Abstract
Chronic Hepatitis B Virus (HBV) infection is a major etiologic factor in hepatocellular carcinoma (HCC) pathogenesis, involving effects of chronic liver inflammation and of the weakly oncogenic HBV X protein (pX). pX-mediated hepatocyte transformation requires Polo-like kinase1 (Plk1) activity, but the mechanism is not fully understood. We identified by a genome-wide shRNA library screen the genes ZNF198 and SUZ12 whose protein depletion rescues pX-expressing cells from DNA damage-induced apoptosis. ZNF198 and SUZ12 are components of chromatin remodeling complexes and associate with promyelocytic leukemia (PML) nuclear bodies. Knockdown of ZNF198 and SUZ12 by siRNA reduced p53 stability and DNA repair, rescued pX-expressing hepatocytes from DNA damage-induced apoptosis, and increased pX-induced polyploidy and oncogenic transformation, suggesting ZNF198 and SUZ12 have a role in pX-mediated transformation. Interestingly, during pX-mediated transformation the protein but not mRNA levels of ZNF198 and SUZ12 progressively decreased, while Plk1 levels increased. Inhibition of Plk1 activity restored protein levels of ZNF198 and SUZ12. In addition, transfected Polo-box-domain (PBD) of Plk1 co-immunoprecipitated with ZNF198 and SUZ12, suggesting these proteins are Plk1 substrates. Elevated Plk1 and reduced protein levels of ZNF198 and SUZ12 were also observed in human liver cancer cell lines derived from HBV-related tumors and in the presence of HBV replication. Importantly, knockdown by siRNA of ZNF198 and SUZ12 enhanced HBV replication.
Conclusion
Reduced protein levels of ZNF198 and SUZ12 and elevated Plk1 occur during pX-mediated hepatocyte transformation, in human liver cancer cell lines, as well as during HBV replication, underscoring the significance of these genes both in HBV-mediated HCC pathogenesis and HBV replication. We propose Plk1 activity down-regulates ZNF198 and SUZ12 thereby enhancing both HBV replication and pX-mediated oncogenic transformation.
Keywords: Hepatitis B Virus X protein, hepatocellular carcinoma, HBV replication, SUZ12, ZNF198, p53, Polo-like kinase 1
Liver cancer has the most rapidly growing mortality rate in the U.S. (1). Chronic HBV infection is a major factor in HCC pathogenesis (2). Despite the HBV vaccine, the World Health Organization estimates that 400 million people are chronically infected with HBV. Current treatments include antiviral nucleoside analogs, eventually resulting in viral resistance (3). Furthermore, high level of viremia in chronic HBV patients is prognostic of the disease advancing to HCC (4).
Contributing factors to HBV-mediated HCC pathogenesis include chronic liver inflammation and regeneration (5), and effects of the weakly oncogenic HBV X protein (pX) (6, 7). In chronic HBV infected patients, viral DNA integrates into the host’s genome, with the majority of tumors displaying continued expression of pX (8). pX is essential for the viral life cycle (9). pX also increases activation of mitogenic pathways (10, 11), resulting in accelerated cell cycle entry (12) and enhanced transcription of cellular genes. Importantly, pX increases expression of replication licencing factors Cdc6 and Cdt1, resulting in DNA re-replication-induced DNA damage (13). Furthermore, pX activates Plk1 in untransformed hepatocytes which mediates checkpoint adaptation, thereby propagating DNA damage and generating partial polyploidy (14). Significantly, expression of Plk1 is elevated during pX-mediated transformation and inhibition of Plk1 suppresses transformation (15).
Plk1 is expressed in proliferating cells (16), and elevated Plk1 expression occurs in many human cancers (17) including virus-induced cancers (18, 19). Plk1 depletion induces apoptosis in cancer cell lines but not normal cells (20). Significantly, Plk1 inhibitors are currently in clinical trials (21, 22).
In this study, we identified by a genome-wide shRNA library screen SUZ12 (23) and ZNF198 (24) as playing a role in pX-mediated hepatocyte transformation. Both proteins associate with PML nuclear bodies (NBs) and are components of chromatin remodeling complexes (23–26). SUZ12 is essential for the activity of the Polycomb Repressive Complex (PRC2), mediating the trimethylation of H3 on lysine 27 (H3K27me3), a mark of silent chromatin (23). Interestingly, a PRC2 target gene is EpCAM (27), expressed in hepatic progenitors (28) and elevated in hepatic cancer stem cells (29). ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex (26) and associates with DNA repair proteins in PML NBs (30).
PML NBs have a role in p53 apoptosis (31), DNA repair (32) and viral replication (33). Viruses employ various mechanisms to disrupt PML NBs in favor of viral replication (33–35). During HBV replication, PML NBs become altered in both number and morphology (36). Significantly, HBV replication depends on histone modifications associated with the HBV covalently closed circular DNA (cccDNA), the template of pregenomic RNA (pgRNA) transcription (37). Moreover, HDAC1 associated with cccDNA correlates with reduction in HBV replication (38).
In this study we show that siRNA knockdown of ZNF198 and SUZ12 results in defective DNA repair and p53 apoptosis, survival of pX-expressing cells from DNA damage-induced apoptosis, enhanced pX-induced polyploidy and accelerated oncogenic transformation. Interestingly, during pX-mediated hepatocyte transformation protein levels of ZNF198 and SUZ12 become reduced, whereas Plk1 levels increase. Intriguingly, down-regulation of the protein levels of ZNF198 and SUZ12 during pX-mediated transformation is dependent on Plk1 activity, suggesting that these proteins are Plk1 substrates. The same inverse relationship occurs in human liver cancer cell lines and during HBV replication. Importantly, siRNA knockdown of ZNF198 and SUZ12 increases HBV replication. We propose Plk1 down-regulates ZNF198 and SUZ12 thus enhancing both pX-mediated transformation and HBV replication.
EXPERIMENTAL PROCEDURES
Cell culture and other assays
Details about growth conditions of 4pX-1 cells (39), flow cytometry, live cell sorting, immunofluorescence microscopy, immunoblots, siRNA transfections, soft agar assays, HBV replication assays (40, 41) and reagents used are provided in supplementary material.
