Abstract
Genomic integrity requires faithful chromosome duplication. Origins of replication, the genomic sites where DNA replication initiate, are scattered throughout the genome Their mapping at a genomic scale in multicellular organisms has been challenging. Here we have profiled origins in Arabidopsis by high-throughput sequencing of newly-synthesized DNA and identified ~1500 putative origins genome-wide. This was supported by ChIP-chip experiments to identify ORC1 and CDC6 binding sites. Origin activity was validated independently by measuring the abundance of nascent DNA strands. The midpoints of most Arabidopsis origin regions are preferentially located within the 5’ half of genes, slightly enriched in G+C, histone H2A.Z, H3K4me2/3 and H4K5ac, and depleted of H3K4me1 and H3K9me2. Our data establish the basis for understanding the epigenetic specification of DNA replication origins in Arabidopsis and have implications for other eukaryotes.
Keywords: DNA replication origin, epigenetic, origin recognition complex (ORC), CDC6, cell cycle, Arabidopsis thaliana, plant
Faithful duplication of the genetic material is crucial to maintain genomic integrity. DNA replication in eukaryotic cells initiates at multiple sites, known as replication origins, which are scattered throughout the genome 1–3. The number of origins ranges from hundreds to thousands depending on the cell type and/or the physiological state 3. One of the key steps for understanding replication origin function is whether and how they are specified in the genome. In S. cerevisiae, a strict sequence-dependent specification occurs whereby the origin recognition complex (ORC) recognizes an 11bp sequence to define the site of each active replication origin 4,5. This mechanism appears to be a rather unique situation because a consensus sequence has not been found in other organisms. For example, in S. pombe, although origins are associated with A+T-rich stretches they are not specified by a known consensus DNA sequence 6,7.
The identification of the molecular nature of replication origins in multicellular organisms has been elusive and only a handful of them have been analyzed 2,8–10. The large genome size of multicellular eukaryotes, their different developmental strategies and the existence of a diversity of proliferating cell populations have led to increased difficulty in determining origin specification, function and spatio-temporal regulation at a genomic scale 3. Local epigenetic modifications can further affect origin selection and usage, e.g. replication timing 3,11–13. Although attempts to obtain genome-wide maps of replication origins in mammalian cells have been reported 14–17, the molecular features defining replication origins in higher eukaryotes and, in particular, their links to epigenetic modifications still remain largely unknown.
In this study, we have identified replication origins, analyzed their organization, and defined their epigenetic signatures at a high-resolution genome-wide scale in the plant Arabidopsis thaliana. Its rather compact genome (~125Mb, ~28,000 protein coding genes), fully sequenced and annotated, and the relatively small amount of repetitive sequences (~17%), largely confined to the pericentromeric areas 18, make Arabidopsis an excellent system to study origins. Furthermore, the comparison of replication origin features in organisms with very different developmental and growth strategies could shed light into the basic principles governing origin specification and function in eukaryotes. In addition, genome-wide maps of epigenetic marks such as DNA methylation and several histone modifications have already been reported 19–21. The use of massive sequencing of short-pulse BrdU-labeled DNA led us to identify ~1500 putative replication origins across the Arabidopsis genome. ORC1 and CDC6 binding regions, which, importantly, are enriched in BrdU-labeled regions, were also identified by chromatin immunoprecipitation and microarray experiments (ChIP-chip). Furthermore, origin activity was validated independently by measurement of nascent DNA strand abundance. Our studies reinforce the idea that some origin features are shared with animal cells whereas others are unique to plants 22,23. The Arabidopsis “originome” reported here provides the basis of identifying the key features of eukaryotic replication origins and delineate their possible regulatory mechanisms.
