Abstract
The detection and characterization of circulating tumor cells (CTCs) holds great promise for personalizing medicine and optimizing systemic therapy. However, low specificity, low sensitivity and the time consuming nature of current approaches have impeded clinical adoption. Here we report a new method using Surface-Enhanced Raman Spectroscopy (SERS) to directly measure targeted CTCs in the presence of white blood cells. SERS nanoparticles with epidermal growth factor (EGF) peptide as a targeting ligand have successfully identified CTCs in the peripheral blood of 19 patients with squamous cell carcinoma of the head and neck (SCCHN), with a range of 1–720 CTCs per milliliter of whole blood. Our technique may provide an important new clinical tool for management of patients with SCCHN and other cancers.
Keywords: CTC, SERS nanoparticle, SCCHN, EGFR, Peripheral blood
Introduction
Circulating tumor cells (CTCs) are a hallmark of invasive cancer cells, which are responsible for the development of metastasis. It is imperative to develop new approaches for the detection and quantification of CTCs, which will significantly contribute to clinical prognosis, diagnosis, individualization and optimization of systemic therapy. Detecting rare CTCs in complex blood samples is a major challenge, requiring an exceptionally specific and sensitive assay for discerning and capturing CTCs with high efficiency. Some progress has been made recently. The U.S. Food and Drug Administration (FDA) approved the use of CellSearch™ based on immunomagnetic separation (1). A potential concern with this method is the impurity of leukocytes, leading to a high false positive rate (2–4). More recently, a CTC chip has been developed by Haber’s group (1, 5–7) for monitoring therapeutic response, but the flow method is time-consuming and requires ~6 hrs to process each sample. The majority of CTC techniques require an initial enrichment step, since CTCs are rare events. After pre-separation, immunofluorescent labeling of captured cells is required to further validate the presence of CTCs.
Here, we report a direct assay based on highly specific and sensitive SERS technology to detect CTCs in peripheral blood, which requires no subsequent separation procedure. One of the key advantages of using SERS spectroscopy rather than a fluorescence technique is that SERS gives a sharp fingerprint-like spectral pattern, which is distinct from other interferences within the complex biological milieu. In contrast, fluorescence spectra could be disguised by strong scattering signals from cells and protein clusters at low signal intensities, such as rare CTC events. The distinctive SERS spectral pattern eliminates the need for tedious separation procedures.
Materials and Methods
Preparation of EGF-conjugated SERS nanoparticles
The bioconjugation of EGF-SERS nanoparticle was similar to previously reported procedures (8). 60 nm citrate-stabilized gold particles (2.6×1010 particles/mL) (Ted Pella Inc) were used. Briefly, the nanoparticles were encoded with QSY reporter molecules which were adsorbed to the negatively charged Au nanoparticle surface through electrostatic interaction. Au-QSY was then functionalized with a mixed layer of polymers. 85% of the mixed layer is thiolated polyethylene glycol (HS-PEG), which ensures minimal non-specific interaction with blood cells. Without the closely-packed PEG protection layer, nanoparticles are prone to aggregate and induce false positive signals. The remaining 15% of the mixed layer is a bi-functional HS-PEG-COOH; one end is tethered to the gold nanoparticle surface, the other has a carboxyl function group for conjugation with the N-terminus of the EGF peptide. Details of each step and characterization are shown in supplemental materials.
Cell lines
Tu212 cell line provided by Dr. Gary L. Clayman (University of Texas) has been tested (genotyping) and authenticated by Research Animal Diagnostic Laboratory (Columbia MO). H292, MDA-MB-231 and H460 cell lines obtained from the American Type Culture Collection.
Patient and mouse blood sample preparation and CTC detection
Blood samples (7.5–15 ml) were collected from patients with different stages of SCCHN and from healthy donors in BD Vacutainer® CPT™ cell preparation tubes (BD Franklin Lakes, NJ), and were processed within 2 hrs of collection. With constant mixing for 30 minutes at room temperature, cells were incubated with 10 pM EGF-SERS nanoparticles, followed by three PBS washes. The SERS spectra from each sample were measured and analyzed using 785 nm laser excitation on a handheld Raman system (DeltaNu, Wyoming). Mouse blood collection and white blood cell separation were performed according to the manufacturer’s protocols. For details, see supplemental materials.