Construction of 4pX-1-derived knockdown cell line
4pX-1 cells were stably transfected by a retrovirus-based pGIPZ vector containing shRNAmir sequence for SUZ12, ZNF198, Topors, and PML (Open Biosystems), as described (42). Clonal cell lines were isolated by selection with puromycin (1.0 μg/ml) and screened by immunoblots.
shRNA library screen
A stable population of 4pX-1 cells was generated following infection with a lentiviral shRNA library (System Biosciences, SBI) and selection by puromycin. The shRNA library represents 39,000 mouse transcripts in the pFIV-H1-Puro lentivirus vector. Each RNA transcript is targeted by 3–5 different siRNAs (one shRNA per vector). 4pX-1 cells (2.5×107) infected with lentiviral shRNA library, were grown for 10 days in presence of doxorubicin (0.25 μg/ml) and pX expression. Clones of infected 4pX-1 cells surviving apoptosis for 10 days were propagated separately. The shRNA insert from each surviving cell clone was cloned, sequenced, and the depleted gene identified.
Statistical analyses performed by Student’s t-test.
RESULTS
To gain insight into the mechanism by which pX mediates hepatocyte transformation, we identified genes whose protein depletion rescued pX-expressing hepatocytes from DNA damage-induced apoptosis. We performed a genome-wide shRNA library screen employing the immortalized, tetracycline-regulated pX-expressing 4pX-1 cell line (39) infected with a lentiviral shRNA library, as described in EXPERIMENTAL PROCEDURES. The most frequently genes identified by this screen were ZNF198, SUZ12 and Topors (43), known to associate with PML NBs (24, 25 and Supplementary figs 1 and 2).
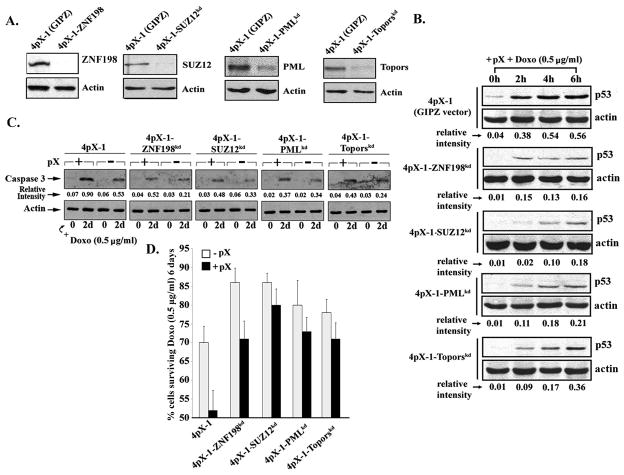
To validate the shRNA library screen, we constructed stable knockdown cell lines in 4pX-1 cells for ZNF198 and SUZ12, as well as Topors and PML (31, 44) serving as controls for PML NB function (Fig. 1A). To determine the role of ZNF198 and SUZ12 in apoptosis, knockdown cell lines were grown in apoptotic conditions induced by co-treatment with pX and doxorubicin (Fig. 1B–D and supplementary fig. 3). To exclude clonal artifacts, additional clonal isolates from each cell line were analyzed obtaining the same results (supplementary fig. 4). All knockdown cell lines exhibited significantly decreased p53 levels in comparison to 4pX-1 cells (Fig. 1B and supplementary fig. 3), suggesting SUZ12 and ZNF198, similar to PML and Topors, modulate p53 apoptosis. Indeed, on day 2 of treatment the level of active caspase3 was reduced in all knockdown cell lines, relative to 4pX-1 cells (Fig. 1C). Furthermore, cell viability of the knockdown cell lines, on day-6 of treatment with pX and doxorubicin, was greatly enhanced in comparison to pX-expressing 4pX-1 cells (Fig. 1D). Since knockdown of SUZ12 and ZNF198 rescued pX-expressing cells from p53 apoptosis, these results validate the shRNA library screen.
Fig. 1.
Knockdown of ZNF198 and SUZ12 increases cell survival in conditions of pX-mediated apoptosis. A. Immunoblots of ZNF198, SUZ12, Topors, and PML, using lysates from the respective knockdown cell lines. 4pX-1 cells with stable integration of pGIPZ empty vector were constructed in parallel. B. and C. Lysates from indicated cell lines grown ± pX and doxorubicin (0.5 μg/ml) for 6 h or 2 days, were immunoblotted for p53 in B., and active caspase3 in C. Actin is loading control. D. Quantification of cell survival of indicated cell lines grown ± pX and doxorubicin (0.5 μg/ml) for 6 days.
Knockdown of ZNF198 and SUZ12 abrogates DNA repair
PML NBs are involved in DNA repair (32), and both ZNF198 and SUZ12 associate with PML NBs (24, 25). Accordingly, we examined the effect of diminished ZNF198 and SUZ12 proteins on DNA repair. Immunofluorescence microscopy for Mre11, a component of the DNA repair Mre11-Rad50-Nbs1 complex, was used to monitor formation of DNA repair foci after doxorubicin treatment (Fig. 2A). 4pX-1 cells treated with doxorubicin displayed the characteristic Mre11 immunostaining of DNA repair foci (45). By contrast, depletion of ZNF198 and SUZ12 disrupted this distinct nuclear localization of Mre11, suggesting defective DNA repair, similar to PML knockdown (32). Furthermore, the DNA repair potential of each knock down cell line was assessed by tranfection of linearized pGL2-Luciferase vector digested with EcoRI within the luciferase gene (46). Restoration of luciferase activity requires precise joining of the EcoRI ends by DNA repair. Indeed, luciferase activity was significantly reduced in ZNF198, SUZ12 and PML knockdown cells (Fig. 2B), indicating that DNA repair is compromised. Furthermore, employing the comet assay (13), nearly 80% of ZNF198 and SUZ12 knockdown cells exhibit DNA tailing independent of pX expression, with significantly increased length of DNA tails (supplementary fig. 5A–C). Immunoblots of γ-H2AX, a marker of DNA damage, directly confirmed that depletion of SUZ12 and ZNF198 enhanced DNA damage (supplementary fig. 5D).
