SUMMARY
The chaperone/usher (CU) pathway is a conserved bacterial secretion system that assembles adhesive fibers termed pili or fimbriae. Pilus biogenesis by the CU pathway requires a periplasmic chaperone and an outer membrane (OM) assembly platform termed the usher. The usher catalyzes formation of subunit-subunit interactions to promote polymerization of the pilus fiber and provides the channel for fiber secretion. The mechanism by which the usher catalyzes pilus assembly is not known. Using the P and type 1 pilus systems of uropathogenic Escherichia coli, we show that a conserved N-terminal disulfide region of the PapC and FimD ushers, as well as residue F4 of FimD, are required for the catalytic activity of the ushers. PapC disulfide loop mutants were able to bind PapDG chaperone-subunit complexes, but did not assemble PapG into pilus fibers. FimD disulfide loop and F4 mutants were able to bind chaperone-subunit complexes and initiate assembly of pilus fibers, but were defective for extending the pilus fibers, as measured using in vivo co-purification and in vitro pilus polymerization assays. These results suggest that the catalytic activity of PapC is required to initiate pilus biogenesis, whereas the catalytic activity of FimD is required for extension of the pilus fiber.
Keywords: pili, fimbriae, usher, pilus assembly, protein secretion
INTRODUCTION
Pili (fimbriae) are polymeric surface fibers expressed by a wide variety of bacteria. Pili generally function as adhesive organelles and have roles in colonization of both abiotic and biotic surfaces, biofilm formation, interactions with host cells, and pathogenesis. P and type 1 pili are prototypical pilus fibers expressed by uropathogenic Escherichia coli. P pili promote adhesion to the kidney and the development of pyelonephritis; type 1 pili mediate adhesion to and invasion of the bladder epithelium, initiating a series of events leading to the development of cystitis (Roberts et al., 1994; Wright et al., 2007). P and type 1 pili are assembled by the chaperone/usher (CU) secretion pathway, which is used by many Gram-negative bacteria for the biogenesis of virulence-associated surface structures (Sauer et al., 2004; Li and Thanassi, 2009). In the CU secretion pathway, a dedicated periplasmic chaperone controls the folding of pilus subunit proteins and an integral outer membrane (OM) protein termed the usher catalyzes the assembly of subunits into the pilus fiber and provides the channel for secretion of the fiber to the cell surface (Nishiyama et al., 2008; Remaut et al., 2008).
P and type 1 pili are encoded by the chromosomal pap and fim gene clusters. Both pili consist of a rigid helical rod that is anchored in the bacterial OM and a flexible, linear tip fiber that is located at the distal end of the rod (Fig. 1A) (Kuehn et al., 1992; Jones et al., 1995). In P pili, the PapG adhesin is located in single copy at the distal end of the pilus tip. PapG is linked by the PapF adaptor subunit to PapE, which is present in several copies in the tip fiber (Kuehn et al., 1992; Jacob-Dubuisson et al., 1993). The P pilus tip is terminated by PapK, which links the tip to the pilus rod (Fig. 1A). The rod contains over 1,000 copies of PapA and is terminated by PapH (Baga et al., 1987; Verger et al., 2006). For type 1 pili, the pilus rod is composed of repeated copies of the FimA major subunit protein, and the tip fiber contains single copies of the FimH adhesin and the FimG and FimF adaptor subunits (Fig. 1A) (Jones et al., 1995; Saulino et al., 2000; Hahn et al., 2002).
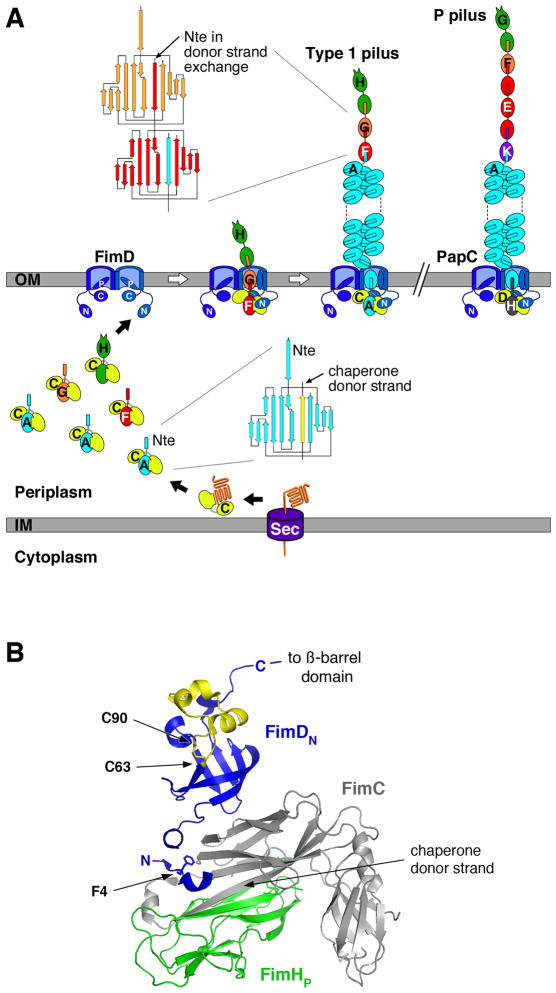
Fig. 1. Assembly of Type 1 and P pili by the CU pathway.
A. Model for pilus assembly. The assembly steps for type 1 pili are shown, together with models for the fully assembled type 1 and P pili. The Fim and Pap proteins are indicated by single letters (H, FimH; etc.). Pilus subunits enter the periplasm as unfolded polypeptides via the Sec general secretory pathway. The subunits fold upon interaction with the periplasmic chaperone, forming stable complexes via donor strand complementation. Assembly and secretion of the pilus fiber occurs at the OM usher, where chaperone-subunit interactions are replaced with subunit-subunit interactions via the donor strand exchange reaction. During donor strand exchange, the Nte of an incoming chaperone-subunit complex displaces the chaperone donor strand from the preceding subunit. Topology diagrams are shown depicting the donor strand complementation and exchange reactions occurring with the chaperone and in the pilus fiber, respectively. The dimeric ushers are depicted with the central ß-barrel domain forming a channel that spans the OM, the N- and C-terminal domains (labeled N and C, respectively) located in the periplasm, and the plug domain (labeled P) gating the channel shut. Chaperone-adhesin complexes have highest affinity for the usher and initiate pilus assembly by binding to the usher N-terminal domain. The pilus tip fiber is assembled first, followed by the pilus rod.
B. Structure of the FimDN-CHp complex (PDB ID: 1ZE3). FimC is shown in gray and FimHp in green. FimDN is shown in blue, with the disulfide loop region colored yellow. FimD residue F4 and the disulfide bond between residues C63 and C90 are shown in stick representation. The donated ß-strand of the FimC chaperone that occupies the FimHp subunit groove is indicated. The FimDN structure represents usher N-terminal domain cartooned in (A). The C-terminal end of the FimDN domain is connected by a linking region of 14 residues to the transmembrane ß-barrel domain of the usher. The structure was generated using PyMOL (http://www.pymol.org).
Biogenesis of P and type 1 pili by the CU pathway requires the periplasmic chaperone (PapD and FimC, respectively) and OM usher (PapC and FimD, respectively). Pilus subunits enter the periplasm as unfolded polypeptides via the Sec general secretory pathway (Driessen and Nouwen, 2008). Upon entering the periplasm, the subunits form stable, binary complexes with the periplasmic chaperone (Fig. 1A). The chaperone is required for proper folding of pilus subunits, to prevent off-pathway interactions, and to maintain subunits in an assembly-competent state (Choudhury et al., 1999; Sauer et al., 1999; Sauer et al., 2002; Zavialov et al., 2003). Pilus subunits contain an incomplete immunoglobulin (Ig)-like fold termed the pilin domain. The chaperone donates a β-strand to complete the Ig fold of the subunit in a mechanism termed donor strand complementation (Fig. 1) (Choudhury et al., 1999; Sauer et al., 1999).