RESULTS
Genome-wide mapping of Arabidopsis DNA replication origins
Functional origins mark the sites where the synthesis of nascent DNA strands occurs. Thus, our strategy was to sequence purified DNA labeled in vivo with a pulse of BrdU and confirm these data with the mapping of pre-RC binding (Supplementary Fig. 1). To obtain sufficient amounts of BrdU-labeled DNA, we used Arabidopsis cultures that contain a substantial amount of proliferating cells. We synchronized cells in G0 using sucrose deprivation and labeled them with BrdU a few hours after release from the block when cells are just entering the S-phase 24,25 (Fig. 2 and Supplementary Fig. 1). DNA was extracted, fractionated by CsCl gradient centrifugation and the BrdU-labeled material was purified and used to generate genomic libraries for sequencing using the Solexa (Illumina) technology. We obtained a total of ~4 million high quality reads that uniquely mapped to the Arabidopsis genome. Likewise, a sample of unlabeled DNA was processed as a control (see Methods). This BrdU-seq method rendered a comprehensive list of genomic locations with a significant enrichment in BrdU-labeled DNA strands (Fig. 1a). To define origin regions using the BrdU-labeled DNA sequencing data we merged BrdU positive regions separated <10kb, as described in Methods (see also Supplementary Fig. 3). An alignment of DNA sequences of ±100bp around the midpoint of BrdU-labeled regions did not render any consensus sequence. To corroborate the analysis of BrdU-labeled regions and deal with possible experimental variations, we carried out an independent assay of cell synchronization, BrdU-labeling and CsCl purification followed by massively parallel sequencing. Significantly, 78.2% (p<1.0e-6) of the BrdU-labeled regions overlapped with the regions defined in the previous experiment, supporting the reproducibility of the two independent experiments.
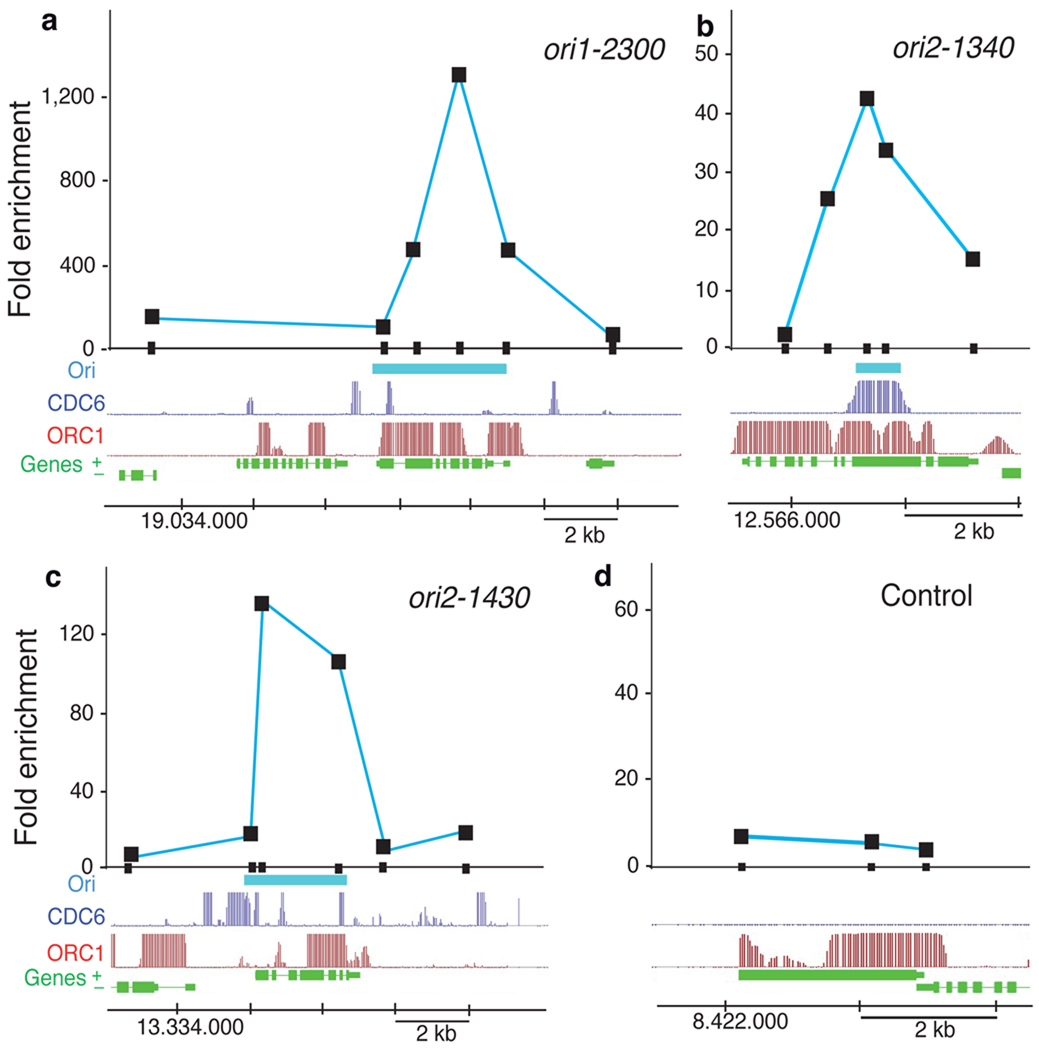
Figure 2. DNA replication origin activity determined by nascent DNA strand abundance.