Fig. 2.
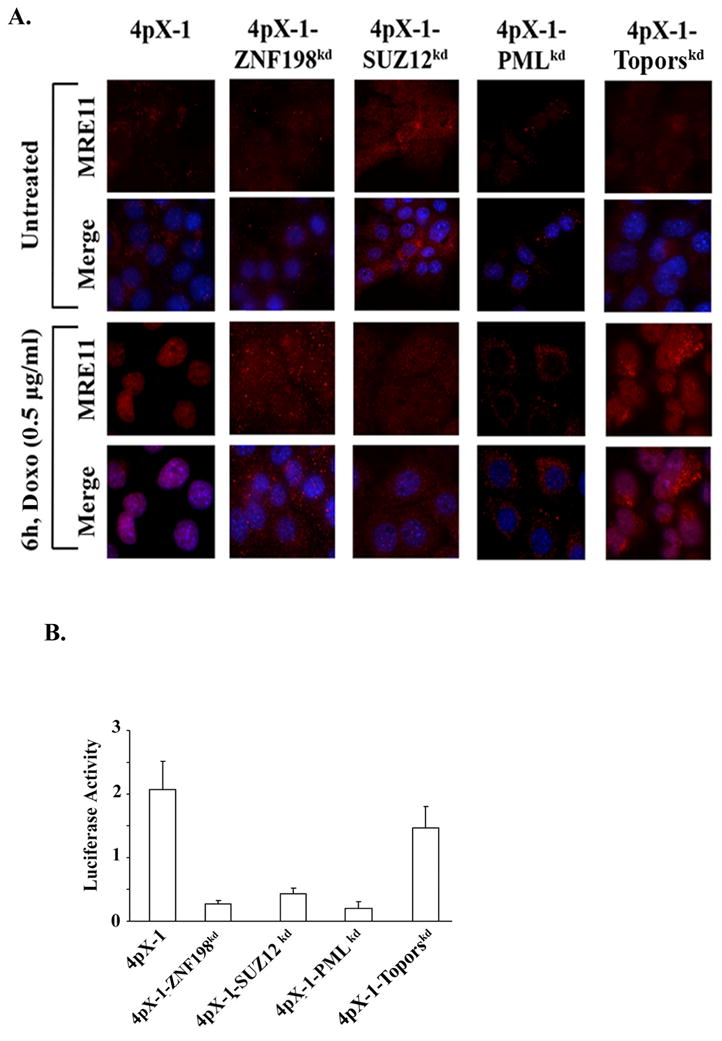
Defective DNA repair in ZNF198 and SUZ12 knockdown cells. A. Immunofluorescence microscopy of Mre11 performed as described (45), using the indicated cell lines treated with or without doxorubicin (0.5 μg/ml) for 6 h. 60× magnification. B. EcoRI-linearized pGL2-Luciferase plasmid DNA transfected for 24 h. Luciferase activity quantified per μg of protein.
Down-regulation of ZNF198 and SUZ12 increases pX-induced polyploidy, leading to transformation
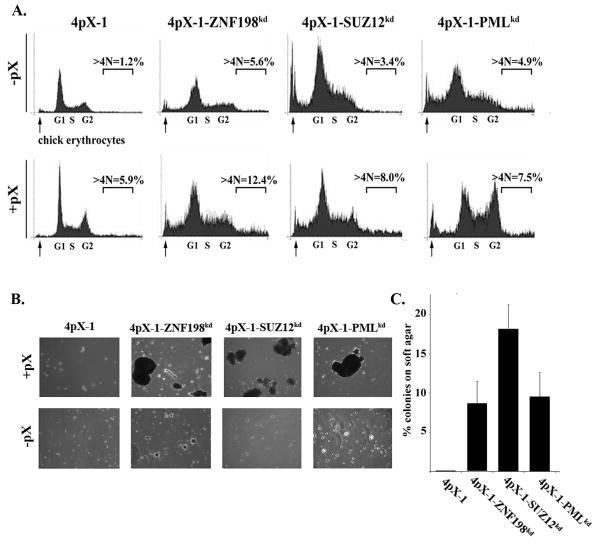
Since knockdown of ZNF198 and SUZ12 suppresses both p53 apoptosis (Fig. 1B–D) and DNA repair (Fig. 2), and pX allows propagation of DNA damage (14), we investigated the effect of diminished protein levels of ZNF198 and SUZ12 on pX-induced polyploidy. ZNF198 and SUZ12 knockdown cells exhibit a 50% increase in polyploidy in the presence of pX, quantified by flow cytometry (Fig. 3A and Table I). Furthermore, pX-induced polyploid cells from ZNF198 and SUZ12 knockdown cells formed robust colonies on soft agar immediately after isolation by live cell sorting (Fig. 3B and C). By contrast, pX-induced polyploid cells from 4pX-1 cells require an additional 40 cell doublings for growth on soft agar (15). Knockdown of PML also resulted in anchorage independent growth with pX expression, in agreement with the oncogenic potential of PML (47). Since growth on soft agar is a criterion of oncogenic transformation, these results suggest that ZNF198 and SUZ12 are tumor suppressors of pX-mediated transformation.
Fig. 3.
Knockdown of ZNF198 and SUZ12 increases pX-induced polyploidy and oncogenic transformation. A. Flow cytometric profiles of indicated cell lines grown ± pX for 16 h; nocodazole (300 nM) was added during the last 6 h as described (13). Arrows indicate chick erythrocytes used for calibration. Quantification of polyploid cells (>4N DNA) from indicated cell lines from three independent flow cytometric experiments is shown in Table I. B. pX-induced polyploid cells were isolated by live cell sorting as described (15) and grown in soft agar for 12 days ± pX. C. Quantification of colonies formed in soft agar from three independent experiments.
Table 1.
