Abstract
Estimates are that double-strand breaks (DSBs) arise in dividing cells about ten times per cell per day. Causes include replication across a nick, free radicals of oxidative metabolism, ionizing radiation, and inadvertent action by enzymes of DNA metabolism (such as failures of type II topoisomerases or cleavage by recombinases at pseudo sequences that look sufficiently similar to the physiologic ones). There are two major double-strand break repair pathways. Homologous recombination (HR) can repair double-strand breaks, but only during S phase and only if there is sufficient homology, usually more than 100 bp. The more general and commonly used pathway is nonhomologous DNA end joining, abbreviated NHEJ. NHEJ can repair a DSB at any time during the cell cycle and does not require any homology, though a few nucleotides of terminal microhomology are often utilized by the NHEJ enzymes, if present. The enzymes of NHEJ include Ku, DNA-PKcs, Artemis, polymerase mu, polymerse lambda, XLF (also called Cernunnos), XRCC4, and DNA ligase IV. These enzymes constitute what some call the classical NHEJ pathway, and in wild type cells, the vast majority of joining events appear to proceed using these components. In rare mammalian cell mutants of ligase IV, for example, much less efficient joining occurs using one of the remaining two ligases, ligase I or III, in a manner that is much more reliant on longer terminal microhomology lengths, such as 7 to 15 nt; this is usually called alternative NHEJ, back-up NHEJ, or microhomology-mediated end joining (MMEJ). NHEJ is present in many prokaryotes, as well as all as eukaryotes, and very similar mechanistic flexibility evolved both convergently and divergently. When two double-strand breaks occur on different chromosomes, then the rejoining is almost always done by NHEJ. The causes of the DSBs in lymphomas most often involve the RAG or AID enzymes that function in the specialized processes of antigen receptor gene rearrangement.
Keywords: double-strand DNA breaks, DNA repair, DNA recombination, NHEJ, Ku, Artemis, DNA-PKcs, DNA-dependent protein kinase, polymerase mu, polymerase lambda, XLF, Cernunnos, XRCC4, DNA ligase IV, incompatible DNA ends, polymerase slippage, template-independent synthesis
1. FREQUENCY AND CAUSES OF DOUBLE-STRAND BREAKS
In dividing primary initial passage mammalian fibroblasts, estimates are that there are about ten double-strand breaks (DSBs) per day per cell, based on metaphase chromosome and chromatid breaks (Lieber et al., 2003, Lieber and Karanjawala, 2004, Martin et al., 1985). Estimates in nondividing cells are difficult to make because methods for assessing DSBs outside of metaphase are subject to even more caveats of interpretation.
The causes of double-strand breaks in wild type cells include replication across a nick, which would give rise to chromatid breaks during S phase (Fig. 1). A second cause is oxidative free radicals. During the course of normal oxidative respiration, mitochondria convert about 0.1 to 1% of the oxygen to superoxide (O2-) (Chance et al., 1979), and superoxide dismutase in the mitochondrion (SOD2) or cytosol (SOD1) can convert this to hydroxyl free radical, which ma react with DNA to cause single-strand breaks. It is not entirely clear whether one free radical can trigger a DSB, but clearly two closely space lesions on anti-parallel strands can cause a DSB. About 1022 free radicals are produced in the human body each hour.
Figure 1. Diagram of the Causes and Known Repair Pathways for Double-Strand Break Repair.
Four prominent causes of pathologic double-strand breaks in living cells are listed. Among multicellular eukaryotes, physiologic double-strand breaks are found only the vertebrate immune system. V(D)J recombination is present in all true vertebrates and is initiated by an endonucleases complex composed of RAG1 and 2 (Lieber et al., 2006). Class switch recombination is present in only a subset of these vertebrates and is initiated by cytidine deaminase called activated-induced deaminase (AID). DSBs that arise in late S or G2 of the cell cycle are often repaired at long regions (> 100 bp) of homology using homologous recombination (though single-strand annealing also can occur) (Sonoda et al., 2006). However, the dominant pathway for the repair of double-strand breaks is called NHEJ, and this repair pathway can function at any time during the cell cycle. NHEJ does not use long stretches of homology, but the processing of the DNA ends can, in a minority of cases, be influenced by alignment of a few nts of homology called terminal microhomology (typically 1 to 4 nt in length). It should be noted that NHEJ proceeds even if there is no terminal microhomology. Important protein components of the repair pathways are listed.
A third cause is inadvertent action by nuclear enzymes of DNA metabolism. These include failures of type II topoisomerases, which transiently break both strands of the duplex. If the topoisomerase fails to rejoin the strands, then a DSB results. Failures of type I topoisomerases (spontaneous or due to inhibition) cause single-strand breaks (SSBs), which can be result in a DSB upon DNA replication.
Still within this same category of inadvertent action by nuclear enzymes are the lymphoid-specific enzymes for antigen receptor gene rearrangement, which include the RAG complex (composed of RAG1 and 2) and activation-induced deaminase (AID). In humans, these account for about half of all of the chromosomal translocations that results in lymphoma, as will be discussed later.
A fourth cause of DSBs is natural ionizing radiation of the enviroment. These include gamma rays and X-rays. At sea level, each person is hit by about 300 million ionizing radiation particles per hour. As these traverse the body, they create free radicals primarily from water along their path. When the particle traverses close to a DNA duplex, clusters of free radical damage in damage to double- and single-stranded DNA at a ratio of about 25 to 1. About half of the ionizing radiation that strikes each of us comes from outside the earth and is leftover radiation from the inception of the universe (the Big Band). Orders of magnitude more such radiation would reach the surface of the earth were it not for the atmosphere, because most of the particles encounter the ozone of the upper atmosphere first. Another half of the radiation that strikes us comes from the decay of radioactive elements, primarily metals) in the earth.