(a-d) Several putative origin-containing regions were chosen for detailed measurement by real-time PCR of nascent strand abundance in a sample of short DNA molecules containing an RNA primer at their 5’end (see Methods). The genomic region under study is indicated at the bottom of each panel and shows the location of genes (green), the ORC1 binding (red) and CDC6 binding (dark blue) signals, and the putative origin location (light blue), defined by direct sequencing of the BrdU-labeled DNA sample (see Methods). DNA fragments (~200bp in length) amplified by primer pairs scanning each region are indicated by the small black rectangles on the X-axis. The coordinates in each chromosome are also indicated at the bottom of each panel. Results correspond to PCR amplifications using fraction #5 (see Methods). (a-c) Data for origins ori1-2300, ori2-1340 and ori2-1430. (d) Data for a region used as a negative control around gene at4g14700 that lacks BrdU-labeled DNA sequences.
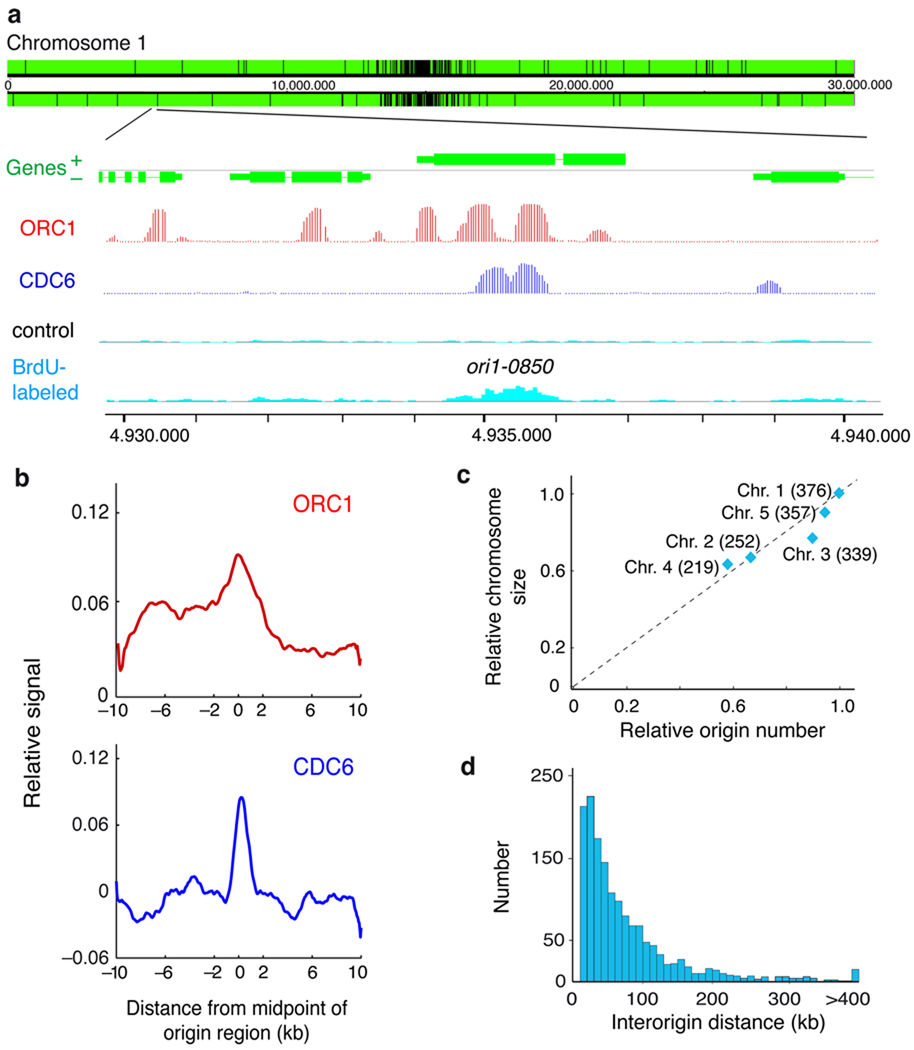
Figure 1. Identification of DNA replication origins in the Arabidopsis genome.
(a) Representative genome-browser view of a region in chromosome 1. Genes (green) transcribed from each strand are shown along the chromosome above and below the position scale. The panels shown in the lower part of the figure correspond to an enlarged region containing a replication origin, determined as a region enriched for BrdU-labeled DNA strands (light blue) relative to the unlabeled control DNA (black), together with the ORC1 (red) and CDC6 (dark blue) binding patterns (posterior probabilities for ORC1 and CDC6 data sets). Origins, e.g. ori1-0850 shown here, are named based on their chromosomal location (ori1- through ori5-) followed by the four digits that indicate the origin number within each chromosome. They are named consecutively starting at the left tip of each chromosome, i.e. for chromosome 1 where we identified 376 origins, the leftmost origin is ori1-0010 and the rightmost one is ori1-3760. (b) The pattern of ORC1 and CDC6 binding over origin regions was obtained by plotting their relative binding signal ±10kb from the origin region midpoint (0) using 50bp-sliding windows (smoothed). The p-values (two-sided) of the