Flow cytometric quantification of pX- polyploidy
| 4pX-1 | 4pX-1-ZNF198kd | 4pX-1-SUZ12kd | 4pX-1-PMLkd | |
|---|---|---|---|---|
| −pX | 1.36±0.68 | 6.02±1.65 | 3.55±0.85 | 4.66±0.85 |
| +pX | 6.67±1.02 | 11.85±1.78 | 9.25±2.55 | 8.24±1.87 |
ZNF198 and SUZ12 are down-regulated during pX-mediated transformation and in human liver cancer cell lines
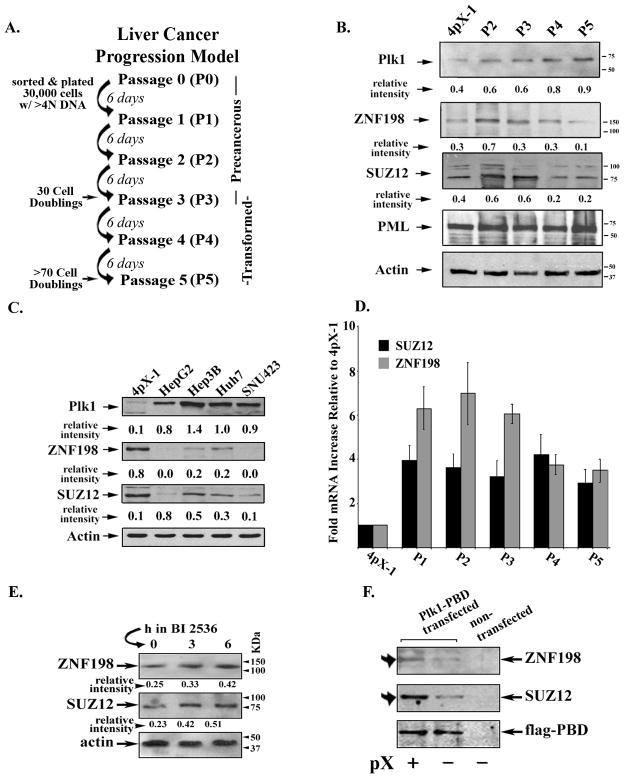
To elucidate the role of ZNF198 and SUZ12 in pX-mediated transformation, we examined by immunoblots ZNF198 and SUZ12 protein levels in our liver cancer progression model of P1-P5 cultures (ref. 15 and Fig. 4A). Protein levels of ZNF198 and SUZ12 progressively decreased, while Plk1 which is required for pX-mediated transformation, increased as pX-induced polyploid cells become transformed (Fig. 4B). This inverse relationship in protein levels of ZNF198 and SUZ12 vs. Plk1 was also observed in human liver cancer cell lines, including those derived from HBV-induced HCCs (Hep3B and SNU423) (Fig. 4C and supplementary fig. 6).
Fig. 4.
Elevated Plk1 and reduced protein levels of ZNF198 and SUZ12 during pX-mediated transformation and in human liver cancer cell lines. A. Diagram shows the in vitro pX-mediated transformation model previously described in (15). B. and C. Immunoblots of ZNF198, SUZ12 and Plk1, using lysates from B., 4pX-1 cells grown without pX, and P2-P5 cultures grown with pX; and C., indicated human liver cancer cell lines. “Relative intensity” quantified vs. actin. D. Real-time PCR quantification of ZNF198 and SUZ12 mRNA, using RNA isolated from 4pX-1 cells without pX and P1-P5 cultures with pX expression. E. Immunoblots of ZNF198 and SUZ12, using lysates from P4 cultures treated with 500 nM BI 2536 for 0–6 h. F. Immunoblots of ZNF198, SUZ12 and FLAG, using FLAG immunoprecipitates from 4pX-1 cells transfected with PBD-FLAG plasmid DNA ± pX.
The observed down-regulation of ZNF198 and SUZ12 during pX-mediated transformation could be due to transcriptional or post-transcriptional mechanisms. Accordingly, we quantified by real-time PCR ZNF198 and SUZ12 mRNA levels in P1-P5 cultures (Fig. 4D). We observe increased mRNA levels for both ZNF198 and SUZ12 during pX-mediated transformation, thus excluding transcriptional effects. Since Plk1 is elevated during pX-mediated transformation (15) and Plk1-mediated phosphorylation usually signals proteasomal degradation of its substrate (16, 22), we investigated the effect of Plk1 inhibition on ZNF198 and SUZ12 protein levels. P4 cultures treated for 3–6h with the Plk1 inhibitor BI 2536 (21) exhibit increased protein levels for ZNF198 and SUZ12 (Fig. 4E). Furthermore, since Plk1 interacts with its substrates via the Polo-box-domain, PBD (48), we investigated the interaction of ZNF198 and SUZ12 with Plk1. PBD-FLAG transiently transfected in 4pX-1 cells, co-immunoprecipitated increased amounts of ZNF198 and SUZ12 in the presence of pX (Fig. 4F), suggesting that ZNF198 and SUZ12 are Plk1 substrates.