Periplasmic chaperone-subunit complexes next must interact with the OM usher for release of the chaperone and assembly of subunits into the pilus fiber. Subunit-subunit interactions form at the periplasmic face of the usher by a mechanism termed donor strand exchange (Sauer et al., 2002; Zavialov et al., 2003). All pilus subunits except the adhesin contain a conserved N-terminal extension (Nte) in addition to the pilin domain (Fig. 1A). The adhesin contains an adhesin domain in place of the Nte. In donor strand exchange, the Nte from an incoming chaperone-subunit complex replaces the donated chaperone ß-strand from the preceding chaperone-subunit complex that is bound at the usher, completing the Ig fold of the preceding subunit and displacing the chaperone to form a subunit-subunit interaction. Thus, the pilus fiber is built from an array of Ig folds, with each subunit bound to the preceding subunit by donor strand exchange (Fig. 1A). Pili are assembled in a defined order, with the adhesin incorporated first, followed by the rest of the tip and finally the rod. Both the pilus tip and rod are built from the same donor strand exchange reaction, but pilus subunits in the rod undergo an additional quaternary interaction upon exiting the usher that promotes coiling of the rod into a helix on the bacterial surface (Fig. 1A). Each subunit specifically interacts with its appropriate neighbor subunit in the pilus, with the specificity of binding determined by the donor strand exchange reaction (Lee et al., 2007; Rose et al., 2008). In addition, the usher ensures ordered and complete pilus assembly by differentially recognizing chaperone-subunit complexes according to their final position in the pilus (Dodson et al., 1993; Saulino et al., 1998; Li et al., 2010). Pilus fibers assemble on the order of minutes in the presence of the usher, but on the order of hours in the absence of the usher (Remaut et al., 2006; Vetsch et al., 2006; Nishiyama et al., 2008), suggesting that the usher functions to increase the rate of fiber assembly. In agreement with this, the FimD usher was recently shown to function as a catalyst to accelerate the rate of polymerization of the type 1 pilus rod by a factor of greater than 1000 (Nishiyama et al., 2008).
Ushers are large, integral OM proteins containing four domains: a central transmembrane ß-barrel domain that forms the secretion channel, a middle domain located within the ß-barrel region that forms a channel gate or plug, and soluble N- and C-terminal domains that are located in the periplasm (Fig. 1A) (Nishiyama et al., 2005; Shu Kin So and Thanassi, 2006; Remaut et al., 2008; Huang et al., 2009; Mapingire et al., 2009; Ford et al., 2010). The N-terminal domain (PapC residues 1–131, FimD residues 1–125) provides the initial binding site for chaperone-subunit complexes (Ng et al., 2004; Nishiyama et al., 2005), whereas the C-terminal domain (PapC residues 641–809, FimD residues 664–833) stabilizes the binding of chaperone-subunit complexes to the usher and may participate in controlling access to the usher channel (Thanassi et al., 2002; Shu Kin So and Thanassi, 2006; Ford et al., 2010). Both the N- and C-terminal domains also participate in the differential affinity of the usher for chaperone-subunit complexes (Li et al., 2010). The usher is present as a dimeric complex in the OM, but only one channel is used for secretion of the pilus fiber (Li et al., 2004; Shu Kin So and Thanassi, 2006; Remaut et al., 2008). The usher dimer may facilitate recruitment of chaperone-subunit complexes from the periplasm and help position subunits for donor strand exchange (Remaut et al., 2008; Li and Thanassi, 2009).
In a previous analysis of the usher N-terminal domain, we found that a PapC mutant deleted for residues 2–11 was unable to bind chaperone-subunit complexes or assemble P pili (Ng et al., 2004). Alanine scanning mutagenesis of this region identified only residue F3 as critical for the binding activity of the PapC N terminus; the PapC F3A mutant was defective for pilus biogenesis and did not bind chaperone-subunit complexes (Ng et al., 2004; Ng et al., 2006). The structural basis for these findings was revealed by Nishiyama and colleagues (Nishiyama et al., 2005), who solved the structure of the FimD N-terminal domain (FimDN, residues 1–125) bound to a FimC chaperone-FimH pilin domain complex (FimCHp). This structure revealed that the first 24 residues of FimD specifically interact with the bound FimCHp complex and that residue F4, which corresponds to PapC F3, directly contacts the FimC chaperone (Fig. 1B) (Nishiyama et al., 2005). Similar to the PapC F3A mutant, a FimD F4A mutant was defective for pilus biogenesis (Nishiyama et al., 2005). In addition to the chaperone-subunit binding region, the usher N-terminal domain contains a conserved pair of cysteines (PapC C70 and C97; FimD C63 and C90) that form a disulfide bond (Fig. 1B) (Henderson et al., 2004; Nishiyama et al., 2005). We previously demonstrated that PapC ushers containing mutations to either or both of the N-terminal cysteines were unable to assemble pili (Ng et al., 2004). However, in contrast to the PapC Δ2–11 and F3A mutants, the PapC cysteine mutants maintained the ability to bind PapDG chaperone-adhesin complexes (Ng et al., 2004). This suggested that the N-terminal cysteines function at a step distinct from the initial binding of chaperone-subunit complexes. In the FimDN-CHp co-crystal structure (Nishiyama et al., 2005), the FimD region containing the conserved N-terminal cysteines makes no interactions with the bound FimCHp complex (Fig. 1B) and therefore the structure does not provide insight into the function of this region.
In this study, we investigated the mechanism of the conserved disulfide region of the usher N terminus in pilus biogenesis. We present evidence that this region is critical for the catalytic activity of both PapC and FimD in polymerizing pilus subunits. Differences were detected between the two ushers, which belong to separate clades of the usher superfamily (Nuccio and Baumler, 2007). In contrast to the essential role of PapC residue F3 in binding chaperone-subunit complexes, we found that residue F4 of FimD functions in the catalytic activity of the usher and is not strictly required for chaperone-subunit binding. In addition, the PapC and FimD mutants were blocked at different stages of pilus biogenesis, suggesting distinct requirements for the catalytic activity of the ushers in initiation and extension of the pilus fiber.
RESULTS AND DISCUSSION
The conserved disulfide region in the PapC N terminus functions to promote subunit-subunit interactions
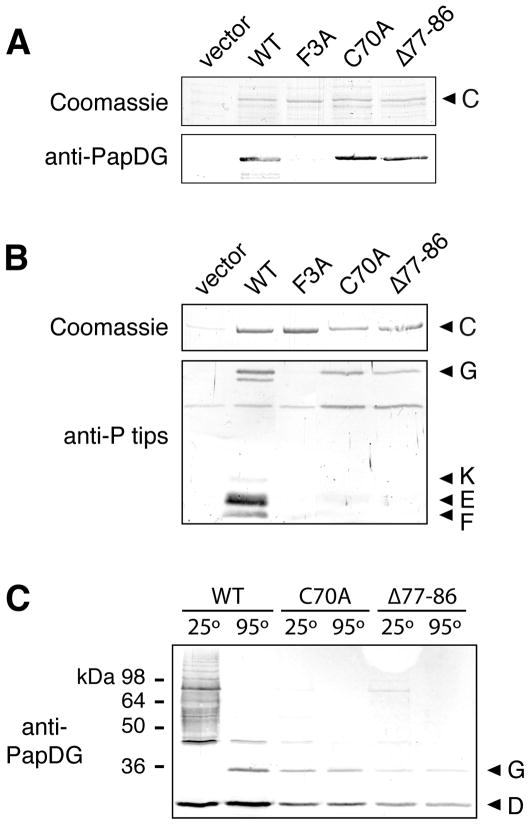
Previous studies demonstrated that the conserved N-terminal cysteines of PapC (C70 and C97) are required for P pilus biogenesis but function at a step distinct from the role of the usher N terminus in the initial binding of chaperone-subunit complexes (Ng et al., 2004). The N-terminal cysteines form a disulfide bond (Henderson et al., 2004; Nishiyama et al., 2005). Thus, the phenotype of the cysteine mutants might be an indirect effect caused by loss of the disulfide bond and resulting structural changes in the loop region bounded by the cysteines (Fig. 1B). In initial studies, we were unable to identify single residues that, when mutated to alanine, were required for function of the loop region (T. W. Ng, I. Talukder, and D. G. Thanassi; unpublished data). Therefore, we constructed a PapC mutant deleted for residues 77–86, which occur in the middle of the loop region. This internal deletion was chosen to avoid disturbing the disulfide bond and because it encompasses several conserved residues. PapC Δ77–86 was present at a similar level in the OM compared to wild-type (WT) PapC (Fig. 2A, upper panel) and the mutation did not affect the overall folding of the usher, as assessed by resistance to denaturation by SDS (data not shown). Similar to a PapC C70A mutant, PapC Δ77–86 was unable to complement a plasmid bearing a ΔpapC pap operon (papAHDJKEFG) for assembly of adhesive P pili on the bacterial surface, as assessed by the hemagglutination (HA) assay (Table 1). Also similar to the C70A mutant, PapC Δ77–86 retained the ability to bind PapDG complexes, as measured using an in vitro overlay assay (Fig. 2A). This is in contrast to a PapC F3A mutant, which was unable to bind PapDG (Fig. 2A), as shown previously (Ng et al., 2004; Ng et al., 2006). The identical behavior of the PapC Δ77–88 and C70A mutants suggests that the loop region between the conserved N-terminal cysteines is functionally important. In addition, the fact that both mutants retain the ability to bind PapDG complexes indicates that loss of the disulfide bond or deletion within the loop region does cause global structural changes to the usher N terminus.