The discussion above relates to mitotic cells. Meiotic cells have an additional form of double-strand breakage which is physiologic and is caused by an enzyme called Spo11 (Zickler and Kleckner, 1999), which creates DSBs in order to cause cross-overs between homologues during meiotic prophase I. But it is not clear that NHEJ occurs in vertebrate meiotic cells because one group reports the lack of one of Ku70 in spermatogonia. Human spermatogonia remain in meiotic prophase I for about 3 weeks, and human eggs remain in meiotic prophase I for 12 to 50 years; hence, these cells can rely on homologous recombination (HR) during these long periods. Given the error-prone nature of NHEJ, reliance on HR may be one way to minimize alterations to the germ cell genome at frequencies that may be deleterious.
2. VERTEBRATE NONHOMOLOGOUS DNA END JOINING
The discussion of NHEJ here relates to the mechanism in vertebrate organisms. In most invertebrate organisms, there is no DNA-PKcs and no Artemis, though the evolutionary emergence of these two components remains an active area of study. The nuclease functions of Artemis seem to be accomplished by the RAD50:MRE11:NBS1 complex in invertebrate animals, yeast, and in plants (Chen et al., 2001).
In vertebrate cells, the first protein thought to bind at a DSB is Ku, because it is abundant (~400,000 molecules per cell) and it binds tighter to duplex DNA ends than any other protein in the cell (KD ~ 1 nM) (Falzon et al., 1993). Ku consists of Ku70 and Ku86 and forms a toroidally-shaped heterodimer that has a hole large enough for duplex DNA (Walker et al., 2001). Ku can load onto DNA only at DNA termini and can thread onto the duplex to internal positions (deVries et al., 1989).
Ku has no confirmed enzymatic function, though claims of helicase activity, ATPase activity, or end bridging have been made at various points without confirmation. The primary function of Ku seems to be to improve the stability of the NHEJ enzymes at the DNA termini so that they can carry out their functions for longer periods (Lieber, 2008). Improvements in stability have been reflected in improved equilibrium dissociation constants determined using surface plasmon resonance and electrophoretic mobility shift assays (EMSA) on acrylamide gels as well as with qualitative methods such as co-immunoprecipitation and atomic force microscopy (Yaneva et al., 1997, West et al., 1998). Hence, Ku seems to function by marking the DNA terminus as a node for the polymerases, nuclease and ligase of NHEJ. In that sense, it functions somewhat like tool belt proteins such as PCNA (Lieber, 2008).
Most DNA repair processes involve a nuclease to remove damaged DNA, a polymerase to fill-in new DNA, and a ligase to restore the integrity of each strand of the duplex. In vertebrate NHEJ, the ligase is DNA ligase IV (Grawunder et al., 1997, Wilson et al., 1997, Schar et al., 1997, Teo and Jackson, 1997), and it is supported by two apparently nonenzymatic components called XRCC4 and XLF (or Cernunnos) (Ahnesorg et al., 2006, Buck et al., 2006). The polymerases are polymerse mu and polymerase lambda, though in double mutants of these two, it appears that other polymerases can also participate (Bertocci et al., 2006, Lieber, 2006). The nuclease functions appear largely covered by a complex of Artemis and DNA-PKcs (Ma et al., 2002). though DNA-PKcs appears to have additional functions related to cell cycle components, chromatin, and others.
2.1 The DNA ligase IV Complex
In the absence of DNA ligase IV in S. cerevisiae, NHEJ does not occur (Wilson et al., 1997, Daley et al., 2005b, Schar et al., 1997, Teo and Jackson, 1997). Hence, the other ligase in S. cerevisiase, DNA ligase I, can not substitute for ligase IV. However, if there are several nucleotides of terminal microhomology between the two ends, then some joining can occur. The amount of joining may depend on the length of terminal microhomology, but it is usually greater than 6 nucleotides (Ma et al., 2003). In the nonrepetitive yeast genome, such microhomology is quite unusual. Hence, the alternative or backup role of such microhomology-mediated end joining (MMEJ) is likely to be of limited survival utility for naturally-occurring DSBs.
In vertebrates, XRCC4 seems to stabilize ligase IV. Other possible roles may be possible, but are still under investigation. XLF (or Cernunnos) seems to stimulate the ability of ligase IV to ligate a more diverse array of DNA end configurations (Gu et al., 2007a, Gu et al., 2007b)(see below). The stoichiometry of the XLF:XRCC4:ligase IV complex is thought to be 2:2:1 (Li et al., 2008, Ahnesorg et al., 2006), but this is difficult to measure, and additional studies are warranted.
DNA ligase IV can ligate fully incompatible DNA ends, namely DNA ends that do not share even one nucleotide of terminal microhomology (e.g., a blunt end joined to a 5′ overhang) (Gu et al., 2007a). Though the efficiency of this joining is low, ligase I and III do not appear able to do this at all. Importantly, XLF can markedly stimulate the ability of the XRCC4:ligase IV complex to carry out incompatible DNA end joining (Gu et al., 2007b).