difference in the ChIP-chip signals in origins (midpoint ± 300bp), using a two-tailed Welch test, were 7.25e-6 and 1.29e-10 for ORC1 and CDC6, respectively. (c) Number of origins relative to chromosomal size. Chromosome size (relative to chromosome 1) was plotted against the number of origins identified in each chromosome (relative to origin number in chromosome 1). The number of origins identified in each chromosome is indicated in parenthesis. (d) Distribution of interorigin distances, measured as the distance between the midpoints of two contiguous origins (median = 51.1kb; average = 77.2kb; s.d. = 83.4kb).
To identify pre-RC binding sites, in the absence of specific antibodies, we used plants expressing constitutively tagged versions of two pre-RC components, ORC1 (ref. 26) and CDC6 (ref. 27). ORC1- and CDC6-bound DNA fragments were purified by chromatin immunoprecipitation (ChIP) (Supplementary Fig. 1) and hybridized to whole-genome Arabidopsis tiling arrays to identify their genome-wide binding sites (Fig. 1a). ORC1 binding was spread over numerous sites (Supplementary Fig. 4) whereas CDC6 binding sites were less abundant (Supplementary Fig. 5). First, we determined the fraction of the BrdU-labeled regions that contained bound pre-RC components. We found that ~76.7% and 17.0% of BrdU-labeled regions overlapped with ORC1 and CDC6 regions, respectively (midpoint of BrdU region ±2.1 kb, p<0.001; see colocalization range in Fig. 1b). More importantly, the midpoints of these regions significantly colocalized with both ORC1 and CDC6 binding sites within ±2kb regions (Fig. 1b). Therefore, the 1543 regions rendered by our approach were considered bona fide replication origins (Supplementary Table 1). They appear uniformly distributed across the genome, although it is possible to identify clusters of more closely spaced origins in some genomic locations (Supplementary Fig. 6). The number of origins varies for different chromosomes but they roughly correlate with chromosome size (Fig. 1c). The distribution of distances between origin region midpoints gave a median of 51.1 kb, with a mean of 77.2 kb (Fig. 1d).