Elevated Plk1 and reduced ZNF198 and SUZ12 protein levels in the presence of HBV replication
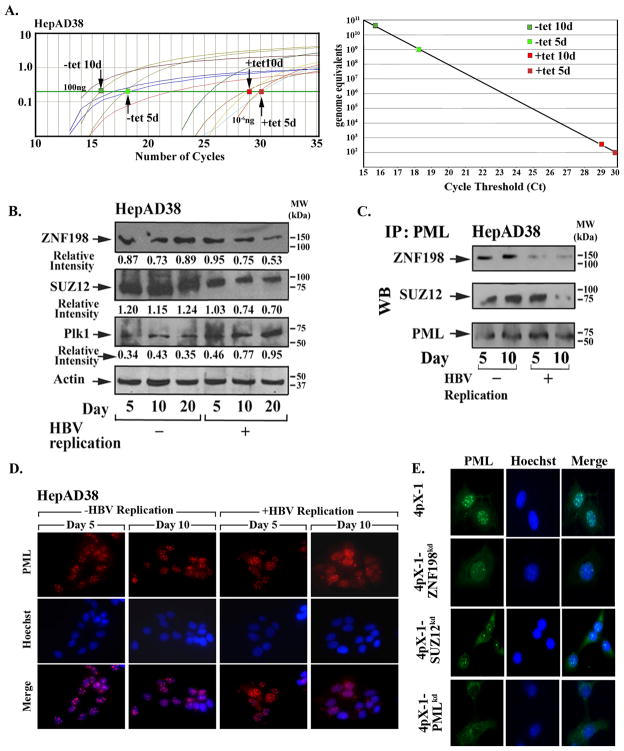
ZNF198 (24) and SUZ12 (25) associate with PML NBs which are frequently disrupted by viral infection (33–35). Therefore, we examined the protein levels of ZNF198 and SUZ12 in the context of HBV replication. In the HepAD38 cell line, the HBV genome is under control of the tet-off promoter (40). Tetracycline removal permits transcription of pgRNA which serves as template for HBV replication. Employing quantitative PCR for HBV replication with template DNA from isolated intracellular virion particles (41), we confirmed HBV replication in HepAD38 cells within one week after removal of tetracycline (Fig. 5A), as previously described (40). Interestingly, in the context of increasing HBV replication the protein levels of ZNF198 and SUZ12 became progressively reduced, while those of Plk1 increased (Fig. 5B). Since ZNF198 and SUZ12 associate with PML NBs (24. 25), we examined by immunoblots the amount of these proteins that co-immunoprecipitated with PML. In agreement with the reduced protein levels of ZNF198 and SUZ12 during HBV replication, ten days following tetracycline removal, i.e., in the presence of HBV replication, the amount of ZNF198 and SUZ12 associated with PML protein was also significantly reduced (Fig. 5C).
Fig. 5.
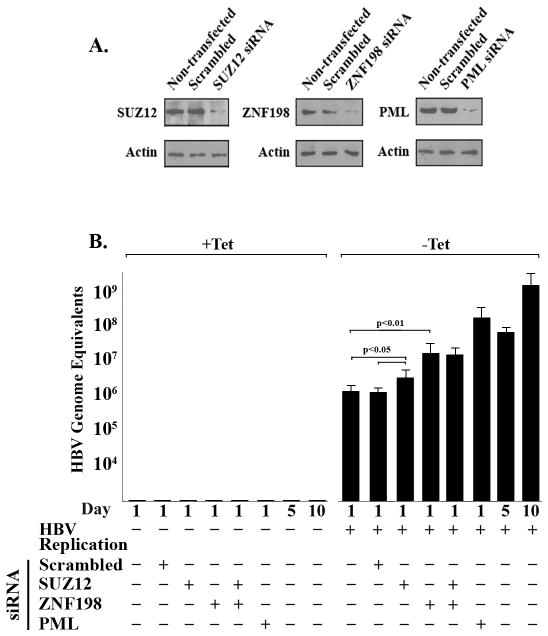
Elevated protein levels of Plk1 and reduced ZNF198 and SUZ12 during HBV replication. A. Quantification of HBV replication by real-time PCR exactly as described (41). (Left) Ct values of serially diluted HBV1.2 ayw plasmid DNA and template DNA from isolated intracellular virions obtained from HepAD38 cells grown ± tetracycline for 5 and 10 days. (Right panel) Quantification of HBV genome equivalents from data shown in left panel. B. Immunoblots of ZNF198, SUZ12, and Plk1 using lysates of HepAD38 grown ± tetracycline for 5, 10, and 20 days. C. PML immunoprecipitates of lysates from B., immunoblotted for ZNF198 and SUZ12. D. and E., PML immunofluorescence microscopy of indicated cell lines (60× magnification).
Therefore, we investigated whether this reduction in the protein levels of ZNF198 and SUZ12 during HBV replication had an effect on PML NBs. Employing immunofluorescence microscopy for PML, we observe that the morphology and number of PML NBs in HepAD38 cells were altered 10 days following tetracycline removal, with fewer PML NBs per cell and diffuse PML immunostaining (Fig. 5D). Similar experiments were performed using 4pX-1, 4pX-1-PMLkd, 4pX-1-SUZ12kd, and 4pX-1-ZNF198kd cells, as controls. Specifically, in 4pX-1-ZNF198kd and 4pX-1-SUZ12kd cells, immunofluorescence microscopy of endogenous PML (Fig. 5E) or transfected PML-RFP demonstrated that knockdown of ZNF198 and to a lesser extent of SUZ12 had an effect on formation of PML NBs (supplementary fig. 7).
To determine the functional significance of the down-regulation of ZNF198 and SUZ12 during HBV replication and their association with PML NBs, we knocked-down each of these proteins by siRNA transfection in HepAD38 cells. Importantly, siRNA knockdown of ZNF198 or PML significantly increased HBV replication (p<0.01) on day 1 after tetracycline removal. Specifically, upon siRNA knockdown of PML or ZNF198, HBV replication in HepAD38 cells, grown without tetracycline for 1 day, increased to nearly the same level as in untransfected HepAD38 cells grown without tetracycline for 5 days. Likewise, SUZ12 knockdown exerted a small but statistically significant increase (p<0.05) in HBV replication (Fig. 6). These results establish that down-regulation of ZNF198 and SUZ12 and disruption of PML NBs enhance HBV replication in HepAD38 cells.
Fig. 6.
siRNA knockdown of ZNF198, SUZ12 and PML enhances HBV replication. A. Immunoblots of SUZ12, ZNF198 and PML using HepAD38 cells transfected with indicated siRNAs grown as follows: tetracycline addition for 24 h after siRNA transfection (−HBV replication), followed by tetracycline removal for 1 day (+HBV replication). B. Quantification of HBV genome equivalents using template DNA from isolated intracellular virions from HepAD38 cells transfected with indicated siRNAs, grown ± tetracycline for 1, 5 and 10 days.