Two ends are partially compatible when they they share one or more nucleotides of terminal microhomology but they can not not be ligated without resection of some excess DNA flaps or fill-in of some gaps on one or both DNA strands of the duplex. Joining by the XLF:XRCC4:ligase IV complex is more efficient when there is one or more nucleotides of microhomology shared between the DNA ends. Ku is able to make joining much more efficient if there is no microhomology and more efficient if there are one or two nucleotides of microhomology (Gu et al., 2007a, Gu et al., 2007b). If there are 4 or more nucleotides of microhomology, then the stimulation by Ku is relatively small (<2-fold in some cases).
In addition to the flexibility mentioned above, ligase IV can ligate one strand when the other strand is in a configuration that cannot be ligated, and it can ligase some single-stranded DNA sequences (Ma et al., 2002). Overall, the ligase IV complex is perhaps the most flexible ligase known for tolerating a wide range of substrates.
2.2 The Polymerases of NHEJ
In S. cerevisiae, POL4 is responsible for a substantial amount of fill-in synthesis at NHEJ junctions (Wilson and Lieber, 1999, Daley et al., 2005a, Daley and Wilson, 2007), though other polymerases such as POL3 (pol delta) also contribute. POL4 is a member of the POLX polymerase family, a family that is a subset of all DNA polymerases (Tseng and Tomkinson, 2002, Burgers et al., 2001).
The human homologues of POL4 are both polymerase mu and polymerase lambda. Both pol mu and pol lambda have BRCT domains located within their N-terminal portions. The other two members of the POLX family in humans are terminal transferase (TdT) and pol beta. TdT also has a BRCT domain within its N-terminal portion. Pol beta is distinctive among POLX polymerases for its lack of a BRCT domain. Ku is able to recruit pol mu and pol lambda by binding to their BRCT domains, and removal of this BRCT domain eliminates the ability of Ku to recruit either polymerase (Gu et al., 2007a).
Pol mu and pol lambda are both capable of fill-in synthesis. But remarkably, pol mu also has a robust ability for template-independent synthesis (Ramadan et al., 2003, Ramadan et al., 2004, Gu et al., 2007a). This template-independent synthesis is useful for generating terminal microhomology when none exists.
Pol mu and pol lambda are both prone to slippage on the template strand, which would generate direct repeats (Bebenek et al., 2003). In addition, when pol mu generates overhangs via template-independent synthesis, it is able to fold that strand onto itself and continue polymerization. This would generate inverted repeats. At NHEJ junctions, both direct and inverted repeats are commonly seen (Jaeger et al., 2000).
There is some evidence that pol mu, in the context of Ku, may be able to cross the discontinuity in the template strand between two DNA ends (NickMcElhinny et al., 2005, Moon et al., 2007). Importantly, without Ku, there is no evidence that pol mu can use a discontinuous template; that is, cross from one DNA end to another DNA end.
Therefore pol mu and lambda are both very flexible. Pol mu has the additional flexibility of template-independent synthesis and possibly of synthesis across a discontinuous template strand. These features make pol mu and pol lambda among the most flexible polymerases known.
2.3 Artemis, DNA-PKcs and the Nuclease of NHEJ
In vertebrates, most and perhaps all of the nuclease activity for NHEJ is due to Artemis (Ma et al., 2004). Artemis forms a complex with DNA-PKcs. and this complex is relatively stable even at 500 mM NaCl (Ma et al., 2002). DNA-PKcs is a serine/threonine protein kinase, but it is only active when bound to duplex DNA ends (Anderson and Carter, 1996). The range of DNA ends that active DNA-PKcs kinase activity is quite diverse (Falzon et al., 1993). Purified Artemis has 5′ exonuclease activity. But Artemis additionally acquires 5′ endonuclease, 3′ endonuclease, and hairpin opening activity when it is in a complex with DNA-PKcs:DNA complexes (Ma et al., 2002). The 5′ endonuclease activity of the Artemis:DNA-PKcs complex prefers to nick 5′ overhangs at the junction of the single- to double-strand portions. The 3′ endonuclease activity of the Artemis:DNA-PKcs complex nicks 3′ overhangs preferentially ~4 nt into the single-stranded overhang from the double- to single-strand transition (Ma et al., 2002). The hairpin opening activity of the Artemis:DNA-PKcs complex preferentially nicks the hairpins two nucleotides past the tip of the hairpin. These diverse endonuclease properties are somewhat unified by a model in which the Artemis:DNA-PKcs complex binds four nucleotides of single-stranded DNA adjacent to the single- to double-strand transition, and nicks 3′ of it (Ma et al., 2002). This explains the hairpin nicking preference because the last two base pairs of a perfect hairpin remain unpaired, thereby providing the four nucleotides of single-strandedness at the hairpin tip.
The primary phosphorylation target of DNA-PKcs is itself. DNA-PKcs autophosphorylates itself at more than 13 sites. This causes a conformational change of DNA-PKcs while bound to a DNA end based on studies of DNA end accessibility to other enzymes, such as ligases.
DNA end activated DNA-PKcs also phosphorylates Artemis at 11 sites within the C-terminus, and removal of the C-terminus of Artemis gives it endonuclease activity on some substrates even in the absence of DNA-PKcs (Ma et al., 2005, Niewolik et al., 2006). Based on this, it appears that the conformational change in Artemis that converts it from an exonuclease to an endonuclease is conconmittant with Artemis phosphorylation. However, if one interferes with the Artemis phosphorylation by adding a DNA-PKcs inhibitor (after the DNA-PKcs has already phosphorylated itself), then the Artemis still acquires its endonucleolytic properties (Goodarzi et al., 2006). Therefore, the conformational change in Artemis induced by DNA-PKcs appears to be the primary event that permits Artemis to acquire endonucleolytic activity.