Fig. 2. Analysis of the PapC N-terminal disulfide loop mutants.
A. Overlay assay for binding of PapDG to PapC. OM fractions were isolated from strain SF100 expressing vector only, WT PapC, or the indicated PapC mutant. Duplicate samples were subjected to SDS-PAGE and either stained with Coomassie blue (upper panel) or transferred to a PVDF membrane for the overlay assay. Binding of PapDG to the usher (lower panel) was determined by immunoblotting with anti-PapDG antibodies.
B. Co-purification of pilus tip subunits with PapC. OM fractions were isolated from strain SF100/pPAP58 (papDJKEFG) expressing vector only, WT PapC, or the indicated PapC mutant. His-tagged PapC was purified from the OM fractions, subjected to SDS-PAGE, and either stained with Coomassie blue to show the amount of PapC present (upper panel) or immunoblotted with anti-P pilus tips antiserum to detect pilus tip subunits that co-purified with the usher (PapG, K, E and F, lower panel).
C. Assembly of PapG into pilus tip fibers. Co-purification of pilus tip subunits with WT PapC or the indicated PapC mutant was performed as in (B). The samples were incubated at 25 or 95°C in SDS-PAGE sample buffer, subjected to SDS-PAGE and immunoblotted with anti-PapDG antibodies.
Table 1.
Ability of the PapC and FimD usher mutants to assemble adhesive pili or pilus tips.
| PapC | HA titera– pili (papAHDJKEFG) | HA titera– tips (papDJKEFG) |
|---|---|---|
| vector | 0 | 0 |
| WT | 64 | 256 |
| F3A | 0 | 0 |
| C70A | 0 | 0 |
| Δ77–86 | 0 | 0 |
| FimD | HA titera– pili (fimAICFGH) | HA titera– tips (fimCGH) |
|---|---|---|
| vector | 0 | 0 |
| WT | 128 | 256 |
| F4A | 0 | 0 |
| C63A | 128 | 128 |
| Δ70–79 | 0 | 0 |
HA titer is the highest fold dilution of bacteria able to agglutinate human red blood cells.
To understand why the PapC C70A and Δ77–86 mutants were defective for pilus biogenesis despite being able to bind PapDG chaperone-adhesin complexes, we co-expressed the PapC mutants with P pilus tip subunits (papDJKEFG) and tested the ability of the tip subunits to form stable assembly intermediates with the ushers in vivo using a co-purification assay. Immunoblotting with an antibody that recognizes P pilus tips showed that whereas PapG, F, E and K co-purified with WT PapC, only PapG co-purified with the C70A and Δ77–86 mutants (Fig. 2B). In comparison, no pilus subunits co-purified with the PapC F3A mutant (Fig. 2B), as expected since this mutant is defective for binding chaperone-subunit complexes. A very low level of additional chaperone-subunit complexes may have co-purified with the PapC mutants, as indicated by faint bands present in the F3A and C70A lanes in Fig. 2B, suggesting some remaining functionality. Nevertheless, the data show that even though the PapC C70A and Δ77–86 mutants are able to perform the first step in pilus biogenesis, which is binding to PapDG, the mutants are defective for carrying out subsequent pilus assembly steps. In agreement with this, the PapC C70A and Δ77–86 mutants were unable to assemble adhesive P pilus tip fibers on the bacterial surface, as measured by the HA assay, in contrast to WT PapC (Table 1).
If the PapC cysteine loop mutants are indeed blocked at the first assembly step, then the PapG that co-purified with the usher mutants should not be engaged in subunit-subunit interactions. To test this, we incubated the proteins that co-purified with WT PapC, PapC C70A or PapC Δ77–86 in SDS sample buffer at 95 or 25°C and performed an immunoblot analysis with anti-PapDG antibodies. Subunit-subunit but not chaperone-subunit interactions are stable to SDS at low temperatures, so any PapG engaged in donor strand exchange with other pilus subunits (i.e., assembled into a pilus fiber) will shift to higher molecular weight when incubated at 25°C (Soto et al., 1998; Saulino et al., 2000). As shown in Fig. 2C, the PapG that co-purified with WT PapC ran at the expected monomer molecular weight of 36 kDa when incubated at 95°C. When incubated at 25°C, the monomer PapG band disappeared and a ladder of higher molecular weight species appeared, demonstrating that all of the PapG was polymerized into tip fibers of various lengths. In contrast, the PapG that co-purified with the C70A or Δ77–86 mutants remained as a monomer when incubated at either 25 or 95°C, with only very faint ladders of higher molecular weight species visible at 25°C (Fig. 2C). These data indicate that the PapC disulfide loop mutants are indeed largely blocked at the first stage of pilus assembly and are defective for promoting the polymerization of PapG into pilus fibers.
The FimD N terminus functions to promote subunit-subunit interactions
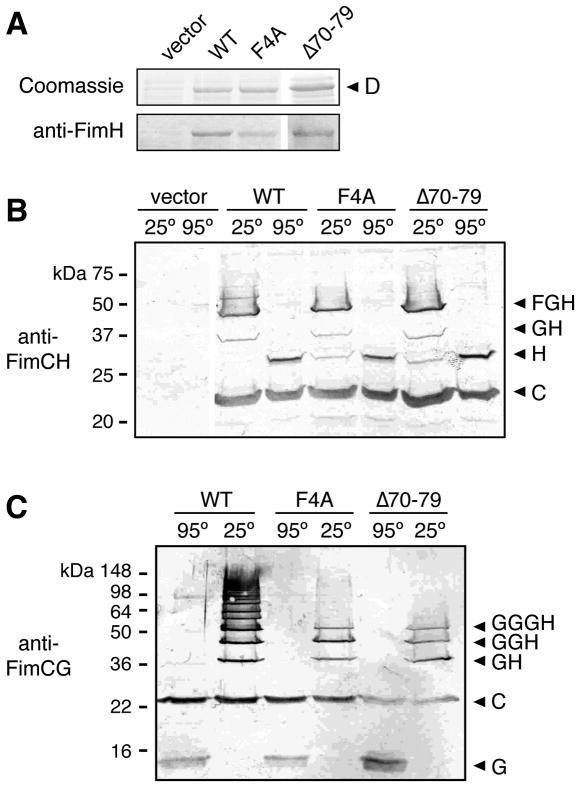
To determine if the N-terminal disulfide region of the FimD usher functions similarly to PapC in promoting subunit polymerization, we constructed a FimD Δ70–79 deletion mutant, which corresponds to the PapC Δ77–86 mutation. For comparison, we also constructed a FimD F4A mutation, which corresponds to the PapC F3A mutation. The FimD mutants were present at similar levels in the OM compared to WT FimD (Fig. 3A, upper panel) and the mutations did not affect the overall folding of the ushers, as assessed by resistance to denaturation by SDS (data not shown). We first tested the ability of the FimD mutants to complement a ΔfimD mutation in the chromosomal fim gene cluster for assembly of type 1 pili, using the HA assay. As found for PapC, the FimD F4A and Δ70–79 mutants were unable to assemble adhesive pili on the bacterial surface (Table 1).
Fig. 3. Analysis of the FimD F4A and Δ70–79 mutants.
A. Overlay assay for binding of FimCH to FimD. OM fractions were isolated from strain SF100 expressing vector only, WT FimD, or the indicated FimD mutant. Duplicate samples were subjected to SDS-PAGE and either stained with Coomassie blue (upper panel) or transferred to a PVDF membrane for the overlay assay. Binding of FimCH to the usher (lower panel) was determined by immunoblotting with anti-FimH antibody.
B. Co-purification of FimCFGH pilus tip fibers with FimD. OM fractions were isolated from strain AAEC185/pNH235 (fimCFGH) expressing vector only, WT FimD, or the indicated FimD mutant. His-tagged FimD was purified from the OM fractions, incubated at 25 or 95°C in sample buffer, subjected to SDS-PAGE, and immunoblotted with anti-FimCH antibodies. The identities of the complexes that co-purified with FimD are indicated on the right using single letters to represent the Fim proteins (H, FimH; etc.).
C. Co-purification of FimCGH pilus tip fibers with FimD. The assay was performed as in (B), except the host strain was AAEC185/pNH222 (fimCGH) and the samples were immunoblotted with anti-FimCG antibodies.
