to the various regulatory sites within LCR, we further investigated this control region by focusing on the methylation status of the inhibitory E2BS as well as the activating E6 enhancer region. We found that the E6 enhancer region was hypomethylated in advanced stage head and neck cancers, which facilitates binding of transcription factors to this region and, thus, allows for overexpression of the viral oncogenes. Interestingly, we found that the E2BS was also significantly hypomethylated. Previous studies reported that methylation of CpG dinucleotides within the binding site of HPV-16 E2 protein in LCR can directly inhibit the binding of E2 to the cognate DNA sequences in vitro; therefore, hypomethylation in this region would facilitate binding of E2 to the E2BS and, subsequently, inhibit E6 and E7 expression, which is contrary to what one would expect in advanced stage cancers. However, an explanation for our findings might be found when reviewing the events which occur upon viral integration into the host genome. Specifically, the E2 gene is often disrupted or lost upon viral DNA integration. Without the inhibitor influence of the E2 protein, the selective pressure to methylate this site is lost, which is consistent with our findings of hypomethylation of this region in the primary tumors (29).
We found a trend between the methylation status of LCR, E6 enhancer and E2BS to levels of E6 and E7 expression within the tumors. However, a statistical significance was not found which could be attributed to the low number of tumor samples available for the analysis. In addition, other potential factors may influence E6 and E7 expression levels including the integrated viral copy number. Experimental transformation of keratinocytes with HPV type 16 reveals a consistent tendency toward reducing the number of transcriptionally active HPV genomes to one or two during the passages and re-activation of the silent viral copies can occur by growth in the DNA methylation inhibitor 5-azacytidine (14). The silencing of the redundant copies may be advantageous as well as critical in clonal selection during carcinogenesis. One could speculate that limiting the number of actively transcribed viral oncogenes can prevent the accumulation of excess genomic instability of the host genome that could affect the growth and survival of the transformed cell. Our findings suggest this exact phenomenon may be occurring in vivo. We found that the majority of head and neck tumors harbored between 24-283 integrated viral copies/genome, and expressed between 51-282 copies of E6/viral load and 5-249 copies of E7/viral load. However, we found one patient who harbored 1866 copies of HPV-16 DNA, but expressed only 7 copies of the E6/viral load and 5 copies of the E7/viral load. In contrast, a patient who harbored 3 copies of HPV-16 DNA, expressed 2220 copies of E6/viral load and 1530 copies of E7/viral load. We can hypothesize that cancers which contain a low number of viral genomes must keep them in an unmethylated form for continued oncogene transcription and maintenance of the carcinogenic phenotype; whereas, those tumors with significant numbers of viral copies may limit the number of transcribed viral oncogenes to maintain cell survival. Support for this hypothesis is found in a study by Cohen et al which evaluated the viral load in 35 HPV-16 associated HNSCC. They found that the patients with the highest viral loads had improved overall and disease-free survival (30). This finding is counter-intuitive; however, based on our study, we propose that those tumors with the highest viral loads could have significant silencing of the redundant copies through methylation and correspondingly low E6 and E7 expression levels, which could account for improved survival rates.
An alternative factor which may influence E6 and E7 expression, in addition to viral copy number and viral methylation status, may be the site of integration of the viral genome, which may be permissive to either high or low levels of viral gene transcription. Over 200 selected HPV-16 and HPV-18 integration sites have been reported. These are widely distributed across the genome; however, there seems to be preferential integration near common fragile sites (CFS), specific chromosomal loci that are particularly prone to forming double-strand breaks (31-32). It has been reported that CaSki harbors between 500-600 integrated copies of HPV-16 DNA. CaSki was found to be significantly methylated with only one active papillomaviral transcriptional center per cell, which maps to a low tandem copy integration site at chromosome 14. SiHa is significantly unmethylated and has 2 integrated copies of HPV-16 DNA at the 13q21 locus of the homologous chromosomes and both viral copies are transcriptionally active. Therefore, for patients with low viral copy numbers, such as our patient with 3 viral copies, it may have integrated into a site which is permissive for the high levels of E6 and E7 expression.
A limitation to our study is that the current methodology does not allow the selective detection of the methylation pattern for the transcriptionally active copies of the viral genome among the inactive ones. Rather, our assay reflects the cumulative status of all of the viral genomes, both active as well as inactive, within the tumor. Despite this limitation, our data demonstrates that we are able to detect certain regions within the viral epigenome that are more likely to be methylated or unmethylated and there is a trend toward hypomethylation of LCR with corresponding detectable levels of E6 and E7 expression in advanced stage OPSCC. Another limitation to the study is the lack of evaluation of early stage disease which would allow us to determine how the HPV-16 DNA methylation status may evolve with progression of disease and/or stage. Since HPV-HNSCC localizes to the tonsil and base of tongue which are areas of the head and neck that are more difficult to routinely evaluate without a directed physical examination, HPV-HNSCC patients are typically diagnosed after lymphatic spread to the cervical nodes, resulting in a diagnosis at an advanced stage of disease. Thus, we were not able to evaluate the HPV-16 methylation status of early stage I or II HPV-HNSCC because patients rarely present with these early stage lesions and tissue was not available for analysis.