Immunohistochemistry staining of CTCs and primary tumor section
Immunohistochemical analysis for cytokeratin and EGFR expression on slides was performed using Dako kit (Dako, Carpinteria, CA). The slides were incubated with anti-cytokeratin and anti-EGFR antibody (1:100 dilution, Dako, Carpinteria, CA) according to the manufacturer’s protocols.
Results and Discussion
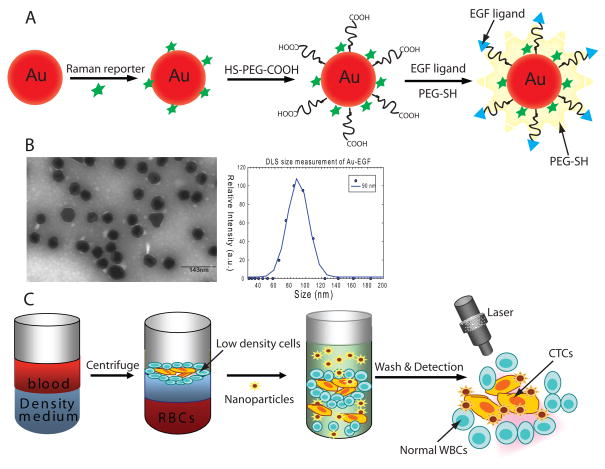
Figure 1A illustrates the preparation of SERS nanoparticles and their conjugation with EGF peptide. SERS sensitivity was optimized by careful titration of reporter molecules. The N-terminus of EGF peptide was conjugated to the outer layer of the gold nanoparticle polymer coating. Figure 1B shows the characterization of EGF-SERS nanoparticles. The PEG coating with targeting ligand is clearly observed as a thin “white” layer of ~5 nm by transmission electron microscopy (TEM) negative staining (Left). Dynamic light scattering (DLS) indicated a hydrodynamic size increase of ~30nm upon PEG-SH addition and peptide conjugation (Right). Figure 1C shows the design of using EGF-SERS nanoparticle for labeling and detecting CTCs. Briefly, lower density CTCs and leukocytes were isolated from whole blood by density-gradient centrifugation, and then incubated with EGF-SERS particles. Free excess nanoparticles were removed by 3 runs of gentle centrifugation (~15 min). The cell pellet was illuminated by 785 nm laser and a SERS spectrum was taken to record the level of targeted nanoparticles. SERS signal intensity corresponds to the number of CTCs in the presence of WBCs.
Figure 1. Design of EGF-SERS nanoparticle for labeling and detection of CTCs.
(A) Preparation and schematic structures of Raman-encoded, PEG-stabilized, and EGF-peptide–functionalized SERS nanoparticle. (B)TEM image and DLS measurement, (C) assay principle of CTC detection from whole blood using EGF-SERS nanoparticles.
Detection sensitivity and specificity of EGF-SERS nanoparticles
It is well known that SERS spectroscopy can achieve single molecule and single nanoparticle detection sensitivity (8–11). The main challenge of this assay was to minimize non-specific binding of nanoparticles to host blood cells, which outnumber CTCs by 5–6 orders of magnitude.