We next tested the FimD F4A and Δ70–79 mutants for ability to bind FimCH chaperone-adhesin complexes, using the in vitro overlay assay. Despite being unable to assemble pili, FimD Δ70–79 bound to FimCH similar to WT FimD (Fig. 3A), matching the phenotype of the ΔPapC 77–86 mutant. Surprisingly, the FimD F4A mutant also bound to FimCH in the overlay assay (Fig. 3A). This is in contrast to the PapC F3A mutant, which was unable to bind chaperone-subunit complexes, and different from the findings of Nishiyama and colleagues, who reported that the F4A mutation prevented the isolated FimDN domain from binding FimCHp (Nishiyama et al., 2005). This difference with Nishiyama et al. is likely due to the fact that we used full-length FimD usher instead of only the N-terminal domain, and full-length FimH adhesin instead of only the pilin domain, providing additional surfaces for the usher to bind the adhesin. Indeed, previous studies showed that FimCH makes stable interactions with a C-terminal region of the usher in addition to the N-terminal domain, and that the FimH adhesin domain by itself binds to the usher (Saulino et al., 1998; Barnhart et al., 2003; Munera et al., 2007). Based on these results, we propose that FimD F4 contributes to, but is not essential for, the binding of chaperone-subunit complexes to the usher. These results reveal differences in the mechanism by which the PapC and FimD ushers bind pilus subunits, with PapC residue F3 playing a more critical role compared to the corresponding FimD F4 residue.
To investigate why the FimD F4A and Δ70–79 mutants were unable to assemble type 1 pili despite being able to bind FimCH complexes, we expressed the usher mutants in bacteria together with type 1 pilus tip subunits (fimCFGH) and performed co-purification assays. The purified proteins were incubated at 25 or 95°C in SDS sample buffer and immunoblotted with anti-FimCH antibodies. Analysis of the samples treated at 95°C showed that similar levels of FimC and FimH co-purified with WT FimD and the F4A and Δ70–79 mutants (Fig. 3B). This confirms the findings from the overlay assay (Fig. 3A) and demonstrates that the FimD mutants are able to form stable complexes with FimCH in vivo. Unexpectedly, analysis of the samples incubated at 25°C revealed that the F4A and Δ70–79 mutants were also able to assemble FimH into pilus tip fibers (Fig. 3B). This is in contrast to the PapC C70A and Δ77–86 mutants, which were highly defective for assembling the adhesin into pilus fibers (Fig. 2B and C). For the FimD samples treated at 25°C, bands at sizes expected for FimGH and FimFGH complexes were present for both the WT and mutant ushers. The composition of the FimGH and FimFGH bands was confirmed using anti-FimCG and anti-FimCF antibodies (data not shown). However, subtle differences between the WT and mutant FimD ushers were apparent. For WT FimD, the FimH monomer band (29 kDa) completely disappeared in the samples treated at 25°C, indicating incorporation of all the FimH into tip fibers, and a faint but distinct ladder of higher molecular weight bands was visible above the FimFGH band (Fig. 3B). For the FimD F4A and Δ70–79 mutants, some monomer FimH remained in the 25°C-treated samples and distinct bands migrating above the FimFGH band were absent (Fig. 3B). The faint ladder of higher molecular weight bands observed with WT FimD corresponds to a low level of type 1 pilus tip fibers containing multiple FimG subunits (see below), as confirmed by blotting with anti-FimCG antibodies (data not shown).
To explore further the differences between the WT and mutant FimD ushers, we performed a modified co-purification experiment in which the ushers were expressed in bacteria together with only fimCGH (omitting the FimF tip subunit). FimG is capable of interacting with itself to form polymers. However, type 1 pilus tip fibers normally contain only single copies of FimG because FimF has high affinity for FimG and binding of FimF prevents FimG-FimG interactions (Saulino et al., 2000). Omitting the FimF subunit allowed the assembly of type 1 tip fibers consisting of FimH bound to various numbers of FimG. Analysis of the samples that co-purified with WT FimD using anti-FimCG antibodies at 25 and 95°C clearly illustrates the polymerization of FimG into a ladder of higher molecular weight species (Fig. 3C). Immunoblotting with anti-FimCH antibody confirmed the incorporation of the adhesin into these fibers (data not shown). Interestingly, compared to WT FimD, the F4A and Δ70–79 mutants were greatly impaired in polymerizing FimG, with only distinct bands corresponding to FimGH, GGH and GGGH fibers visible (Fig. 3C). Thus, the FimD mutants are able to bind chaperone-subunit complexes and initiate fiber assembly, but appear to be defective in extending the pilus fibers. This behavior of the FimD F4A and Δ70–79 mutants is similar to the PapC C70A and Δ77–86 mutants, demonstrating that the N-terminal domains of both ushers function to promote subunit-subunit interactions. However, the subunit-polymerizing activities of the PapC and FimD N termini appear to be required for different stages of pilus biogenesis, with the PapC mutants defective for initiation of fiber assembly and the FimD mutants defective for fiber elongation.
Given the ability of the FimD F4A and Δ70–79 mutants to assemble FimFGH pilus tip fibers (Fig. 3B), it was surprising that these mutants were unable to complement the ΔfimD fim gene cluster for assembly of adhesive pili (Table 1). The FimD mutants should have been able to assemble type 1 pilus tips, exposing the FimH adhesin to the extracellular side of the OM and allowing for HA activity (Saulino et al., 2000; Remaut et al., 2008). A possible explanation for these results is that the FimFGH tip fibers may be too short to sufficiently expose FimH beyond the OM to provide robust agglutination. Indeed, we found that the HA activity of bacteria expressing only type 1 pilus tips (WT FimD co-expressed with FimCFGH) was very low and close to background levels (HA titer = 8), compared to bacteria expressing complete pili (HA titer = 128). In contrast, expression of FimD together with only FimCGH (omitting FimF) led to very strong agglutination activity (HA titer = 256; Table 1), presumably because the longer FimGn-FimH fibers extended sufficiently beyond the bacterial surface. Notably, bacteria in which the FimD F4A and Δ70–79 mutants were expressed together with only FimCGH still lacked HA activity (Table 1). Therefore, even though the F4A and Δ70–79 mutants are able to initiate assembly of pilus tip fibers, the fibers assembled by the mutants are presumably too short to mediate adhesive activity. These results, together with the results of the co-purification experiments (Fig. 3C), also indicate that the defect of the FimD mutants in polymerizing pilus fibers is not due to an inability to transition from assembly of the pilus tip to the pilus rod; pilus fibers were not elongated even in the absence of the FimA rod subunit.
The FimD N terminus is required for the catalytic activity of the usher
The defects of the FimD and PapC N-terminal domain mutants in promoting subunit-subunit interactions suggest that in addition to providing the initial binding site for chaperone-subunit complexes, the usher N terminus may be important for the catalytic activity of the usher. The catalytic activity of FimD is activated by the binding of FimCH chaperone-adhesin complexes to the usher (Nishiyama et al., 2008). Binding of FimCH causes a conformational change in FimD, detectable by a change in the sensitivity of the usher to digestion by extracellularly added trypsin, resulting in the appearance of a protected C-terminal fragment of the usher (Saulino et al., 1998). To determine if the defect of the FimD F4A and Δ70–79 mutants in polymerizing pilus subunits was due to a loss of activation of the usher by FimCH, we co-expressed FimD in bacteria together with FimCH and examined sensitivity of the usher to trypsin digestion. As observed previously (Saulino et al., 1998), co-expression of FimCH with WT FimD resulted in the appearance of a trypsin-protected FimD fragment. Similar trypsin-protected fragments also appeared for the FimD F4A and Δ70–79 mutants (data not shown). Thus, the FimD mutants maintain the ability to be activated by FimCH, in agreement with our findings above that the FimD mutants are able to bind FimCH both in vitro and in vivo (Fig 3). In contrast to FimD, PapC does not require interaction with the adhesin to initiate pilus assembly (Li et al., 2010); it remains to be determined whether the catalytic activity of PapC might be activated by some other mechanism.