The implications of our findings are several-fold. We report that HPV-related head and neck cancers have regulated mechanisms of methylation of the viral epigenome. Our findings demonstrate that LCR is preferentially hypomethylated in the majority of advanced stage HPV-16 associated head and neck cancers. Hypomethylation of LCR corresponds to detectable expression levels of E6 and E7 within head and neck cancers, supporting the feasibility of targeting these antigens for novel immunotherapeutic strategies (34, 35). We demonstrate the feasibility of detecting methylated HPV genes in body fluids such as serum and saliva. Since viral epigenetic changes occurs only in the setting of viral integration into the human genome, the detection of methylated HPV genes in the serum and/or saliva may help to identify the presence of viral integration as compared to episomal forms, and can serve as a biomarker for HPV integration and may potentially allow assessment of risk for cancer development in high risk individuals. Further studies which distinguish preferential sites of methylation within the HPV epigenome may be relevant and complement current assays for HPV integration.
Supplementary Material
Supplemental Figure 1. Expression levels of E7/viral load within the tumors. Real-time PCR was performed with cDNA from 9 OPSCC patients. Relative expression of E7/viral load was calculated by comparing the ratios of mRNA expression of E7 to viral load. Numbers represent the patient samples.
Supplemental Figure 2. HPV-16 viral copy number within the tumors. Real-time PCR was performed with gDNA from 9 OPSCC patients. gDNA from the CaSki cell line was used to develop standard curves for the HPV-16 viral load. The viral load was calculated by multiplying by 2000 to normalize against CaSki which harbors 600 copies of HPV-16 DNA/genome equivalent. Experiments were done in triplicate, and repeated twice. C, CaSki; S, SiHa. Numbers represent the patient samples.
Acknowledgments
Grant Support: NIH P50 CA19032 (WW, DS, SIP) and The Milton J. Dance, Jr. Head and Neck Center at the Greater Baltimore Medical Center (JRS, SIP).
Footnotes
No potential conflict of interest to disclose.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 3.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:675–7. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 4.Nasseri M, Gage JR, Lorincz A, Wettstein FO. Human papillomavirus type 16 immortalized cervical keratinocytes contain transcripts encoding E6, E7, and E2 initiated at the P97 promoter and express high levels of E7. Virology. 1991;184:131–40. doi: 10.1016/0042-6822(91)90829-z. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz E, Freese UK, Gissmann L, et al. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. 1985;314:111–4. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 6.Peitsaro P, Johansson B, Syrjanen S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol. 2002;40:886–91. doi: 10.1128/JCM.40.3.886-891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butel JS. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405–26. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- 8.Zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–50. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 9.Gatza ML, Chandhasin C, Ducu RI, Marriott SJ. Impact of transforming viruses on cellular mutagenesis, genome stability, and cellular transformation. Environ Mol Mutagen. 2005;45:304–25. doi: 10.1002/em.20088. [DOI] [PubMed] [Google Scholar]
- 10.Frattini MG, Hurst SD, Lim HB, Swaminathan S, Laimins LA. Abrogation of a mitotic checkpoint by E2 protines from oncogenic human papillomaviruses correlates with increased turnover of the p53 tumor suppressor protein. EMBO J. 1997;16:318–31. doi: 10.1093/emboj/16.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howley PM, Münger K. Human papillomaviruses and squamous cell carcinomas. In: Parsonnet J, editor. Microbes and Malignancy: Infection as a Cause of Human Cancers. Oxford University Press; Oxford, UK: 1999. pp. 157–179. [Google Scholar]
- 12.Wise-Draper TM, Wells SI. Papillomavirus E6 and E7 proteins and their cellular targets. Front Biosci. 2008;13:1003–17. doi: 10.2741/2739. [DOI] [PubMed] [Google Scholar]
- 13.Steenbergen RD, Walboomers JM, Meijer CJ, et al. Transition of human papillomavirus type 16 and 18 transfected human foreskin keratinocytes towards immorality: activation of telomerase and allele losses at 3p, 10p, 11q and/or 18q. Oncogene. 1996;13:1249–57. [PubMed] [Google Scholar]