Assessing origin activity by nascent strand abundance
The BrdU-labeled regions identified in our study and the marked colocalization with ORC1- and CDC6-binding sites strongly support the notion that they represent active DNA replication origins. To assess origin activity directly we measured the relative abundance of nascent DNA strands of various putative origin regions relative to adjacent regions in a sample of short DNA molecules purified by sucrose gradient centrifugation and containing a RNA primer at their 5’ end 28,29. Thus, origin activity was determined by real-time PCR methods using primer pairs spanning 5–16kb around putative origin regions. In all cases analyzed, we could demonstrate a high enrichment of origin sequences in the short nascent DNA strand sample (Fig. 2a-c). Importantly, one of the BrdU-labeled regions included in this analysis was one showing a relatively low CDC6 signal in the ChIP-chip experiment (Fig. 2a). In spite of this, it showed a high abundance of nascent DNA strands measured by qPCR, demonstrating the activity of this region as a functional origin as well as the robustness of our approach. A control region, lacking BrdU-labeled DNA sequences, did not show any appreciable enrichment (Fig. 2d). These data together led us to conclude that the set of origins identified here provides a solid starting point to define their molecular landscape.
Genomic location of Arabidopsis DNA replication origins
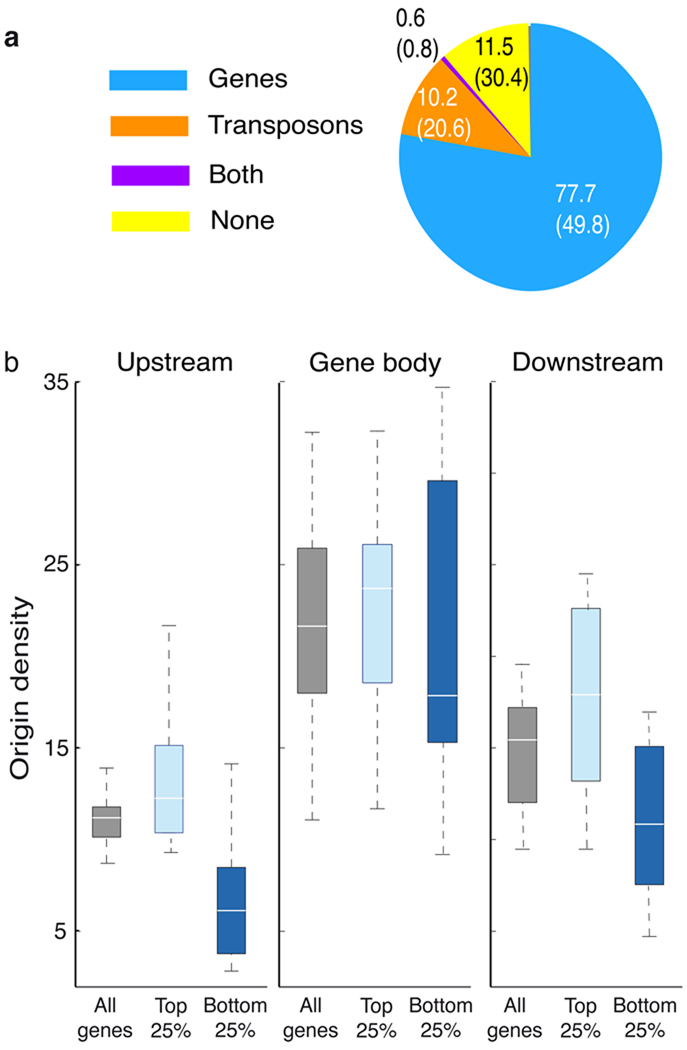
To test whether origins are randomly distributed along the genome or show a preferential location we estimated origin location relative to various genomic elements. We found that 77.7% and 10.2% of origins colocalized with gene units and transposons, respectively. These percentages are significantly different from the proportion of the Arabidopsis genome represented by these elements (Fig. 3a). Next, we analyzed origin density across genes and their 5’ and 3’ upstream regions. We observed that most origins were identified within gene bodies (Fig. 3b), but preferentially towards their 5’ ends (Supplementary Fig. 7). Origin localization to the bodies of genes did not correlate with gene expression levels (Fig. 3b), according to expression data obtained from cell suspensions at the same synchornization time used for BrdU labeling 30. However, highly expressed genes, compared to lowly expressed genes, tended to have more origins in regions immediately upstream (Wilcoxon ranksum test, p<0.005) or downstream (Wilcoxon ranksum test, P<0.01) of genes (Fig. 3b).