2.4 Concluding Comments on Vertebrate NHEJ
The nuclease, polymerase, and ligase components of NHEJ bind to the Ku;DNA end, and in that sense, the Ku marks the DNA end as a node for recruitment of these other NHEJ proteins. The nuclease, polymerases and ligase are all among the most flexible enzymes of their type. This flexibility permits NHEJ to join a very diverse range of DNA ends that have various sequences, various overhang configurations, and various degrees of oxidative damage.
3. CHROMOSOMAL TRANSLOCATIONS
3.1 Types of Translocations and Relation to Cancer
Genetic changes in neoplasms can arise from a variety of causes including numerical changes in chromosome number; structural changes in chromosomes (translocations, inversions and deletions); focal mutations (e.g., point mutations); DNA methylation, which can cause transcriptional shutdown; and reduction to homozygosity due to homologous recombination or due to loss of a chromosomal segment.
Changes in chromosome structure (translocations, inversions, or deletions) are among the most common change in many neoplasms, particularly neoplasms of the hematopoietic system. In acute leukemias, oncogenic translocations often result in the formation of fusion proteins with capabilities beyond those of the original constituent proteins (Gelb and Medeiros, 2002). Examples include fusions of the MLL gene in 3 to 5% of acute myeloid leukemia with many different partner genes (Gelb and Medeiros, 2002), and the E2A-Pbx1 fusion in 6% of acute lymphoblastic leukemia.
In chronic leukemias and lymphomas, oncogenic translocations often activate a gene that drives cell proliferation (e.g., an oncogene)(Gelb and Medeiros, 2002). Many of these translocations involve one of the antigen receptor loci. While these generalizations are useful for formulating a framework, there are many exceptions.
In order for a chromosomal translocation to occur, two independent breaks typically occur on different chromosomes, thereby generating two DNA ends at each break, or four DNA ends total (Lieber, 1998). If the correct DNA ends get rejoined, then no translocation occurs. But if ends from different chromosomes are joined, then a translocation arises.
Interstitial deletions are mechanistically similar to translocations, but in this case, both breakpoints are on the same chromosome, and the region between the breaks is deleted. Chromosomal inversions can also arise this way, but where the DNA ends of the chromosomal segment in the middle of two breaks are joined.
3.2 NHEJ in Chromosomal Structural Changes
Nearly all chromosome structural changes involve double-strand DNA breaks. When the breaks become rejoined, NHEJ is responsible for the vast majority of the joining (Ferguson and Alt, 2001). If a key component of NHEJ is missing, such as DNA ligase IV, then one of the other two ligases (ligase I or III) can do the joining (Ferguson and Alt, 2001), although the efficiency of joining seems to be lower and there appears to be more nucleotide loss from the junctions (Han and Yu, 2008). However, in human spontaneous neoplasms, NHEJ is nearly always intact, and apparently responsible for the vast majority of joining.
3.3. Causes of DSBs That Initiate Translocations
As mentioned in the first section of this review, there are many causes of DSBs. In considering the causes underlying a particular chromosomal translocation, it is useful to examine the sequence features and distribution of the translocation. For translocations that create a fusion protein (such as a novel transcription factor that changes the regulation of the cell), the fusion protein is often a specific set of exons from each original protein. In these cases, the DSB often can occur anywhere within the intron following the last required exon. Upon examination of many patient breakpoints, if breakpoints are distributed evenly across the entire intron, then the simplest explanation involves sequence-nonspecific causes of DSBs, such as oxidative free radicals or natural ionizing radiation.
However, even for some fusions, such as E2A joined to Pbx1, a subset of the breakpoints can be tightly clustered in a manner that is 50 to 300-fold more focused than random (Wiemels et al., 2002) (Fig. 2).
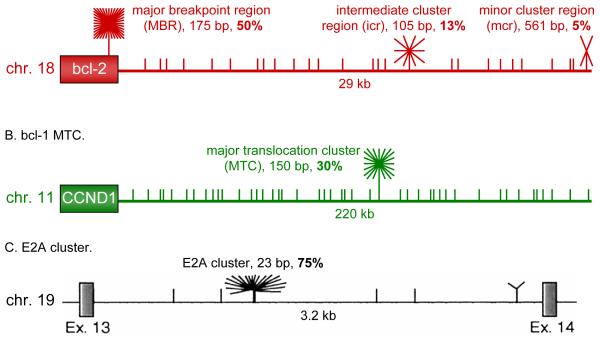
Figure 2. Schematics of breakpoint cluster regions.
Schematics of the bcl-2, bcl-1, and E2A regions illustrate clustering of breakpoints within the various identified cluster regions. The breakpoints that do not fall into cluster regions are plotted randomly for illustrative purposes, as most of them are never sequenced and often only mapped to general regions by Southern blotting.
(A) depicts relative proportions of breakpoints at the bcl-2 MBR, icr, and mcr cluster regions. The third exon of the bcl-2 gene, boxed, contains the MBR (within the 3′ UTR region), while the centromeric 29 kb contains the icr and mcr. Short lines above the gene diagram mark the approximate locations and relative abundance of patient breakpoints. The 175 bp MBR, 105 bp icr, and 561 bp mcr account for about 50%, 13%, and 5% of bcl-2 translocation breakpoints, respectively.
The bcl-1 MTC, represented in (B), is located about 110 kb from CCND1, the gene for the cyclin D1 oncoprotein. The 150 bp MTC contains about 30% of breakpoints, whereas the remaining 70% of events are distributed widely over the surrounding 220 kb.