DISCUSSION
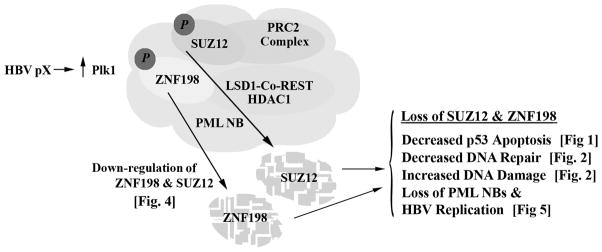
In this study, we identified ZNF198 and SUZ12 as novel negative regulators of pX-mediated hepatocyte transformation. We show that during pX-mediated transformation, protein levels of SUZ12 and ZNF198 are down-regulated, whereas those of Plk1 required for pX-mediated transformation (15), increase. We provide evidence suggesting that ZNF198 and SUZ12 are Plk1 substrates. Importantly, increased protein levels of Plk1 and reduced levels of ZNF198 and SUZ12 occur in liver cell lines derived from HBV-induced tumors, and in the presence of HBV replication. We propose that Plk1 down-regulates ZNF198 and SUZ12 thereby enhancing both HBV replication and pX-mediated transformation (Fig. 7).
Fig. 7.
Diagram shows effects of down-regulation of ZNF198 and SUZ12 identified by our studies. We propose Plk1-mediated down-regulation of ZNF198 and SUZ12 links oncogenic transformation and HBV replication.
Loss of ZNF198 and SUZ12 accelerate pX-mediated transformation
SUZ12 and ZNF198 were identified by a genome-wide shRNA library screen, searching for proteins whose depletion rescues pX-expressing hepatocytes from DNA damage-induced apoptosis. ZNF198 and SUZ12 modulate chromatin remodeling and associate with PML NBs (23–26). Independent studies have shown that deregulation of the function of ZNF198 and SUZ12 is linked to cancer (49, 50). Interestingly, the ZNF198 gene located on chromosome 13q12.11 is frequently affected by loss of heterozygosity in early onset HCC, correlated with high tumor grade and HBV-induced tumors (51, 52). In our studies, knockdown of ZNF198 and SUZ12 increased survival of pX-expressing cells from DNA damage-induced apoptosis (Fig. 1) and suppressed DNA repair (Fig. 2) likely by disruption of PML NBs (Fig. 5E). Indeed, in 4pX-1 cells, PML knockdown abrogates both p53 apoptosis and DNA repair (Figs. 1 and 2), in agreement with earlier studies by others (31, 32). Reduction in p53 apoptosis and DNA repair by knockdown of ZNF198 or SUZ12 increased polyploidy and accelerated transformation by pX (Fig. 3), suggesting ZNF198 and SUZ12 are tumor suppressors of pX-mediated transformation.
Interestingly, in our liver cancer progression model, as P1-P5 cultures undergo pX-mediated transformation, we observed that protein levels of ZNF198 and SUZ12 become progressively reduced, whereas those of Plk1 required for pX-mediated transformation, increase (Fig. 4B). We hypothesized that during pX-mediated transformation, Plk1 by phosphorylation down-regulates ZNF198 and SUZ12. In support of this hypothesis we demonstrate: i) mRNA levels of ZNF198 and SUZ12 are not reduced during pX-mediated transformation (Fig. 4D), thereby excluding down-regulation at the transcriptional level; ii) inhibition of Plk1 by treatment with BI 2536 restored protein levels of ZNF198 and SUZ12 in P4 cultures (Fig. 4E), suggesting that Plk1 activity induces their degradation. iii) ZNF198 and SUZ12 co-immunoprecipitate with PBD of Plk1 in the presence of pX (Fig. 4F), similar to other Plk1 substrates (48). Studies are in progress to identify Plk1 phosphorylation sites in ZNF198 and SUZ12, and determine their functional significance in pX-mediated transformation.
The mechanism by which down-regulation of ZNF198 and SUZ12 contributes to pX-mediated transformation is not yet understood. ZNF198 stabilizes the LSDI-Co-REST-HDAC1 complex (26) thereby suppressing transcription. The telomerase reverse transcriptase (hTERT) gene, activated in more than 90% of human cancers, is a target of the LSD1-Co-REST-HDAC1 complex (53). Regarding the SUZ12/PRC2 complex, gene targets of the silencing H3K27me3 modification include among others (54), EpCAM (27) and IGF II (55) elevated in HBV-mediated HCCs (56, 57). Significantly, EpCAM-positive HCC cells are tumor initiating cells (29). Additional genes transcriptionally silenced by the SUZ12/PRC2 complex and relevant to HBV-mediated HCC are presented in our recent review (58). Specifically, in addition to EpCAM and IGFII, SUZ12 target genes relevant to HCC include DKK1,2, BAMBI, MYC, MCM5, RPA2, CCNA1 and CCND2. Importantly, EpCAM, IGFII, DKK1, 2 and BAMBI are up-regulated in hepatic cancer stem cells (29) and poor prognosis HCCs derived from HBV-positive patients (56, 57). The genes MYC, MCM5, RPA2, CCNA1 and CCND2 that comprise the proliferation gene cluster are upregulated in poor prognosis HCC (18, 59–60). We reason that a cellular mechanism that down-regulates SUZ12 enables re-expression of these genes that are elevated in HBV-induced and poor prognosis HCCs.
Down-regulation of ZNF198 and SUZ12 during HBV replication
Viruses disrupt by various mechanisms PML NBs, enhancing viral replication (33–35). In HepAD38 cells undergoing HBV replication, the protein levels of ZNF198 and SUZ12 become reduced while those of Plk1 increase. In addition, PML NBs become disrupted (Fig. 5). Importantly, siRNA knockdown of ZNF198 and PML enhances HBV replication, indicating the functional involvement of ZNF198 and PML NBs in HBV replication (Fig. 6). In HSV1 infected cells, ICP0 disrupts nuclear localization of LSD1-CoREST-HDAC complex (34, 35) shown to be stabilized by ZNF198 (26). Whether the same occurs in HBV replication is unknown. Likewise, SUZ12 knockdown exerts a small but statistically significant (p<0.05) effect on HBV replication (Fig. 6). Since the SUZ12/PRC2 complex mediates transcriptional repression of cellular genes via the H3K27me3 modification, it is plausible that it has a similar effect on the transcription of the viral pgRNA. Further studies are needed to elucidate the mechanism by which ZNF198 and SUZ12 modulate HBV replication. Importantly, the suggested down-regulation of ZNF198 and SUZ12 by Plk1 together with the role of these proteins in HBV replication identifies Plk1 as a likely antiviral target. Recent studies have also identified involvement of Plk1 in HCV replication (61).