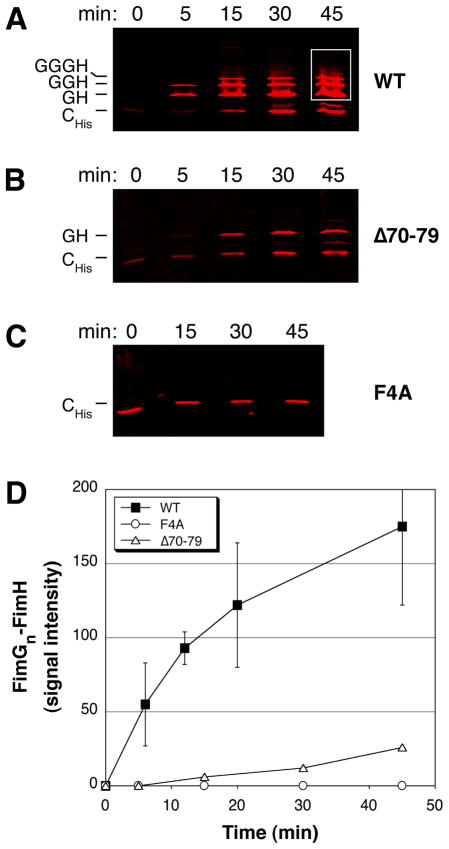
To directly assess the catalytic activity of the usher in polymerizing pilus subunits, we developed an in vitro reconstitution assay for type 1 pilus biogenesis. For this assay, we separately purified OM fractions expressing the FimD usher and periplasm fractions expressing either FimCH or His-tagged FimC (FimCHis)-FimG complexes. The periplasm and OM fractions were mixed together and incubated at room temperature (25°C). At various time points, the samples were placed on ice and centrifuged to collect the OM fragments. The OM was then solubilized with non-denaturing detergent and the extracted OM proteins were passed over a metal affinity column to capture pilus tip fibers assembling through the FimD usher. Fractions eluted from the metal affinity column were then incubated in SDS sample buffer at 25°C and analyzed by quantitative immunoblotting using anti-FimCG antibodies. Note that only OM extracts were run over the affinity column and that only the FimC chaperone expressed together with FimG contained a His-tag. These measures allowed us to select for active pilus assembly intermediates in which the His-tagged chaperone was bound to the last incorporated FimG subunit at the usher (Remaut et al., 2008).
We first tested the catalytic activity of WT FimD in the in vitro reconstitution assay. As shown in Fig. 4A, FimGn-FimH complexes appeared within 5 min of incubation and increased in both intensity and number of FimG subunits over the 45 minute time course of the experiment. The incorporation of FimH into the various complexes was confirmed using anti-FimCH antibodies (data not shown). No FimGn-FimH complexes were obtained if OM lacking FimD was used for the reconstitution (data not shown). Quantitation of the increase in intensity of the FimGn-FimH complexes over time with WT FimD is shown in Fig. 4D. We next tested the catalytic activity of the FimD F4A and Δ70–79 mutants in the in vitro reconstitution assay. As shown in Fig. 4C and D, the FimD F4A mutant was unable to assemble FimGn-FimH complexes over the course of the experiment. In contrast, some FimGn-FimH complexes were assembled by the FimD Δ70–79 mutant, although to a much lower extent compared to WT FimD (Fig. 4B and D). In addition, the complexes assembled by the Δ70–79 mutant appeared to be largely stalled at the FimG1-FimH stage (Fig. 4B). These data show that residues F4 and 70–79 are indeed critical for the catalytic activity of the usher in promoting subunit-subunit interactions, as suggested by the co-purification experiments (Fig. 3C). The more severe defect of the FimD F4A mutant compared to the Δ70–79 mutant in the reconstitution assay may be a reflection of the involvement of residue F4 in binding chaperone-subunit complexes as well as in catalyzing fiber assembly. Although the F4A mutant showed no catalytic activity in the reconstitution assay, the co-purification assay clearly showed that this mutant was able to initiate assembly of type 1 tip fibers in vivo similar to the Δ70–79 disulfide loop mutant (Fig. 3B and C). The stronger phenotype obtained in the reconstitution assay likely reflects the more stringent conditions present in this vitro system, including lower temperature, limited time course, and differences in protein concentrations compared to pilus biogenesis in vivo.
Fig. 4. In vitro reconstitution assay for polymerization of type 1 pili.
A–C. Reconstitution assay for pilus assembly. OM fractions were isolated from strain Tuner expressing WT FimD (A), FimD Δ70–79 (B), or FimD F4A (C). The OM fractions were mixed together with separately isolated periplasm fractions containing FimCH and FimCHisG and incubated at 25°C for the indicated time points. Pilus assembly intermediates purified from the OM fractions were incubated in sample buffer at 25°C, subjected to SDS-PAGE, and immunoblotted using anti-FimCG antibodies. The blots were analyzed using the Odyssey Infrared Imaging System.
D. Quantitation of FimGn-FimH complex formation. The amount of FimGn-FimH complexes assembled at each time point in the reconstitution assay, preformed as in (A–C), was determined by measuring the signal intensity within identical areas boxed as shown in (A) for the 45 min time point. The values for WT FimD represent means ± standard deviation of at least 5 separate experiments; the values for the FimD Δ70–79 and F4A mutants represent means of 2 separate experiments.
To measure fiber assembly, both the in vitro reconstitution assay and in vivo co-purification assay took advantage of the resistance of subunit-subunit interactions (donor strand exchange), but not chaperone-subunit interactions (donor strand complementation), to dissociation by SDS. Moreover, both assays captured pilus fibers in stable association with the usher; these complexes presumably represent bona fide assembly intermediates in the process of secretion through the usher. In comparison, previous in vitro polymerization assays have relied on incubating chaperone-subunit complexes with the usher and using electron microscopy to measure the lengths of pilus fibers formed (Nishiyama et al., 2008; Huang et al., 2009). It is possible in these assays that a portion of the fibers formed might be the result of off-pathway subunit-subunit interactions triggered by interaction with usher domains and the high concentrations of subunits present, rather than the result of assembly and secretion of the fibers through the usher. For example, Huang et al. (Huang et al., 2009) reported that the PapC N domain was dispensable for fiber polymerization in their in vitro assay, despite its known requirement for pilus biogenesis in vivo. In contrast, our FimD results show that a functional usher N terminus is required for pilus assembly in vitro as well as in vivo. We have not yet succeeded in directly testing the catalytic activity of PapC using an in vitro reconstitution assay similar to the assay we used for FimD. However, the co-purification experiments (Fig. 2C) clearly showed defects of the PapC C70A and Δ77–86 mutants in polymerizing pilus fibers, and support a catalytic role for the N-terminal disulfide region of PapC, as found for FimD. An alternative explanation for the failure of the PapC disulfide loop mutants to assemble pili is that these mutants are altered in their affinity for chaperone-subunit complexes or unable to bind subunits other than PapG. However, we found that the binding affinity of the PapC C70A mutant for PapDG is unchanged compared to WT PapC, and that this mutant is able to bind PapDF chaperone-subunit complexes in addition to PapDG (Q. Li and D. G. Thanassi, unpublished results). Taken together, these results indicate a conserved function of the usher N-terminal domain in promoting subunit-subunit interactions.
CONCLUSIONS
The usher acts as a multifunctional assembly and secretion platform in the bacterial OM. The usher N-terminal domain was previously shown to provide the initial binding site for chaperone-subunit complexes and participate in the differential affinity of the usher for chaperone-subunit complexes (Ng et al., 2004; Nishiyama et al., 2005; Li et al., 2010). Our findings reported here reveal a third function for the usher N terminus: catalyzing the exchange of chaperone-subunit for subunit-subunit interactions to promote polymerization of the pilus fiber. In particular, we identified the conserved N-terminal disulfide region of the PapC and FimD ushers, as well as residue F4 in the chaperone-subunit binding region of FimD, as important for the catalytic activity of the usher.
The mechanism by which the usher catalyzes pilus assembly is not known. The formation of subunit-subunit interactions at the usher occurs by a concerted strand displacement mechanism (Remaut et al., 2006; Vetsch et al., 2006), in which the Nte of an incoming chaperone-subunit complex displaces the chaperone donor strand from the preceding chaperone-subunit complex to allow donor strand exchange between the subunits (Fig. 1A). Nucleation of the donor strand exchange reaction is critically dependent on insertion of the Nte into a binding pocket in the subunit groove occupied by the chaperone donor strand (Remaut et al., 2006). Therefore, the catalytic activity of the usher is likely related to the positioning of the incoming Nte relative to the binding pocket of the preceding chaperone-subunit complex to favor the donor strand exchange reaction. The usher may also act to weaken the chaperone-subunit interaction, thereby facilitating invasion of the incoming Nte. The FimDN-CHp co-crystal structure (Nishiyama et al., 2005) shows that the N-terminal disulfide region of the usher does not directly interact with the bound chaperone-subunit complex (Fig. 1B), and we found that mutations to the disulfide loop region do not affect the binding of chaperone-subunit complexes to the usher. In the context of the full-length usher, the disulfide loop region would be located adjacent to the strand connecting the N-terminal domain to the ß-barrel translocation domain of the usher (Fig. 1). Taking these findings together, we propose that the disulfide loop region may influence positioning of the chaperone-subunit complex relative to the rest of the usher-pilus assembly complex; i.e., the function of the disulfide loop region in catalysis may be to ensure optimal placement of the Nte of the incoming chaperone-subunit complex relative to the subunit groove of the preceding chaperone-subunit complex to promote donor strand exchange. In contrast to the disulfide loop region, residue F4 of FimD directly contacts the FimC chaperone (Fig. 1B). Therefore, FimD F4 may act to weaken the binding of the chaperone to its subunit and thus promote donor strand exchange by facilitating invasion of the incoming Nte.