Figure 3. Genomic location of Arabidopsis replication origins.
(a) Percentage of origins colocalizing with various genomic elements, as indicated. Numbers in parenthesis indicate the proportion of the Arabidopsis genome represented by each class. (b) Origin densities were computed for regions upstream, downstream, and within genes of different expression levels (all genes vs highest 25% vs lowest 25%). Regions 2kb upstream and downstream of genes, as well as the bodies of genes were each divided into 10 bins, and the origin densities (origins per 106bp) were calculated for each bin and represented as boxplots. White lines represent the median, the edges of the boxes represent the 25th (bottom) and 75th percentiles (top), and the whiskers stretch out to the minimum and maximum points that fell within the 1.5xIQR range below the 25th percentile or above the 75th percentile.
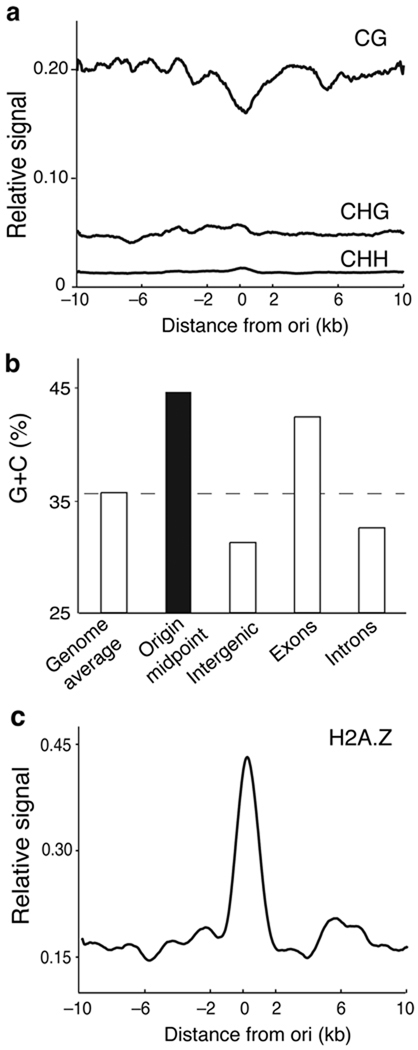
The body of highly expressed genes in Arabidopsis is enriched in CG methylation whereas the three types of C methylation (CG, CHG and CHH, where H is A, T or C) are highly enriched in the repeat-rich pericentromeric regions of the Arabidopsis genome 19,31. Interestingly, we found a slight decrease in CG methylation levels around origin midpoints compared to regions flanking them (Fig. 4a). Furthermore, we observed that regions ±0.1kb around the origin midpoints showed higher G+C contents (44.5%), compared to the whole Arabidopsis genome (Fig. 4b). It is known that the histone variant H2A.Z is preferentially deposited near the 5’ end of target genes and anticorrelates with CG methylation 32. We found a strong correlation between the presence of H2A.Z within ±1kb and the origin midpoints (Fig. 4c).