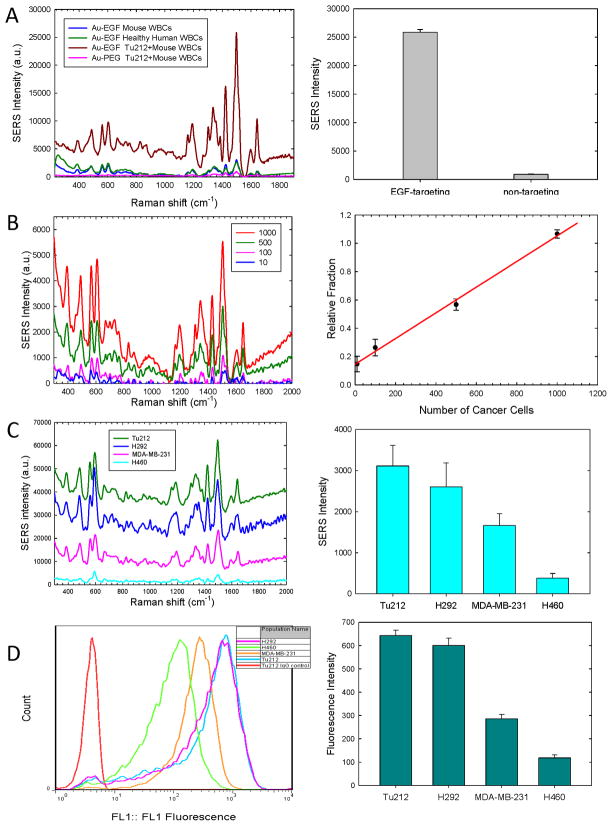
In a proof-of-principle experiment, the SCCHN cell line Tu212, which overexpresses EGFR, was used to test the targeting specificity of EGF-SERS nanoparticles. Tu212 cells (10,000) were spiked into 2ml of mouse WBCs (~1×107) and incubated with EGF-conjugated or pegylated-SERS nanoparticles. Targeting was confirmed by strong SERS signals detected in samples incubated with EGF-SERS nanoparticle (Figure 2A, brown), whereas only negligible SERS signals were detected in samples incubated with pegylated-SERS nanoparticles (Figure 2A, pink), indicating minimal non-specific binding. A further control experiment was carried out to test the non-specific binding of EGF-SERS nanoparticles to healthy mouse and human WBCs (1×107). Figure 2A shows that SERS intensities in healthy mouse and human blood cells (blue and green, respectively) were reproducible and much lower compared with that in the presence of tumor cells (brown).
Figure 2. Detection sensitivity and specificity.
(A) Left SERS signals from EGF-SERS (brown) and non-targeted SERS (pink) nanoparticles in Tu212 cells (1×104) spiked in 2ml mouse blood. Controls tested non-specific binding of EGF-SERS nanoparticles to healthy blood cells of mouse (blue) and human (green); Right: Comparing the SERS intensities for targeted and non-targeted nanoparticles in (A). (B) Left: SERS spectra of 10, 100, 500, and 1000 Tu212 cells labeled with EGF-SERS tags in 1×107 WBCs. Right: The correlation between relative SERS-signal intensity and the number of spiked Tu212 cells. (C) Left: SERS spectra of 1×104 Tu212, MDA-MB-231, H292, H460 cells in 5×106 WBCs. (D) Left: Examination of EGFR-expression by FACS. Right: Various levels of EGFR-expression measured by SERS (C) and fluorescence (D) intensities. Error bars represent the standard deviation of three replicates.
To determine assay sensitivity, 10, 100, 500, and 1000 Tu212 cells were spiked into 2ml of mouse WBCs (~1×107 cells) and each incubated with 10pM EGF-SERS nanoparticle. Figure 2B left shows SERS spectra measured in cell pellets (50 μL) with 785 nm laser excitation and background corrected by subtracting the spectrum of pure WBCs (negative control lacking Tu212 tumor cells). The detection specificity was greater than 104:1. Figure 2B right demonstrates the correlation between relative SERS signal intensity and the number of spiked Tu212 cells. The relative fraction was obtained by taking the SERS signal intensity at 1498 cm−1 from each background-corrected spectrum in Figure 2B left divided by the peak intensity of non-specific SERS signal from the WBC negative control. The linear correlation in Figure 2B indicates the limit of detection is in the range of 5–50 tumor cells per ml of blood for the bulk measurement.