Although the N-terminal domains of both PapC and FimD were found to function similarly in promoting subunit polymerization, we also observed differences between the two ushers. PapC and FimD belong to different clades within the usher superfamily (Nuccio and Baumler, 2007) and these differences likely reflect evolutionary divergence in their assembly mechanisms. Our results showed that PapC and FimD do not bind chaperone-subunit complexes by identical mechanisms, since residue F3 is required for the binding activity of PapC, whereas the corresponding FimD F4 residue is not essential for binding. A structure of the PapC-DG complex will be important to reveal the basis for this difference. Our results also revealed that the catalytic activity of the PapC and FimD ushers is required for different stages of pilus biogenesis. The PapC N-terminal disulfide mutants were almost completely stalled at the stage of PapDG binding to the usher, with only barely detectable levels of PapG polymerization into pilus fibers. Thus, the catalytic activity of PapC appears to be critical for formation of the first subunit-subunit interaction and initiation of fiber assembly (presumably, the PapC catalytic activity is also required for fiber elongation once initiation of assembly takes place). In contrast, the FimD F4A and Δ70–79 mutants behaved similar to WT FimD in initiating pilus assembly, but were defective for assembling higher order pilus fibers. This suggests that the catalytic activity of FimD is required following initiation for subsequent rounds of subunit incorporation necessary to extend the pilus fiber. This agrees with the findings of Nishiyama and colleagues, who examined the catalytic activity of FimD during assembly of the FimA pilus rod and concluded that the usher accelerates polymerization of the rod following incorporation of the first FimA subunit (Nishiyama et al., 2008). Overall, our findings highlight important similarities as well as specific differences in the mechanism of pilus assembly by the PapC and FimD ushers. Our results provide a foundation for exploring the molecular basis of the catalytic activity of the ushers and for understanding virulence factor biogenesis at the bacterial outer membrane.
EXPERIMENTAL PROCEDURES
Strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 2. Unless otherwise indicated, all strains were grown at 37°C with aeration in LB medium with appropriate antibiotics. DH5α was used as the host strain for plasmid manipulations and all mutants were sequenced to verify the intended mutation.
Table 2.
Strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strainsa | ||
| DH5α | hsdR recA endA | (Grant et al., 1990) |
| Tuner | OmpT− Lon− | Novagen |
| BL21 | OmpT− Lon− | (Rosenberg et al., 1987) |
| SF100 | ΔompT | (Baneyx and Georgiou, 1990) |
| AAEC185 | Δfim | (Blomfield et al., 1991) |
| ORN103 | Δfim | (Orndorff et al., 1985) |
| MM294ΔfimD | ΔfimD | (Shu Kin So and Thanassi, 2006) |
| Plasmids | ||
| pMON6235Δcat | vector, Para, Ampr | (Jones et al., 1997) |
| pMMB66 | vector, Ptac, Ampr | (Morales et al., 1991) |
| pMMB91 | vector, Ptac, Kanr | (Dodson et al., 1993) |
| pBAD18-cm | vector, Para, Clmr | (Guzman et al., 1995) |
| pBAD18-kan | vector, Para, Kanr | (Guzman et al., 1995) |
| pMJ3 | PapC with C-terminal His-tag in pMON6235Δcat | (Thanassi et al., 1998) |
| pM05 | PapC F3A in pMJ3 | (Ng et al., 2004) |
| pTN5 | PapC C70A in pMJ3 | (Ng et al., 2004) |
| pTN26 | PapC Δ77–86 in pMJ3 | This study |
| pMJ2 | ΔpapC pap operon in pACYC184, Ptrc, Tetr | (Thanassi et al., 1998) |
| pPAP58 | PapDJKEFG in pMMB91 | (Hultgren et al., 1989) |
| pJP1 | PapDG in pMMB91 | (Dodson et al., 1993) |
| pETS4 | FimD with C-terminal His-tag in pMMB66 | (Saulino et al., 1998) |
| pETS7 | FimD with C-terminal His-tag in pBAD18-cm | (Saulino et al., 2000) |
| pAN2 | FimD (no His-tag) in pMMB91 | (Saulino et al., 1998) |
| pNH227 | FimD F4A in pETS4 | This study |
| pIT2 | FimD F4A in pETS7 | This study |
| pNH238 | FimD F4A in pAN2 | This study |
| pNH228 | FimD C63A in pETS4 | This study |
| pIT4 | FimD C63A in pETS7 | This study |
| pNH236 | FimD Δ70–79 in pETS4 | This study |
| pIT12 | FimD Δ70–79 in pETS7 | This study |
| pNH239 | FimD Δ70–79 in pAN2 | This study |
| pNH222 | FimCGH in pBAD18-kan | This study |
| pNH235 | FimCFGH in pBAD18-cm | This study |
| pETS1007 | FimCH in pMMB66 | (Saulino et al., 2000) |
| pNH221 | FimC (no His-tag) in pBAD18-cm | This study |
| pETS1000 | FimC with C-terminal His-tag in pMON6235Δcat, Specr | (Saulino et al., 1998) |
| pHJ20 | FimH in pMMB66 | (Jones et al., 1995) |
| pETS2A | FimG in pMMB66 | (Saulino et al., 1998) |
All strains are E. coli K-12, except BL21 and Tuner, which are E. coli B.
Ampr, ampicillin resistance; Kanr, kanamycin resistance; Clmr, chloramphenicol resistance; Tetr, tetracycline resistance; Specr, spectinomycin resistance; Para, arabinose-inducible promoter, Ptac or Ptrc, IPTG-inducible promoter.
Plasmid pTN26, encoding PapC Δ77–86 with a C-terminal hexahistadine tag (His-tag) was derived from plasmid pMJ3 using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and the primers listed in Table 3. Plasmids pIT2 (FimD F4A), pIT4 (FimD C63A) and pIT12 (FimD Δ70–79) were derived from plasmid pETS7, which encodes FimD with a C-terminal His-tag, using the QuikChange Site-Directed Mutagenesis Kit and the primers listed in Table 3. Plasmids pNH227 (FimD F4A), pNH228 (FimD C63A) and pNH236 (FimD Δ70–79) were similarly derived from plasmid pETS4, which encodes FimD with a C-terminal His-tag. Plasmids pNH238 (FimD F4A) and pNH239 (FimD Δ70–79) were similarly derived from plasmid pAN2, which encodes FimD (no His-tag).
Table 3.
Primers used in this study.
| Mutation | Sequence of QuikChange forward primer (5’-3’)a |
|---|---|
| PapC Δ77–86 | CCGCAGGCCTGTCTGACATCAGATATGGTCGATAAAGTTG |
| FimD F4A | CTGCCGACCTCTATGCTAATCCGCGC |
| FimD C63A | GGGATTGTTCCCGCCCTGACACGCGCG |
| FimD Δ70–79 | GCGCGCAACTCGCCGGTATGAATCTGC |
The reverse primer is the reverse complement of the forward primer.
Plasmid pNH222 was constructed by subcloning an EcoRI-HindIII fragment containing fimCGH from plasmid pETS1013 (Saulino et al., 2000) into vector pBAD18-Kan. Plasmid pNH235, which contains fimCFGH in vector pBAD18-cm, was constructed by subcloning an EcoRI-KpnI fragment containing fimC from plasmid pETS1007 and a SmaI-HindIII fragment containing fimFGH from plasmid pHJ27 (Hultgren laboratory collection, Washington University, St. Louis). Plasmid pNH221 was constructed by subcloning fimC from pETS1007 into vector pBAD18-cm.
OM isolation and analysis of usher expression and folding
The expression levels of the PapC and FimD usher mutants in the OM were compared with the WT parental usher. Strain SF100 harboring the appropriate PapC or FimD expression plasmid was induced at OD600 = 0.6 with 0.1% L-arabinose (for PapC) or 50 μM isopropyl-ß-D-thiogalactoside (IPTG; for FimD) for 1 h. OM fractions were isolated by French press disruption and Sarkosyl extraction, as previously described (Ng et al., 2004; Shu Kin So and Thanassi, 2006). Expression levels of the ushers in the OM were determined by inspection of Coomassie blue-stained SDS-PAGE gels or immunoblotting with anti-His-tag (Covance), anti-PapC or anti-FimD antibodies. Immunoblots were developed with alkaline phosphatase-conjugated secondary antibodies and BCIP (5-bromo-4-chloro-3-indolylphosphate)-NBT (nitroblue tetrazolium) substrate (KPL). Proper folding of the ushers in the OM was checked by resistance to denaturation by SDS, which provides an indication of the correct folding and stability of the ß-barrel domain (Sugawara et al., 1996). This was determined by heat-modifiable mobility on SDS-PAGE, performed as previously described (Ng et al., 2004; Shu Kin So and Thanassi, 2006).