Figure 4. Relationship of Arabidopsis replication origins to CG methylation and histone H2A.Z.
(a) Relative levels of CG, CHG and CHH methylation were plotted ±10kb relative to the origin midpoint (0) in 50bp sliding windows (smoothed). Methylation data were published elsewhere 19. (b) G+C content (%) of replication origins (blue) and the indicated genomic regions (orange). These values were calculated from the sequence data files available at TAIR web site. (c) Density of the histone variant H2A.Z in a ±10kb region relative to the origin midpoint (0) in 50bp sliding windows (smoothed). The genomic distribution of H2A.Z was published elsewhere 32. The p-value of the difference in the ChIP-chip signals in origins, calculated as in Fig. 1b (see Methods), was 9.34e-34.
The epigenomic landscape of Arabidopsis DNA replication origins
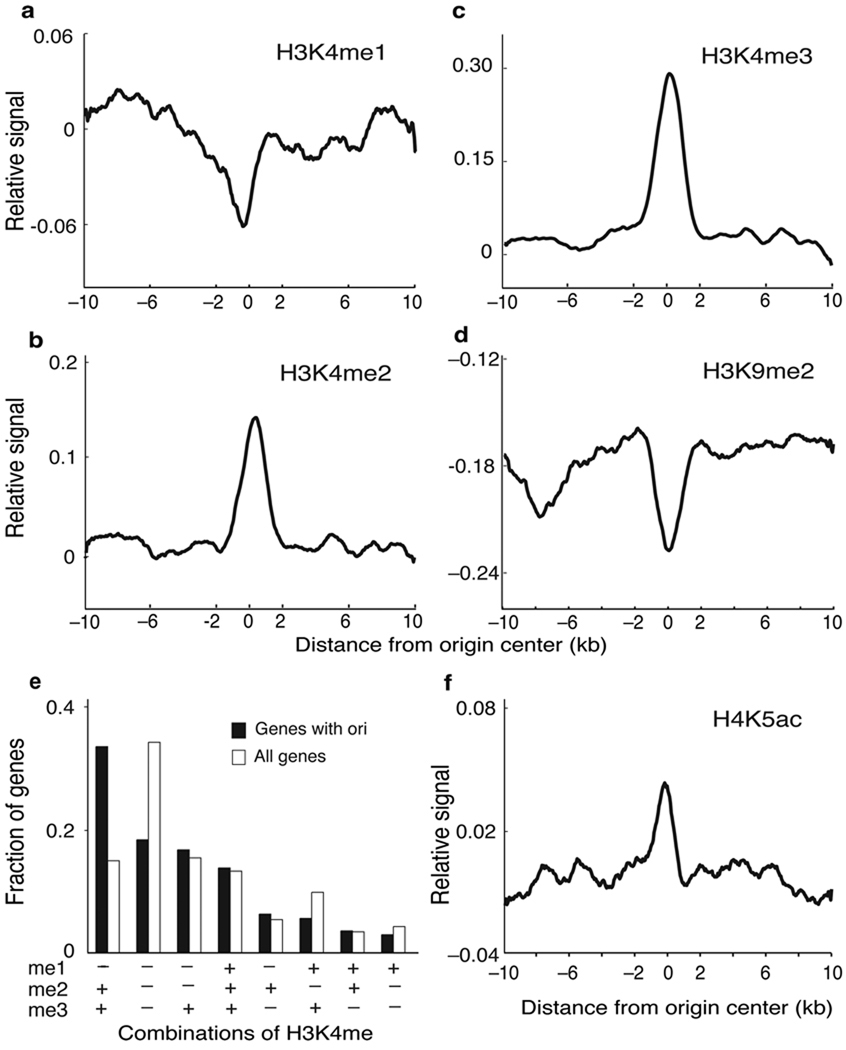
To further determine features defining Arabidopsis replication origins we next sought to profile the landscape of epigenetic histone marks that appear to associate with replication origins. Arabidopsis epigenomics data are already available for dimethylation of histone H3 at lysine 9 (H3K9me2) and for the three methylated forms of H3K4 20,21. We found that most origins tend to be depleted of H3K4me1 (Fig. 5a) but are highly enriched in H3K4me2 and H3K4me3 (Fig. 5b-c). In fact, we observed that H3K4me3 and/or H3K4me2, with or without H3K4me1, appears to be a signature of ~80% of origins associated with genes (Fig. 5e). This is consistent with the preferential localization of origins in 5’ gene body regions observed here and the anticorrelation of these marks and CG methylation 21. Furthermore, H3K9me2 is highly depleted in most of the origins identified in our study (Fig. 5d).