In summary, our studies demonstrate an inverse relationship between elevated protein levels of Plk1 and down-regulation of ZNF198 and SUZ12 during pX-mediated transformation and HBV replication. Diminished protein levels of ZNF198 and SUZ12 enhance both pX-mediated transformation and HBV replication. Our results also suggest that Plk1 mediates the down-regulation of ZNF198 and SUZ12. We propose inhibition of Plk1 will exert both anticancer and antiviral effects.
Supplementary Material
Acknowledgments
FINANCIAL SUPPORT
This work was supported by NIH grants DK044533 and CA135192 to OMA. LS supported by the Bilsland’s graduate student fellowship of Purdue University. The authors acknowledge support of the Purdue University Cytometry Laboratory.
The authors thank Drs. B. Slagle for assistance with HBV replication assays, C. Seeger and M. Bouchard for HepAD38 and HepG2 cell lines, respectively, and R.L. Hullinger for critical review of this manuscript.
LIST OF ABBREVIATIONS
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- WCE
whole cell extract
- H2AX
H2A histone family, member X
- Plk1
Polo-like kinase1
- PCR
Polymerase Chain Reaction
- PRC2
Polycomb repressive complex 2
- SUZ12
suppressor of zeste 12 homolog (Drosophila)
- ZNF198
zinc finger, MYM-type 2
- LSD1-Co-REST-HDAC1
Lysine-Specific Demethylase1-corepressor of RE1-Silencing Transcription Factor-Histone Deacetylase1
- PML NBs
promyelocytic leukemia nuclear bodies
Contributor Information
Wen-Horng Wang, Email: wwang@purdue.edu.
Leo L. Studach, Email: lstudach@purdue.edu.
Ourania M. Andrisani, Email: andrisao@purdue.edu.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Wang XW, Hussain SP, Huo TI, Wu CG, Forgues M, Hofseth LJ, et al. Molecular pathogenesis of human hepatocellular carcinoma. Toxicology. 2002;181/182:43–47. doi: 10.1016/s0300-483x(02)00253-6. [DOI] [PubMed] [Google Scholar]
- 3.Zoulim F, Poynard T, Degos F, Slama A, El Hasnoui A, Blin P, et al. A prospective study of the evolution of lamivudine resistance mutations in patients with chronic hepatitis B treated with lamivudine. J Viral Hepatitis. 2006;13:278–288. doi: 10.1111/j.1365-2893.2005.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 5.Hagen TM, Huang S, Curnutte J, Fowler P, Martinez V, Wehr CM, et al. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc Natl Acad Sci USA. 1994;91:12808–12812. doi: 10.1073/pnas.91.26.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P, Buendia MA. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 7.Madden CR, Finegold MJ, Slagle BL. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J Virol. 2001;75:3851–3858. doi: 10.1128/JVI.75.8.3851-3858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su Q, Schroder CH, Hofmann WJ, Otto G, Pichlmayr R, Bannasch P. Expression of Hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109–1120. doi: 10.1002/hep.510270428. [DOI] [PubMed] [Google Scholar]
- 9.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrisani OM, Barnabas S. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis. Int J Oncol. 1999;15:373–379. doi: 10.3892/ijo.15.2.373. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Tarn C, Wang WH, Chen S, Hullinger RL, Andrisani OM. Hepatitis B virus X protein differentially regulates cell cycle progression in X-transforming versus non-transforming hepatocyte (AML12) cell lines. J Biol Chem. 2002;277:8730–8740. doi: 10.1074/jbc.M108025200. [DOI] [PubMed] [Google Scholar]
- 13.Rakotomalala L, Studach L, Wang W-H, Gregori G, Hullinger RL, Andrisani OM. Hepatitis B virus X protein increases the Cdt1 to geminin ratio inducing DNA re-replication and polyploidy. J Biol Chem. 2008;283:28729–28740. doi: 10.1074/jbc.M802751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Studach L, Wang WH, Weber G, Tang J, Hullinger RL, Malbrue R, Liu X, Andrisani O. Hepatitis B Virus X protein activates Polo-like Kinase-1 inducing checkpoint adaptation, suppression of DNA repair and p53 apoptosis. J Biol Chem. 2010;285:30282–93. doi: 10.1074/jbc.M109.093963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Studach L, Rakotomala L, Wang WH, Hullinger RL, Cairo S, Buendia MA, Andrisani O. Polo-like kinase1 inhibition suppresses Hepatitis B virus X protein-induced transformation, in an in vitro model of liver cancer progression. Hepatology. 2009;50:414–423. doi: 10.1002/hep.22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takaki T, Trenz K, Constanzo V, Pentronczki Polo-like kinase1 reaches beyond mitosis-cytokenisis, DNA damage response and development. Current Opinion in Cell Biology. 2008;20:650–660. doi: 10.1016/j.ceb.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24:287–91. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Incassati A, Patel D, McCance DJ. Induction of tetraploidy through loss of p53 and upregulation of Plk1 by human papillomavirus type-16E6. Oncogene. 2006;25:2444–2451. doi: 10.1038/sj.onc.1209276. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Lei M, Erikson RL. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol Cell Biol. 2006;26:2093–2108. doi: 10.1128/MCB.26.6.2093-2108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krssák M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 22.Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nature Reviews. 2010;9:644–660. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- 23.Pasini D, Bracken AP, Jensen MR, Lazzerini DE, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunapuli P, Kasyapa CS, Chin S-F, Caldas C, Cowell JK. ZNF198, a zinc finger protein rearranged in myeloproliferative disease, localizes to the PML nuclear bodies and interacts with SUMO-1 and PML. Exp Cell Res. 2006;312:3739–3751. doi: 10.1016/j.yexcr.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 25.Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Viré E, et al. Role of Polycomb Repressive Complex 2 in Acute Promyelocytic Leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 26.Gocke CB, Yu H. ZNF198 stabilizes the LSD-1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS ONE. 2008;3:e3255. doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu T-Y, Lu R-M, Liao M-Y, Yu J, Chung C-H, et al. Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J Biol Chem. 2010;285:8719–8732. doi: 10.1074/jbc.M109.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmelzer E, Wauthier E, Reid IM. The phenotypes of pluripotent human hepatic progenitors. Stem Cells. 2006;24:1852–1858. doi: 10.1634/stemcells.2006-0036. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, et al. EpCAM-positive hepatocellular carcinoma cells are tumor initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunapuli P, Somerville R, Still IH, Cowell JK. ZNF198 protein, involved in rearrangement in myeloproliferative disease, forms complexes with the DNA repair-associated HHR6A/6B and RAD18 proteins. Oncogene. 2003;22:3417–3423. doi: 10.1038/sj.onc.1206408. [DOI] [PubMed] [Google Scholar]