To further verify the specificity of this assay, we compared the SERS spectral intensity with flow-cytometry measurements of cell surface EGFR expression on four different cell lines: Tu212 SCCHN cells and H292 lung cancer cells (high EGFR-expression); MDA-MB-231 breast cancer cells (moderate EGFR-expression), and H460 lung cancer cells (low EGFR-expression). As shown in Figure 2C left, the SERS intensities of different cell lines represent the various levels of cell surface EGFR-expression. Figure 2C right shows that the contrast ratio between high (Tu212) and low (H460) EGFR-expression was quantified to be ~10 fold, based on SERS measurements. Figure 2D shows EGFR-expression levels measured by conventional flow-cytometry using pure cancer cells labeled with anti-EGFR antibody. Flow-cytometry analysis revealed the contrast ratio between high (Tu212) and low (H460) EGFR-expression is ~6 fold. Although the two methods reflected a similar pattern, it is worth noting that the flow-cytometry measurement failed to report a meaningful result if a 500-fold excess of WBCs was present in the spiked tumor cells, as in the SERS measurement. These results indicate that the newly developed EGF peptide-conjugated SERS nanoparticle have superior targeting specificity when compared to whole antibody targeting.
Detection of CTCs in blood samples from SCCHN patients
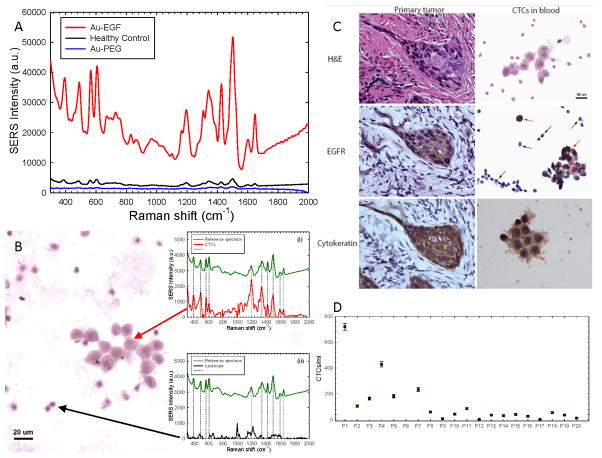
More than 90% of SCCHN cells overexpress EGFR (12–14). An IRB-approved clinical trial was initiated to test whether the above optimized EGF-SERS nanoparticle assay was able to detect CTCs in SCCHN patients’ peripheral blood. Figure 3A shows the blood sample from patient-1 (stage: T4N3M0) incubated with both targeted (red) and non-targeted (blue) nanoparticles. This sample showed a strong targeting effect, 44-fold greater than the non-targeted SERS tag. Blood samples from three healthy donors were tested as controls using EGF-SERS nanoparticle and gave reproducibly low background SERS intensity (black, Figure 3A), which may have resulted from EGF-SERS nanoparticle remnants that were not completely removed through the washing procedure. WBCs may express very low levels of EGFR (15), which may also contribute to the observed background signal. Our flow-cytometry analyses found that expression of EGFR on the surface of normal blood cells from three healthy donors was negligible when compared to that of SCCHN cells (Suppl. Figure 1).
Figure 3. Detection of CTCs in patients’s blood samples and IHC staining for EGFR and cytokeratin.
(A) Blood sample from Patient-1 incubated with both targeted (red) and non-targeted (blue) nanoparticles. Blood samples from three healthy donors were tested as controls using EGF-SERS tags (black). (B) Image of a cluster of CTCs stained with hematoxylin. Insets: Single-cell SERS spectra of CTC (i) and WBC (ii) recorded from the area indicated by the red and black arrow, respectively. (C) Left: Representative IHC images of primary tumor from Patient-1, stained with H&E, anti-EGFR, and pan-cytokeratin antibody. Right: Representative images of CTCs (red arrows) and hematologic cells (black arrows) from Patient-1’s blood sample. (D) CTC counts in peripheral blood samples from 20 SCCHN patients.
To verify that the bulk measurement of the EGF-SERS signal indeed originates from CTCs, we also examined each sample microscopically. A series of blood smear slides was prepared from the same set of SERS nanoparticle-labeled cell pellets. Figure 3B captured a cluster of CTCs stained with hematoxylin. To confirm that the CTCs were indeed labeled with EGF-SERS nanoparticles, we took single-cell SERS spectra for the CTC cluster by switching the microscope to the Raman mode with 785 nm laser excitation. As shown in inset (i) of Figure 3B, the red spectrum was recorded from the areas indicated by the arrow. All the spectral fingerprints matched with the reference spectrum of pure SERS nanoparticles in green. The feature of multiple peak matching serves as a cross-reference method by itself, which gives us high confidence that the cells in this CTC cluster are indeed labeled with EGF-SERS nanotags. As a control experiment, the laser spot was moved to a different area where only leukocytes were present (Figure 3B, black arrow). As expected, the peak positions of the single-cell SERS from host leukocytes (black curve of inset (ii)) did not align with the reference spectrum in dotted lines. The fingerprint-like spectral pattern of SERS-nanotags provides a potential system for quantitative Raman molecular profiling studies of CTCs.