(C) shows a diagram of intron 13 of the E2A gene, taken from (Wiemels et al., 2002a). 75% of breakpoints occur in the 23 bp E2A cluster, while the surrounding 3 kb only account for 25%.
For translocations that do not generate a fusion protein but rather upregulate a gene such as bcl-2 or cyclin D1, many or even most of the breakpoints are focused by a factor of roughly 100-fold over random (Tsai et al., 2008b)(Fig. 3). In neoplasms with such intense focusing, some of the patient breakpoints are scattered across wide zones outside of the focused zone. However, in most cases, as in the bcl-2 or bcl-1 translocations, the clinical features of the neoplasm are not highly dependent on the location of the DSB (Jaeger et al., 2000, Welzel et al., 2001).
Figure 3. Proximity of Patient Chromosome 18 Breakpoints in Follicular Lymphoma Relative to CpG Sites within the Major Breakpoint Region.
Each breakpoint is represented as a triangle adjoining the breakpoint site, with the top strand sequences running telomeric to centromeric, with der(14) breakpoints above, and der(18) breakpoints below.
There are two general mechanisms that have been proposed for intense focusing of translocation breakpoints in hematopoietic neoplasms, particularly those of the lymphoid lineage (Tsai et al., 2008b).
3.3.1 Mistakes of V(D)J Recombination
The RAG complex is expressed in pre-T and pre-B cells, and consists of RAG1, RAG2 and HMGB1. It normally functions in antigen receptor gene rearrangement by cutting at heptamer/nonamer sequences, for which the consensus is flexible. The heptamer/nonamer sequences are often called recombination signal sequences (or RSS). The flexibility of the RSS sequence means that there are locations in the genome that can look similar to an RSS, but not located at the antigen receptor loci. These have been called pseudo RSSs, cryptic RSSs, or misrecognition sites, all of which refer to aberrant, non-receptor sites of RAG complex cutting (Lewis et al., 1997). In some T-cell acute lymphoblastic leukemias/lymphomas, these pseudo RSSs are cut and joined to a T-cell receptor (TCR) locus. The enhancers at the TCR locus then upregulate the gene. If this gene can serve as a first step on a pathway to neoplasia, then the resulting translocation is seen in the resulting tumor. This is precisely what occurs in many types of T-ALL (T-cell acute lymphoblastic lymphoma).
DSBs that arise due to misrecognition of a pseudo RSS are referred to a DSBs of the V(D)J-type. Recall that chromosomal translocations require two DSBs. In most cases, the DSB at the oncogene by a V(D)J-type mechanism joins to a DSB at one of the antigen receptor loci. Therefore, one could think of this as the joining of ends from two DSBs in which both were of the V(D)J-type (Tsai et al., 2008b).
In some cases, both DSBs are sites of pseudo RSS, such as in the interstitial deletion between SCL and SIL. Both DSBs are still V(D)J-type, but the distinction from above is that one V(D)J-type event is at an antigen receptor locus and the other other is at a pseudo RSS (Raghavan et al., 2001).
3.3.2 Sequential Action by AID and the RAG Complex
For many translocations that occur in pre-B cells during V(D)J recombination, translocation breakpoints are often tightly clustered into small zones (e.g., the major breakpoint region (MBR) in bcl-2 translocations, or the major translocation cluster (MTC) in bcl-1 translocations) but do not have features of a pseudo RSS. The mechanism for breakage at these zones has been unclear.
Recently, we reported the striking statistical observation that the peaks of the translocations occur at CpG sequences (Tsai et al., 2008b). CpG is unique because the C in such dinucleotides can be methylated. Methyl C is distinctive from C because when it deaminates spontaneously, regular C becomes a U, but methyl C becomes a T. U:G mismatches are relatively efficiently repaired back to the original sequence. But T:G mismatches are poorly repaired. The poor repair of T:G mismatches that arise from meCpGs explains why 25 to 50% of the point mutations at the p53 gene are located at CpG sites. This also explains the evolutionary depletion of CpG from the vertebrate genome, except in regions where the CpG is unmethylated or of functional importance (i.e., at promoters, which thus give rise to CpG islands).
Our finding of some translocations centered at CpG sites suggests a mechanism (Tsai et al., 2008b). We find this CpG propensity only in lymphoid malignancies where there is RAG expression at the time of the translocation (Fig. 4). Nonlymphoid neoplasms and lymphoid neoplasms where the translocation occurs before or after the B lineage window of RAG expression do not appear to suffer DSBs by this mechanism. In other words, breaks at CpG sites only appear to occur in pro-B and pre-B cells and not in all other B cell stages of differentiation and not in other hematopoietic lineages.
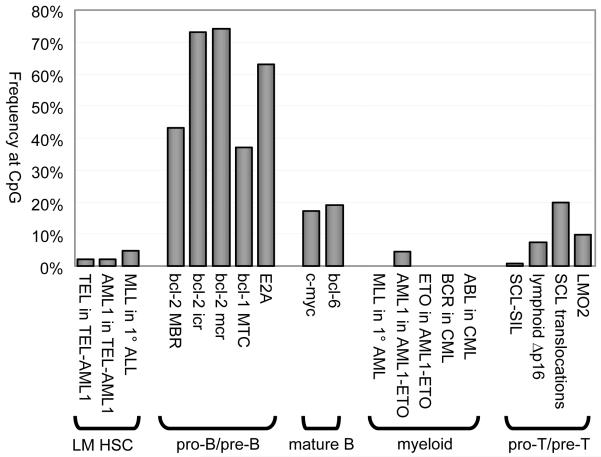
Figure 4. Frequencies of Breakpoints at CpG in Various Chromosomal Rearrangements.