Hemagglutination assay
HA assays were performed by serial dilution in microtiter plates as previously described (Shu Kin So and Thanassi, 2006). HA titers were determined visually as the highest fold dilution of bacteria still able to agglutinate human red blood cells (for P pili) or guinea pig red blood cells (Colorado Serum Company; for type 1 pili). Each assay was performed in triplicate. For analysis of the assembly of complete P pili, AAEC185/pMJ2 (ΔpapC pap operon; papAHDJKEFG) was used as the host strain. For analysis of assembly of P pilus tips, AAEC185/pPAP58 (papDJKEFG) was used as the host strain. Host strains transformed with pMON6235Δcat (vector), pMJ3 (WT PapC with C-terminal His-tag), pM05 (PapC F3A), pTN5 (PapC C70A) or pTN26 (PapC Δ77–86) were induced at OD600 = 0.6 with 0.1% L-arabinose and 0.1 mM IPTG for 1 h. For analysis of assembly of complete type 1 pili, MM294ΔfimD, which contains a fimD deletion in the chromosomal fim operon (fimAICFGH), was used as the host strain. MM294ΔfimD transformed with pMMB66 (vector), pETS4 (WT FimD with C-terminal His-tag), pNH227 (FimD F4A), pNH228 (FimD C63A) or pNH236 (FimD Δ70–79) was grown statically for 24 h and then FimD expression was induced for an additional 3 h by addition of 50 μM IPTG with shaking at 100 rpm. For analysis of assembly of FimFGH or FimGH type 1 pilus tips, AAEC185/pNH235 (fimCFGH) or AAEC185/pNH222 (fimCGH) were used as the host strains, respectively. Host strains transformed with the FimD expression plasmids described above were induced at OD600 = 0.6 with 50 μM IPTG and 0.1% L-arabinose for 1 h.
Overlay assay
Overlay assays were performed as previously described (Shu Kin So and Thanassi, 2006). OM fractions were isolated as described above from strain SF100 harboring the appropriate PapC (pMON6235Δcat, pMJ3, pM05, pTN5 or pTN26) or FimD (pBAD18-cm, pETS7, pIT2, pIT4, pIT5 or pIT12) expression plasmid. The strains were induced at OD600 = 0.6 for 1 h with 0.1 or 0.02% L-arabinose for PapC or FimD, respectively. The OM fractions were subjected to SDS-PAGE, transferred to a PVDF (polyvinylidene difluoride; Osmonics) membrane, and incubated with periplasm fractions containing PapDG (to test binding to PapC) or FimCH (to test binding to FimD). The periplasm fractions were isolated from strain BL21/pJP1 (papDG) or ORN103/pETS1007 (fimCH) as previously described (Ng et al., 2004). Binding of the chaperone-adhesin complexes to the ushers on the PVDF membrane was detected by immunoblotting with anti-PapDG or anti-FimH antibodies. Immunoblots were developed with alkaline phosphatase-conjugated secondary antibodies and BCIP-NBT substrate.
Co-purification of pilus subunits with the usher
Co-purification assays were performed as previously described (Shu Kin So and Thanassi, 2006). For analysis of PapC, OM fractions were isolated as described above from strain SF100/pPAP58 (papDJKEFG) harboring pMON6235Δcat, pMJ3, pM05, pTN5 or pTN26. The OM fractions were solubilized with the nondenaturing detergent dodecyl-maltopyranoside (DDM; Anatrace) and subjected to nickel affinity chromatography. PapC was eluted from the nickel column using an imidazole step gradient. Peak fractions containing PapC were subjected to SDS-PAGE and either stained with Coomassie blue to detect PapC or immunoblotted with anti-P pilus tips antiserum to detect pilus tip subunits that co-purified with the usher. To examine oligomerization of PapG into P pilus tip fibers, the PapC-containing fractions from the nickel column were incubated for 10 min at room temperature (25°C) or 95°C in SDS-PAGE sample buffer, subjected to SDS-PAGE and immunoblotted with anti-PapDG antiserum. For analysis of FimD, OM fractions were isolated as described above from strain AAEC185/pNH235 (fimCFGH) or AAEC185/pNH222 (fimCGH) harboring pMMB66, pETS4, pNH227, pNH228 or pNH236. The OM fractions were solubilized and purified as for PapC, except the affinity column was charged with cobalt instead of nickel. The FimD-containing fractions were incubated for 10 min at 25 or 95°C in SDS-PAGE sample buffer, subjected to SDS-PAGE and immunoblotted with anti-FimCH, anti-FimCG or anti-FimCF antibodies. Immunoblots were developed with alkaline phosphatase-conjugated secondary antibodies and BCIP-NBT substrate.
FimD trypsin sensitivity assay
Sensitivity of FimD to digestion by extracellularly added trypsin was performed as previously described (Saulino et al., 1998). Strains AAEC185/pETS7 (WT FimD with C-terminal His-tag) or AAEC185/pETS1007 (fimCH) + pETS7, pIT2 (FimD F4A) or pIT12 (FimD Δ70–79) were induced at OD600 = 0.6 with 50 μM IPTG and 0.02% L-arabinose for 1 h. The cells were washed and resuspended in 20 mM HEPES (pH 8.5) and equal portions were either mock digested or digested with fresh 100 μg/ml trypsin (Sigma) at 37°C for 2 h with rocking. Digestion was stopped by addition of 0.1 mM phenylmethylsulfonyl fluoride, the cells were harvesting by centrifugation, and OM fractions were isolated as described above. The OM was subjected to SDS-PAGE and immunoblotted with anti-FimD antibodies. The blots were developed with alkaline phosphatase-conjugated secondary antibodies and BCIP-NBT substrate.
In vitro reconstitution assay for pilus assembly
OM fractions were isolated from strain Tuner harboring pMMB91 (vector), pAN2 (WT FimD, no His-tag), pNH238 (FimD F4A) or pNH239 (FimD Δ70–79) as described above. FimD expression was induced at OD600 = 0.6 with 50 μM IPTG for 1 h. Periplasm fractions were isolated as previously described (Ng et al., 2004) from strain Tuner/pNH221 (FimC) + pHJ20 (FimH) or strain Tuner/pETS1000 (FimC with C-terminal His-tag) + pETS2A (FimG). The strains were induced at OD600 = 0.6 with 0.1 mM IPTG and 0.05% L-arabinose for 1 h. The periplasm fractions were dialyzed into buffer A (20 mM Tris-HCl (pH 8), 0.3 M NaCl) to match the buffer used for the OM isolation. OM fractions (6.5 mg protein; determined using the BCA assay (Pierce)) were mixed with the FimCH and FimCHisG periplasm fractions (2.9 mg protein each) and the total volume adjusted to 10 ml with buffer A. The samples were incubated at room temperature (25°C) and the reactions were stopped at the indicated time points by addition of 50 ml cold buffer A and placed on ice. The OM fractions were harvested by centrifugation (100,000 g, 1 h, 4°C), resuspended in 10 ml buffer A and solubilized by rocking overnight with 1% DDM at 4°C. The OM extracts were centrifuged again to remove any unsolubilized material and pilus assembly intermediates were purified by cobalt affinity chromatography as described above for the co-purification assay. Peak fractions from the affinity column were precipitated by addition of 9% final concentration trichloroacetic acid and incubation on ice for 30 min. The precipitated proteins were harvested by centrifugation (10,000 g, 5 min, 4°C), washed twice with cold acetone, and resuspended in 150 μl SDS-PAGE sample buffer. The samples were incubated in sample buffer for 10 min at room temperature, subjected to SDS-PAGE, transferred to PVDF membrane and immunoblotted using anti-FimCG or anti-FimCH antibodies. The blots were developed using infrared-conjugated secondary antibodies (LI-COR Biosciences) and analyzed using the Odyssey Infrared Imaging System (LI-COR Biosciences). Quantitation of FimGn-FimH complexes formed at each time point was done by measuring the signal intensity within identical areas boxed as shown for the 45 min time point in Fig. 4A. For each experiment, the intensity value obtained for the 0 min time point was subtracted from the values obtained for the subsequent time points.
Acknowledgments
We thank Huilin Li (Brookhaven National Laboratory and Stony Brook University) and Wali Karzi (Stony Brook University) for critical reading of the manuscript. This work was supported by National Institutes of Health grant GM062987.