Immunohistochemical studies of primary tumor and CTCs in blood
Figure 3C right shows a typical example of CTCs detected in blood samples from SCCHN patients. Importantly, the nanoparticle-labeled cells with large nuclei were both cytokeratin- and EGFR-positive (stained in brown, red arrows), confirming the malignant characteristics and EGFR-overexpression of CTCs. In contrast, the smaller size hematologic cells were cytokeratin- and EGFR-negative (stained in blue, black arrows). We also examined the control blood samples from healthy donors with very low SERS signals, and consistently, no cytokeratin- or EGFR-positive cells were detected (Suppl. Figure 2). To investigate correlations between the molecular biomarker profiles of CTCs with those of primary tumors, we analyzed serial sections of primary tumors from SCCHN patients in this trial. Specifically for patient-1 with late-stage SCCHN cancer, both cytokeratin- and EGFR-expression were positive for the primary tumor (Figure 3C, left).
We further collected and examined blood samples from 20 SCCHN patients with various histologies. One patient’s blood clotted and was not examined. We successfully detected positive SERS signals that were greater than background in 17 of 19 (~90%) of patients. Among 17 patients who showed positive SERS signals in the peripheral blood, 11 were found to express EGFR in the corresponding primary carcinoma (Table 1). The other 6 patients underwent surgery or biopsy at other sites and tumor tissue blocks were not available. Low SERS signals were detected in the specimens from patients 12 and 17. Alternative procedures using filtration membrane to isolate CTCs confirmed the absence of CTCs in the blood specimens from these two patients. CTC levels among the 19 patients according to disease progression were: 2 of 19 (~10%) patients with localized disease (median: 21 CTCs/ml; range: 7 to 36 CTCs/ml); 17 of 19 (~90%) patients with metastatic or recurrent disease (median: 55 CTCs/ml; range: 1 to 720 CTCs/ml) (Figure 3D). Blood samples from 3 normal controls did not show any CTCs. The data suggest a potential association between SERS intensity and disease progression. Further inclusion of larger numbers of patient specimens may provide valuable information validating the clinical significance of the SERS assay described here.
Table 1.
Patient characteristics and estimated levels of CTCs
| Number | Stage | Site | Number of CTCs per ml | EGFR expression on the primary tumor | Cytokeratin expression on the primary tumor |
|---|---|---|---|---|---|
| P1 | T4BN2CM0 | Larynx | 720 | 2+ | 2+ |
| P2 | T3N3M0 | Tonsil | 104 | N/A | N/A |
| P3 | T4N2CM0 | BOT | 163 | 1+ | 3+ |
| P4 | T4aN1M0 | Maxilla | 429 | N/A | N/A |
| P5 | T2N2bM1 | Larynx | 181 | 3+ | 1+ |
| P6 | T1N3M0 | Tonsil | N/A | 3+ | |
| P7 | T4N2CM0 | Maxilla | 232 | 2+ | 2+ |
| P8 | T4N2CM0 | Soft Palate | 59 | N/A | N/A |
| P9 | T1N0M0 | Oral Tongue | 7 | 3+ | 1+ |
| P10 | T4bN2CM0 | Pyriform Sinus | 43 | N/A | N/A |
| P11 | Recurrent | Oral Tongue | 84 | 3+ | 1~2+ |
| P12 | Recurrent | Larynx | 2 | 1+ | 1+ |
| P13 | T4N1M0 | Nasopharynx | 34 | 2+ | 2+ |
| P14 | Recurrent | FOM | 31 | 2+ | 3+ |
| P15 | T1N2bM0 | BOT | 40 | 1+ | 3+ |
| P16 | T2N2bM0 | Oral Tongue | 25 | 1+ | 3+ |
| P17 | Recurrent | Oral Tongue | 1 | 2+ | 2+ |
| P18 | T3N2cM0 | Supraglottic | 55 | N/A | N/A |
| P19 | T3N0M0 | Larynx | 36 | N/A | N/A |
| P20 | Recurrent | BOT | 11 | 1+ | 3+ |
| Healthy subjects(n=3) | N/A | N/A | 0 | N/A | N/A |
Of note, patient-20 was tested when he developed an isolated, biopsy-proven spinal metastases which was detected by PET/CT scan. At the time of the distant failure, his CTC count was 11 cells/ml. The patient received radiosurgery followed by four cycles of chemotherapy (paclitaxel, carboplatin and ifosfamide). Two months later, PET/CT showed no evidence of disease and the SERS assay showed undetectable levels of CTCs.