Percentages of breakpoints at CpGs are plotted for various chromosomal rearrangements, organized by cell lineage and stage of development when the rearrangement occurs. Frequencies for pro-B/pre-B rearrangements far exceed those from other lineages and stages.
What is this RAG-dependent mechanism? We have shown previously that the RAG complex can nick small bubble structures, and we now know that such nicking applies even to 1 bp mismatches (Tsai et al., 2008b). This propensity of the RAG complex to nick small bubble structures, even ones as small as 1 bp, may have derived from the fact that the RAG complex has to convert a nicked species to a hairpin form during V(D)J recombination. Therefore, the RAG complex may readily bind duplexes that have DNA distortions, and in the course of such binding, it appears that the RAG complex can nick the top or bottom strand of an isolated bubble structure at efficiencies that are within 10-fold of the nicking at a duplex RSS (Tsai et al., 2008b, Raghavan et al., 2005a, Raghavan and Lieber, 2006, Raghavan et al., 2007). The ability of the RAG complex to nick at a 1bp mismatch means that any meC in a meCpG site that deaminates to a T can be subsequently nicked by the RAG complex. We call this a DSB of the CpG-type to distinguish it from other mechanisms
The ability of the RAG complex to nick 1bp sites of deaminated meC would predict that one could have a RAG-dependent mechanism in both pre-B (and pro-B) and pre-T cells (Tsai et al., 2008b). However, we only observe this CpG localization of breakpoints in pro-/pre-B cell translocations. This means that one additional factor must participate. Moreover, this additional factor must overlap with the temporal expression of the RAG complex. There is only one enzyme that converts C to U or methyl C to T and is specific to B cells. This enzyme is activation-induced deaminase (AID). AID is most highly expressed in germinal center B cells located in the lymph nodes, spleen, and Peyer’s patches of the GI tract. The deaminase activity of AID triggers the somatic hypermutation (SHM) and class switch recombination (CSR) processes. Surprisingly, a percentage of mammalian pre-B cells prematurely express substantial levels of AID. Hence, AID and the RAG complex are concurrently present in some pre-B cells (Tsai et al., 2008b).
Our model then is that the B lineage specificity of the CpG-type of DSB is due to AID acting at meC within meCpG. Then the RAG complex nicks both the top and bottom DNA strands, resulting in a DSB (Tsai et al., 2008b)(Fig. 5).
Figure 5. Proposed Mechanism for CpG-type Double-strand Breakage.
Deamination at a methylcytosine within a CpG creates a T:G mismatch which persists due to catalytic inefficiency of methyl-CpG binding domain protein 4 (MBD4) and thymine DNA glycosylase (TDG) in cleaving the thymine glycosidic bond, leaving an abasic site (asterisk). Single-strand breaks are generated either by the normal base excision repair pathway of glycosylase and AP endonuclease (APE) activity, or by RAG nicking the mismatch directly, resulting in a poorly-ligatable flap. Heterogeneous RAG nicking of the remaining strand creates a double-strand break close to the original site of the T:G mismatch. We observe the following features at CpG-type translocation hotspots: (1) an extremely high degree of focusing to CpGs, (2) a breakpoint distribution consistent with a structure-specific endonuclease, (3) specificity to the pro-B/pre-B stage, and (4) specificity to the B-cell lineage. Involvement of the RAG complex explains points 2 and 3, while involvement of AID explains point 4 and is strongly suggestive for point 1.
If one DSB of the translocation occurs by the CpG-type mechanism, how does the other DSB occur? For the t(14;18) translocation of follicular lymphoma, which is the most common translocation in human lymphoma, the other DSB occurs via a V(D)J-type mechanism at an RSS at the immunoglobulin heavy chain (IgH) locus. For the t(11;14) translocation of mantle cell lymphoma, the CpG-type DSB occurs in the major translocation cluster (MTC) about 100kb from the cyclin D1 gene, and the other DSB is a V(D)J-type mechanism at the IgH locus (Tsai et al., 2008b).
For the t(1;19) translocation involving the E2A gene on chromosome 1 and the Pbx1 gene on chromosome 19, the E2A break is of the CpG-type, occurring typically within a 23 bp window on the 3.2 kb intron 13, while the Pbx1 break appears to be random, occuring anywhere within the 232 kb intron 1 (Wiemels et al., 2002).
3.3.3 Source of Single-Strandedness at Sites where AID Acts on meC Sites
In order for AID to convert C to U or meC to T, the DNA must be single-stranded (Bransteitter et al., 2003). So how does single-strandedness arise? We do not know with certainty. However, the bcl-2 MBR and the bcl-1 MTC do contain regions of distinctive reactivity with chemical probes that require either some degree of single-strandedness or substantial departures from the B-form duplex (Raghavan et al., 2004a, Tsai et al., 2008a).
3.3.4 Concluding Comments
Chromosomal translocations and related pathologic genome rearrangements in somatic cells require two DSBs, and the mechanism for the two breaks does not need to be the same. Once the breaks are formed, then NHEJ generally does the joining of the four DNA ends.