- 31.Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP. The function of PML in p53-dependent apoptosis. Nat Cell Biol. 2000;2:730–736. doi: 10.1038/35036365. [DOI] [PubMed] [Google Scholar]
- 32.Dellaire G, Ching RW, Ahmed K, Jalali F, Tse KC, Bristow RG, Bazett-Jones DP. Promyelocyte leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol. 2006;175:55–66. doi: 10.1083/jcb.200604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Everett RD. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20:7266–7273. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 34.Gu H, Roizman B. The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by CoREST/REST/HDAC complex and degradation of PML, are executed in tandem. J Vir. 2009;83:181–187. doi: 10.1128/JVI.01940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu H, Roizman B. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J Vir. 2009;83:4379–4385. doi: 10.1128/JVI.02515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung Y-L, Tsai T-Y. PML NBs link the DNA damage repair pathway to hepatitis B virus replication: implications for hepatitis B virus exacerbation during chemotherapy and radiotherapy. Mol Cancer Res. 2009;7:1672–1685. doi: 10.1158/1541-7786.MCR-09-0112. [DOI] [PubMed] [Google Scholar]
- 37.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4. Gastroenterology. 2006;130:827–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad USA. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarn C, Bilodeau ML, Hullinger RL, Andrisani OM. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J Biol Chem. 1999;274:2327–2336. doi: 10.1074/jbc.274.4.2327. [DOI] [PubMed] [Google Scholar]
- 40.Ladner S, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. Antimicrobial agents and chemotherapy. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keasler VV, Hodgson AJ, Madden CR, Slagle BL. Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J Virol. 2007;81:2656–2706. doi: 10.1128/JVI.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang WH, Hullinger RL, Andrisani OM. Hepatitis B virus X protein via the p38MAPK pathway induces E2F1 release and ATR kinase activation mediating p53 apoptosis. J Biol Chem. 2008;283:25455–25467. doi: 10.1074/jbc.M801934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, et al. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem. 2004;279:36440–36444. doi: 10.1074/jbc.C400300200. [DOI] [PubMed] [Google Scholar]
- 44.de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13:523–535. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 45.Cerosaletti K, Wright J, Concannon P. Active role for Nibrin in the kinetics of Atm activation. Mol Cell Biol. 2006;26:1691–1699. doi: 10.1128/MCB.26.5.1691-1699.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:215–216. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 48.Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc Natl Acad Sci U S A. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao S, Nalabolu S, Aster JC, Ma J, Abruzzo L, Jaffe ES, et al. FGFR1 is fused with novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nature Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 50.Li H, Ma XY, Wang J, Koontz J, Nucci M, Sklar J. Effects of rearrangement and allelic exclusion of JJAZ1/SUZ12 on cell proliferation and survival. Proc Natl Acad Sci. 2007;104:20001–20006. doi: 10.1073/pnas.0709986104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moinzadeh P, Breuhahn K, Stutzer H, Schirmacher P. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade –results of an explorative CGH meta-analysis. British J Cancer. 2005;92:935–941. doi: 10.1038/sj.bjc.6602448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen CF, Yeh SH, Chen DS, Chen PJ, Jou YS. Molecular Genetic Evidence Supporting a Novel Human Hepatocellular Carcinoma Tumor Suppressor Locus at 13q12.11. Genes Chromosomes & Cancer. 2005;44:320–328. doi: 10.1002/gcc.20247. [DOI] [PubMed] [Google Scholar]
- 53.Zhu Q, Liu C, Ge Z, Fang X, Zhang X, et al. Lysine-specific demethylase 1 (LSD1) is required for transcriptional repression of the Telomerase Reverse Transcriptase (HTERT) gene. PLoS ONE. 2008;3:e1446. doi: 10.1371/journal.pone.0001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li T, Hu JF, Qiu X, Ling J, Chen H, Wang A, et al. CTCF regulates allelic expression of Igf2 by orchestrating a promoteor-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 57.Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 58.Andrisani OM, Studach L, Merle P. Gene Signatures in Hepatocellular Carcinoma. Semin Cancer Biol. 2010 doi: 10.1016/j.semcancer.2010.09.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Breuhahn K, Vreden S, Haddad R, Beckebaum S, Strippel D, Flemming P, et al. Molecular profiling of human hepatocellular carcinoma defines mutually exclusive interferon regulation and insulin-like-growth factor II overexpression. Cancer Res. 2004;64:6058–6064. doi: 10.1158/0008-5472.CAN-04-0292. [DOI] [PubMed] [Google Scholar]
- 60.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 61.Chen YC, Su WC, Huang JY, Chao TC, Seng KS, Machida K, Lai MC. Polo-like Kinase 1 is Involved in Hepatitis C Virus replication by hyperphosphorylating NS5A. J Virol. 2010;84:7983–7993. doi: 10.1128/JVI.00068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.