In summary, we have developed a specific and sensitive methodology using EGF-SERS nanoparticles that can rapidly detect CTCs in peripheral blood specimens from SCCHN patients. Further application of this new technology could identify CTCs and analyze their expression of specific prognostic and predictive biomarkers to predict disease progression and monitor patient response to a given therapy, which will provide a novel approach that may be superior to current imaging procedures.
Supplementary Material
Acknowledgments
This work was funded by the National Cancer Institute through CCNE award (U54CA119338), H&N cancer SPORE award (P50CA128613) and P30 CA138292. D.M.S., J.J.B., Z.G.C. and S.N. would like to acknowledge the Georgia Cancer Coalition for support. The authors thank Dr. Anthea Hammond for critical reading and editing of the manuscript.
Footnotes
Supplementary information is available on the Cancer Research website.
References
- 1.Kaiser J. Medicine. Cancer’s circulation problem. Science. 2010;327:1072–4. doi: 10.1126/science.327.5969.1072. [DOI] [PubMed] [Google Scholar]
- 2.Panteleakou Z, Lembessis P, Sourla A, et al. Detection of circulating tumor cells in prostate cancer patients: methodological pitfalls and clinical relevance. Mol Med. 2009;15:101–14. doi: 10.2119/molmed.2008.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maheswaran S, Haber DA. Circulating tumor cells: a window into cancer biology and metastasis. Curr Opin Genet Dev. 2010;20:96–9. doi: 10.1016/j.gde.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–40. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 5.Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stott SL, Lee RJ, Nagrath S, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra3. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian X, Peng XH, Ansari DO, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 9.Moore BD, Stevenson L, Watt A, et al. Rapid and ultra-sensitive determination of enzyme activities using surface-enhanced resonance Raman scattering. Nat Biotechnol. 2004;22:1133–8. doi: 10.1038/nbt1003. [DOI] [PubMed] [Google Scholar]
- 10.Nie S, Emory SR. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science. 1997;275:1102–6. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 11.Tian JH, Liu B, Li X, et al. Study of molecular junctions with a combined surface-enhanced Raman and mechanically controllable break junction method. J Am Chem Soc. 2006;128:14748–9. doi: 10.1021/ja0648615. [DOI] [PubMed] [Google Scholar]
- 12.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53:3579–84. [PubMed] [Google Scholar]
- 13.Kalyankrishna S, Grandis JR. Epidermal growth factor receptor biology in head and neck cancer. J Clin Oncol. 2006;24:2666–72. doi: 10.1200/JCO.2005.04.8306. [DOI] [PubMed] [Google Scholar]
- 14.Xi L, Nicastri DG, El-Hefnawy T, Hughes SJ, Luketich JD, Godfrey TE. Optimal markers for real-time quantitative reverse transcription PCR detection of circulating tumor cells from melanoma, breast, colon, esophageal, head and neck, and lung cancers. Clin Chem. 2007;53:1206–15. doi: 10.1373/clinchem.2006.081828. [DOI] [PubMed] [Google Scholar]
- 15.Chan G, Nogalski MT, Yurochko AD. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc Natl Acad Sci U S A. 2009;106:22369–74. doi: 10.1073/pnas.0908787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.