REFERENCES
- AHNESORG P, SMITH P, JACKSON SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- ANDERSON CW, CARTER TH. The DNA-Activated Protein Kinase-DNA-PK. In: JESSBERGER R, LIEBER MR, editors. Molecular Analysis of DNA Rearrangements in the Immune System. Springer-Verlag; Heidelberg: 1996. [Google Scholar]
- BEBENEK B, GARCIA-DIAZ M, BLANCO L, KUNKEL TA. The frameshift infidelity of human DNA polymerase lambda: implications for function. J. Biol. Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- BERTOCCI B, DESMET A, WEILL J-C, REYNAUD CA. Non-overlapping functions of polX family DNA polymerases, pol m, pol l, and TdT, during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- BRANSTEITTER R, PHAM P, SCHARFF MD, GOODMAN MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCK D, MALIVERT L, DECHASSEVAL R, BARRAUD A, FONDANECHE M-C, XANAL O, PLEBANI A, STEPHAN J-L, HUFNAGEL M, LEDIEST F, FISCHER A, DURRANDY A, VILLARTAY J-PD, REVY P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- BURGERS PM, KOONIN EV, BRUFORD E, BLANCO L, BURTIS KC, CHRISTMAN MF, COPELAND WC, FRIEDBERG EC, HANAOKA F, HINKLE DC, LAWRENCE LW, NAKANISHI M, OHMORI H, PRAKASH L, PRAKASH S, REYNAUD CA, SUGINO A, TODO T, WANG Z, WEILL JC, WOODGATE R. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 2001;276:43487–90. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- CHANCE B, SIES H, BOVERIS A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979;59:527–603. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- CHEN L, TRUJILLO K, RAMOS W, SUNG P, TOMKINSON AE. Promotion of DNA ligase IV-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- DALEY JM, LAAN RLV, SURESH A, WILSON TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 2005a;280:29030–7. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- DALEY JM, PALMBOS PL, WU D, WILSON TE. Nonhomologous end joining in yeast. Ann. Rev. Genet. 2005b;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- DALEY JM, WILSON TE. Evidence that base stacking potential in annealed 3′ overhangs determines polymerase utilization in yeast nonhomologous end joining. DNA Repair (Amst) 2007;7:67–76. doi: 10.1016/j.dnarep.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEVRIES E, VANDRIEL W, BERGSMA WG, ARNBERG AC, VANDERVLIET PC. HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J. Mol. Biol. 1989;208:65–78. doi: 10.1016/0022-2836(89)90088-0. [DOI] [PubMed] [Google Scholar]
- FALZON M, FEWELL J, KUFF EL. EBP-80, a Transcription Factor Closely Resembling the Human Autoantigen Ku, Recognizes Single- to Double-Strand Transitions in DNA. J. Biol. Chem. 1993;268:10546–52. [PubMed] [Google Scholar]
- FERGUSON DO, ALT FW. DNA double-strand break repair and chromosomal translocations: lessons from animal models. Oncogene. 2001;20:5572–5579. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- GELB AB, MEDEIROS LJ. The Molecular Biology of Leukemias. In: COLEMAN WB, TSONGALIS GJ, editors. The Molecular Basis of Human Cancer. Humana Press; Totowa: 2002. [Google Scholar]
- GOODARZI AA, YU Y, RIBALLO E, DOUGLAS P, WALKER SA, YE R, HARER C, MARCHETTI C, MORRICE N, JEGGO PA, LEES-MILLER SP. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. Embo J. 2006;25:3880–9. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRAWUNDER U, WILM M, WU X, KULESZA P, WILSON TE, MANN M, LIEBER MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- GU J, LU H, TIPPIN B, SHIMAZAKI N, GOODMAN MF, LIEBER MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. Embo J. 2007a;26:1010–23. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GU J, LU H, TSAI AG, SCHWARZ K, LIEBER MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007b;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN L, YU K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV--deficient B cells. J Exp Med. 2008;205:2745–53. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAEGER U, BOCSKOR S, LE T, MITTERBAUER G, BOLZ I, CHOTT A, KNEBA A, MANNHALTER C, NADEL B. Follicular lymphomas BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- LEWIS SM, AGARD E, SUH S, CZYZYK L. Cryptic signals and the fidelity of V(D)J Joining. Mol. Cell. Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Y, CHIRGADZE DY, BOLANOS-GARCIA VM, SIBANDA BL, DAVIES OR, AHNESORG P, JACKSON SP, BLUNDELL TL. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. Embo J. 2008;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIEBER MR. Pathologic and Physiologic Double-Strand Breaks: Roles in Cancer, Aging, and the Immune System. Am. J. Path. 1998;153:1323–1332. doi: 10.1016/s0002-9440(10)65716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIEBER MR. The polymerases for V(D)J recombination. Immunity. 2006;25:7–9. doi: 10.1016/j.immuni.2006.07.007. [DOI] [PubMed] [Google Scholar]
- LIEBER MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- LIEBER MR, KARANJAWALA ZE. Ageing, repetitive genomes and DNA damage. Nature Rev. Mol. Cell. Biol. 2004;5:69–75. doi: 10.1038/nrm1281. [DOI] [PubMed] [Google Scholar]