- 14.Van Tine BA, Kappes JC, Banerjee NS, et al. Clonal selection for transcriptionally active viral oncogenes during progression to cancer. J Virol. 2004;78:11172–86. doi: 10.1128/JVI.78.20.11172-11186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgers WA, Blanchon L, Pradhan S, de Launoit Y, Kouzarides T, Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–5. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B, Laribee RN, Klemsz MJ, Roman A. Human papillomavirus type 16 E7 protein increases acetylation of histone H3 in human foreskin keratinocytes. Virology. 2004;329:189–98. doi: 10.1016/j.virol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Badal V, Chuang LS, Tan EH, et al. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression. J Virol. 2003;77:6227–34. doi: 10.1128/JVI.77.11.6227-6234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalantari M, Calleja-Macias IE, Tewari D, et al. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. J Virol. 2004;78:12762–72. doi: 10.1128/JVI.78.23.12762-12772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandsma JL, Sun Y, Lizardi PM, et al. Distinct human papillomavirus type 16 methylomes in cervical cells at different stages of premalignancy. Virology. 2009;389:100–7. doi: 10.1016/j.virol.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan SH, Leong LE, Walker PA, Bernard HU. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J Virol. 1994;68:6411–20. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Carrance A, Theirry F, Yaniv M. Interplay of viral and cellular proteins along the long control region of human papillomavirus type 18. J Virol. 1988;62:4321–30. doi: 10.1128/jvi.62.11.4321-4330.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan WK, Chong T, Bernard HU, Klock G. Transcription of the transforming genes of the oncogenic human papillomavirus-16 is stimulated by tumor promoters through AP1 bindings sites. Nucleic Acids Res. 1990;18:763–9. doi: 10.1093/nar/18.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apt D, Chong T, Liu Y, Bernard HU. Nuclear factor I and epithelial cell-specific transcription of human papillomavirus type 16. J Virol. 1993;67:4455–63. doi: 10.1128/jvi.67.8.4455-4463.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong T, Chan WK, Bernard HU. Transcriptional activation of human papillomavirus 16 by nuclear factor I, AP1, steroid receptors and a possibly novel transcription factor, PVF: a model for the composition of genital papillomavirus enhancers. Nucleic Acids Res. 1990;18:465–70. doi: 10.1093/nar/18.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vernon SD, Hart CE, Reeves WC, Icenogle JP. The HIV-1 tat protein enhances E2-dependent human papillomavirus 16 transcription. Virus Res. 1993;27:133–45. doi: 10.1016/0168-1702(93)90077-z. [DOI] [PubMed] [Google Scholar]
- 26.O'Connor MJ, Tan SH, Tan CH, Bernard HU. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J Virol. 1996;70:6529–39. doi: 10.1128/jvi.70.10.6529-6539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong T, Apt D, Gloss B, Isa M, Bernard HU. The enhancer of human papillomavirus type 16: binding sites for the ubiquitous transcription factors oct-1, NFA, TEF-2, NF1, and AP-1 participate in epithelial cell-specific transcription. J Virol. 1991;65:5933–43. doi: 10.1128/jvi.65.11.5933-5943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stunkel W, Bernard HU. The chromatin structure of the long control region of human papillomavirus type 16 represses viral oncoprotein expression. J Virol. 1999;73:1918–30. doi: 10.1128/jvi.73.3.1918-1930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thain A, Jenkins O, Clarke AR, Gaston K. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J Virol. 1996;70:7233–5. doi: 10.1128/jvi.70.10.7233-7235.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen MA, Basha SR, Reichenbach DK, Robertson E, Sewell DA. Increased viral load correlates with improved survival in HPV-16-associated tonsil carcinoma patients. Act Otolaryngol. 2008;128:583–9. doi: 10.1080/00016480701558880. [DOI] [PubMed] [Google Scholar]
- 31.Dall KL, Scarpini CG, Roberts I, et al. Characterization of naturally occurring HPV16 integration sites isolated from cervical keratinocytes under noncompetitive conditions. Cancer Res. 2008;68:8249–59. doi: 10.1158/0008-5472.CAN-08-1741. [DOI] [PubMed] [Google Scholar]
- 32.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–92. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 33.Thorland EC, Myers SL, Persing DH, et al. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 2000;60:5916–21. [PubMed] [Google Scholar]
- 34.Best SR, Peng S, Juang CM, et al. Administration of HPV DNA vaccine via electroporatino elicits the strongest CD8+ T cell immune response compared to intramuscular injection and intradermal gene gun delivery. Vaccine. 2009;27:5450–9. doi: 10.1016/j.vaccine.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu AA, Niparko KJ, Pai SI. Immunotherapy for head and neck cancer. J Biomed Sci. 2008;15:275–89. doi: 10.1007/s11373-008-9247-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Expression levels of E7/viral load within the tumors. Real-time PCR was performed with cDNA from 9 OPSCC patients. Relative expression of E7/viral load was calculated by comparing the ratios of mRNA expression of E7 to viral load. Numbers represent the patient samples.
Supplemental Figure 2. HPV-16 viral copy number within the tumors. Real-time PCR was performed with gDNA from 9 OPSCC patients. gDNA from the CaSki cell line was used to develop standard curves for the HPV-16 viral load. The viral load was calculated by multiplying by 2000 to normalize against CaSki which harbors 600 copies of HPV-16 DNA/genome equivalent. Experiments were done in triplicate, and repeated twice. C, CaSki; S, SiHa. Numbers represent the patient samples.