References
- Baga M, Norgren M, Normark S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell. 1987;49:241–251. doi: 10.1016/0092-8674(87)90565-4. [DOI] [PubMed] [Google Scholar]
- Baneyx F, Georgiou G. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J Bacteriol. 1990;172:491–494. doi: 10.1128/jb.172.1.491-494.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Sauer FG, Pinkner JS, Hultgren SJ. Chaperone-subunit-usher interactions required for donor strand exchange during bacterial pilus assembly. J Bacteriol. 2003;185:2723–2730. doi: 10.1128/JB.185.9.2723-2730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield IC, McClain MS, Eisenstein BI. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol Microbiol. 1991;5:1439–1445. doi: 10.1111/j.1365-2958.1991.tb00790.x. [DOI] [PubMed] [Google Scholar]
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- Dodson KW, Jacob-Dubuisson F, Striker RT, Hultgren SJ. Outer membrane PapC usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci U S A. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Ford B, Rego AT, Ragan TJ, Pinkner J, Dodson K, Driscoll PC, Hultgren S, Waksman G. Structural homology between the C-terminal domain of the PapC usher and its plug. J Bacteriol. 2010;192:1824–1831. doi: 10.1128/JB.01677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Haner M, Taschner N, Burkhard P, Aebi U, Muller SA. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J Mol Biol. 2002;323:845–857. doi: 10.1016/s0022-2836(02)01005-7. [DOI] [PubMed] [Google Scholar]
- Henderson NS, Shu Kin So S, Martin C, Kulkarni R, Thanassi DG. Topology of the outer membrane usher PapC determined by site-directed fluorescence labeling. J Biol Chem. 2004;279:53747–53754. doi: 10.1074/jbc.M409192200. [DOI] [PubMed] [Google Scholar]
- Huang Y, Smith BS, Chen LX, Baxter RH, Deisenhofer J. Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc Natl Acad Sci U S A. 2009;106:7403–7407. doi: 10.1073/pnas.0902789106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultgren SJ, Lindberg F, Magnusson G, Kihlberg J, Tennent JM, Normark S. The PapG adhesin of uropathogenic Escherichia coli contains separate regions for receptor binding and for the incorporation into the pilus. Proc Natl Acad Sci U S A. 1989;86:4357–4361. doi: 10.1073/pnas.86.12.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Heuser J, Dodson K, Normark S, Hultgren SJ. Initiation of assembly and association of the structural elements of a bacterial pilus depend on two specialized tip proteins. EMBO J. 1993;12:837–847. doi: 10.1002/j.1460-2075.1993.tb05724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Danese PN, Pinkner JS, Silhavy TJ, Hultgren SJ. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Pinkner JS, Roth R, Heuser J, Nicholoes AV, Abraham SN, Hultgren SJ. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MJ, Heuser J, Normark S, Hultgren SJ. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature. 1992;356:252–255. doi: 10.1038/356252a0. [DOI] [PubMed] [Google Scholar]
- Lee YM, Dodson KW, Hultgren SJ. Adaptor function of PapF depends on donor strand exchange in P-pilus biogenesis of Escherichia coli. J Bacteriol. 2007;189:5276–5283. doi: 10.1128/JB.01648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Qian L, Chen Z, Thahbot D, Liu G, Liu T, Thanassi DG. The outer membrane usher forms a twin-pore secretion complex. J Mol Biol. 2004;344:1397–1407. doi: 10.1016/j.jmb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Li H, Thanassi DG. Use of a combined cryo-EM and X-ray crystallography approach to reveal molecular details of bacterial pilus assembly by the chaperone/usher pathway. Curr Opin Microbiol. 2009;12:326–332. doi: 10.1016/j.mib.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ng TW, Dodson KW, So SS, Bayle KM, Pinkner JS, Scarlata S, Hultgren SJ, Thanassi DG. The differential affinity of the usher for chaperone-subunit complexes is required for assembly of complete pili. Mol Microbiol. 2010;76:159–172. doi: 10.1111/j.1365-2958.2010.07089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapingire OS, Henderson NS, Duret G, Thanassi DG, Delcour AH. Modulating effects of the plug, helix and N- and C-terminal domains on channel properties of the PapC usher. J Biol Chem. 2009;284:36324–36333. doi: 10.1074/jbc.M109.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales VM, Backman A, Bagdasarian M. A series of wide-host-range low-copy- number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- Munera D, Hultgren S, Fernandez LA. Recognition of the N-terminal lectin domain of FimH adhesin by the usher FimD is required for type 1 pilus biogenesis. Mol Microbiol. 2007;64:333–346. doi: 10.1111/j.1365-2958.2007.05657.x. [DOI] [PubMed] [Google Scholar]
- Ng TW, Akman L, Osisami M, Thanassi DG. The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J Bacteriol. 2004;186:5321–5331. doi: 10.1128/JB.186.16.5321-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TW, Akman L, Osisami M, Thanassi DG. Author's Correction: The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J Bacteriol. 2006;188:2295. doi: 10.1128/JB.186.16.5321-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Horst R, Eidam O, Herrmann T, Ignatov O, Vetsch M, Bettendorff P, Jelesarov I, Grutter MG, Wuthrich K, Glockshuber R, Capitani G. Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 2005;24:2075–2086. doi: 10.1038/sj.emboj.7600693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Ishikawa T, Rechsteiner H, Glockshuber R. Reconstitution of Pilus Assembly Reveals a Bacterial Outer Membrane Catalyst. Science. 2008;320:376–379. doi: 10.1126/science.1154994. [DOI] [PubMed] [Google Scholar]
- Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orndorff PE, Spears PA, Schauer D, Falkow S. Two modes of control of pilA, the gene encoding type 1 pilin in Escherichia coli. J Bacteriol. 1985;164:321–330. doi: 10.1128/jb.164.1.321-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol Cell. 2006;22:831–842. doi: 10.1016/j.molcel.2006.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut H, Tang C, Henderson NS, Pinkner JS, Wang T, Hultgren SJ, Thanassi DG, Waksman G, Li H. Fiber Formation across the Bacterial Outer Membrane by the Chaperone/Usher Pathway. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Marklund BI, Ilver D, Haslam D, Kaack MB, Baskin G, Louis M, Mollby R, Winberg J, Normark S. The Galα (1–4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc Natl Acad Sci USA. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RJ, Verger D, Daviter T, Remaut H, Paci E, Waksman G, Ashcroft AE, Radford SE. Unraveling the molecular basis of subunit specificity in P pilus assembly by mass spectrometry. Proc Natl Acad Sci U S A. 2008;105:12873–12878. doi: 10.1073/pnas.0802177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AH, Lade BN, Chui DS, Lin SW, Dunn JJ, Studier FW. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Sauer FG, Fütterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- Sauer FG, Pinkner JS, Waksman G, Hultgren SJ. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- Sauer FG, Remaut H, Hultgren SJ, Waksman G. Fiber assembly by the chaperone-usher pathway. Biochim Biophys Acta. 2004;1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Saulino ET, Bullitt E, Hultgren SJ. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9240–9245. doi: 10.1073/pnas.160070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulino ET, Thanassi DG, Pinkner JS, Hultgren SJ. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Kin So S, Thanassi DG. Analysis of the requirements for pilus biogenesis at the outer membrane usher and the function of the usher C-terminus. Mol Microbiol. 2006;60:364–375. doi: 10.1111/j.1365-2958.2006.05111.x. [DOI] [PubMed] [Google Scholar]
- Soto GE, Dodson KW, Ogg D, Liu C, Heuser J, Knight S, Kihlberg J, Jones CH, Hultgren SJ. Periplasmic chaperone recognition motif of subunits mediates quaternary interactions in the pilus. EMBO J. 1998;17:6155–6167. doi: 10.1093/emboj/17.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara E, Steiert M, Rouhani S, Nikaido H. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J Bacteriol. 1996;178:6067–6069. doi: 10.1128/jb.178.20.6067-6069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi DG, Saulino ET, Lombardo MJ, Roth R, Heuser J, Hultgren SJ. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc Natl Acad Sci U S A. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi DG, Stathopoulos C, Dodson KW, Geiger D, Hultgren SJ. Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J Bacteriol. 2002;184:6260–6269. doi: 10.1128/JB.184.22.6260-6269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger D, Miller E, Remaut H, Waksman G, Hultgren S. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 2006;7:1228–1232. doi: 10.1038/sj.embor.7400833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetsch M, Erilov D, Moliere N, Nishiyama M, Ignatov O, Glockshuber R. Mechanism of fibre assembly through the chaperone-usher pathway. EMBO Rep. 2006;7:734–738. doi: 10.1038/sj.embor.7400722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Berglund J, Pudney AF, Fooks LJ, Ibrahim TM, MacIntyre S, Knight SD. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell. 2003;113:587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]