- LIEBER MR, MA Y, PANNICKE U, SCHWARZ K. Mechanism and regulation of human non-homologous DNA end-joining. Nature Rev. Mol. Cell. Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- LIEBER MR, YU K, RAGHAVAN SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- MA JL, KIM EM, HABER JE, LEE SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA Y, LU H, TIPPIN B, GOODMAN MF, SHIMAZAKI N, KOIWAI O, HSIEH C-L, SCHWARZ K, LIEBER MR. A Biochemically Defined System for Mammalian Nonhomologous DNA End Joining. Molecular Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- MA Y, PANNICKE U, LU H, NIEWOLIK D, SCHWARZ K, LIEBER MR. The DNA-PKcs phosphorylation sites of human artemis. J. Biol. Chem. 2005;280:33839–46. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- MA Y, PANNICKE U, SCHWARZ K, LIEBER MR. Hairpin opening and overhang processing by an Artemis:DNA-PKcs complex in V(D)J recombination and in nonhomologous end joining. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- MARTIN GM, SMITH AC, KETTERER DJ, OGBURN CE, DISTECHE CM. Increased chromosomal aberrations in first metaphases of cells isolated from the kidneys of aged mice. Israel J. Med. Sci. 1985;21:296–301. [PubMed] [Google Scholar]
- MOON AF, GARCIA-DIAZ M, BATRA VK, BEARD WA, BEBENEK K, KUNKEL TA, WILSON SH, PEDERSEN LC. The X family portrait: Structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NICKMCELHINNY SA, HAVENER JM, GARCIA-DIAZ M, JUAREZ R, BEBENEK K, KEE BL, BLANCO L, KUNKEL TA, RAMSDEN DA. A gradient of template dependence defines distinct biological roles for family x polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- NIEWOLIK D, PANNICKE U, LU H, MA Y, WANG LC, KULESZA P, ZANDI E, LIEBER MR, SCHWARZ K. DNA-PKcs dependence of artemis endonucleolytic activity: differences between hairpins and 5′ or 3′ overhangs. J. Biol. Chem. 2006;281:33900–33909. doi: 10.1074/jbc.M606023200. [DOI] [PubMed] [Google Scholar]
- RAGHAVAN SC, GU J, SWANSON PC, LIEBER MR. The structure-specific nicking of small heteroduplexes by the RAG complex: implications for lymphoid chromosomal translocations. DNA Repair (Amst) 2007;6:751–9. doi: 10.1016/j.dnarep.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAGHAVAN SC, KIRSCH IR, LIEBER MR. Analysis of the V(D)J recombination efficiency at lymphoid chromosomal translocation breakpoints. J. Biol. Chem. 2001;276:29126–29133. doi: 10.1074/jbc.M103797200. [DOI] [PubMed] [Google Scholar]
- RAGHAVAN SC, LIEBER MR. DNA structures at chromosomal translocation sites. BioEssays. 2006;28:480–494. doi: 10.1002/bies.20353. [DOI] [PubMed] [Google Scholar]
- RAGHAVAN SC, SWANSON PC, MA Y, LIEBER MR. Double-strand break formation by the RAG complex at the bcl-2 Mbr and at other non-B DNA structures in vitro. Mol. Cell. Biol. 2005a;25:5904–5919. doi: 10.1128/MCB.25.14.5904-5919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAGHAVAN SC, SWANSON PC, WU X, HSIEH C-L, LIEBER MR. A non- B-DNA structure at the bcl-2 major break point region is cleaved by the RAG complex. Nature. 2004a;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- RAMADAN K, MAGA G, SHEVELEV IV, VILLANI G, BLANCO L, HUBSCHER U. Human DNA polymerase lambda possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J. Mol. Biol. 2003;328:63–72. doi: 10.1016/s0022-2836(03)00265-1. [DOI] [PubMed] [Google Scholar]
- RAMADAN K, SHEVELEV IV, MAGA G, HUBSCHER U. De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu, and terminal deoxynucleotidyl transferase. J. Mol. Biol. 2004;339:395–404. doi: 10.1016/j.jmb.2004.03.056. [DOI] [PubMed] [Google Scholar]
- SCHAR P, HERRMANN G, DALY G, LINDAHL T. A newly identified DNA ligase of S. cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes & Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SONODA E, HOCHEGGER H, SABERI A, TANIGUCHI Y, TAKEDA S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–9. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- TEO SH, JACKSON SP. Identification of S. cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAI AG, ENGELHART AE, HATMAL MM, HOUSTON SI, HUD NV, HAWORTH IS, LIEBER MR. Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites. J Biol Chem. 2008a doi: 10.1074/jbc.M806866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAI AG, LU H, RAGHAVAN SC, MUSCHEN M, HSIEH CL, LIEBER MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008b;135:1130–42. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSENG HM, TOMKINSON AE. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 2002;277:45630–45637. doi: 10.1074/jbc.M206861200. [DOI] [PubMed] [Google Scholar]
- WALKER JR, CORPINA RA, GOLDBERG J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- WELZEL N, T TL, MARCULESCU R, MITTERBAUER G, CHOTT A, POTT C, KNEBA M, DU MQ, KUSEC R, DRACH J, RADERER M, MANNHALTER C, LECHNER K, NADEL B, JAEGER U. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001:1629–1636. [PubMed] [Google Scholar]
- WEST RB, YANEVA M, LIEBER MR. Productive and Nonproductive Complexes of Ku and DNA-PK at DNA Termini. Mol. Cell. Biol. 1998;18:5908–5920. doi: 10.1128/mcb.18.10.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIEMELS JL, LEONARD BC, WANG Y, SEGAL MR, HUNGER SP, SMITH MT, CROUSE V, MA X, BUFFLER PA, PINE SR. Site-specific translocation and evidence of postnatal origin of the t(1;19) E2A-PBX1 fusion in childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2002;99:15101–6. doi: 10.1073/pnas.222481199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILSON TE, GRAWUNDER U, LIEBER MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- WILSON TE, LIEBER MR. Efficient processing of DNA ends during yeast nonhomologous end joining: evidence for a DNA polymerase beta (POL4)-dependent pathway. J. Biol. Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- YANEVA M, KOWALEWSKI T, LIEBER MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZICKLER D, KLECKNER N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]