Figure 5. Histone modification landscape around replication origins.
(a-d) The relative level of the indicated histone mark is plotted ±10kb relative to the center of origins (0) in 50bp sliding windows (smoothed). Data for H3K4me and H3K9me2 were reported elsewhere 20,21. The p-values of the difference in the ChIP-chip signals in origins, calculated as in Fig. 1b (see Methods), were 0.86, 3.52e-28, 1.07e-41 and 7.33e-14 for H3K4me1, H3K4me2, H3K4me3 and H3K9me2, respectively. (e) Relationship between H3K4 methylation status and the presence of origins. We calculated the fraction of genes containing origins and different combinations of H3K4 methylation, as indicated, and compared it with the fraction of all genes containing the same H3K4me combinations 21. Different classes are ordered with decreasing values of the fraction of “genes with origins”. (f) The relative level of H4K5ac is plotted ±10kb relative to the center of origins (0) in 50bp sliding windows (smoothed). Calculations are based on the ChIP-chip dataset generated in this work. The p-value of the difference in the ChIP-chip signals in origins, calculated as in Fig. 1b (see Methods), was 1.23e-23.
A correlation exists between histone hyperacetylation and origin activation in Xenopus 11 and Drosophila cells 33–35. Consistent with this, immunofluorescence data obtained in several plant species indicate that increases in histone acetylation occurs during S-phase 22,23. Recently, ChIP experiments have revealed that H4K5 and H4K12 (also H4K8 to a lesser extent) but not H4K16 need to be acetylated by the HBO1 histone acetylase at origins in human cells to overcome geminin inhibition and facilitate MCM loading 36. Thus, we profiled H4K5ac over the genome by ChIP-chip and found an enrichment of this mark at the origin midpoint (Fig. 5f).
DISCUSSION
Initiation of DNA replication in eukaryotes depends on the assembly of pre-replication complexes (pre-RC) in G1 of the cell cycle at certain chromosomal locations and its further activation to initiate DNA replication in S-phase. Both steps must be tightly coordinated to ensure that the genome is duplicated once per cell cycle 2. We have found that ORC1 binding sites tend to form clusters, a situation similar to Drosophila cells 37 but highly different from that of S. cerevisiae5. The presence of ORC1 binding sites across the genome may represent not only broad initiation zones with several potential initiation sites but also reflect the function of ORC1 in other processes, e.g. heterochromatin silencing 38, transcriptional control 26,39 or chromatid cohesion 40. In any case, detection of CDC6 in BrdU regions is highly valuable taking into account the release of CDC6 from the pre-RC once an origin fired 41.
The distribution of distances between origin region midpoints rendered values that fall within the range estimated for other eukaryotes 42 and roughly match estimations of replicon size in Arabidopsis 13,43. It is possible that a fraction of the putative origin regions identified here correspond to elongating forks rather than to initiation events. However, our direct measurements of origin activity by abundance of RNA primer-containing nascent strands support the idea that the “originome” reported here is a bona fide list of putative Arabidopsis DNA replication origins. Future analysis should address this point individually. The abundance of origin sequences and the width of the peak of amplified fragments varied for different origins analyzed, suggesting differences in the efficiency of origin usage or in the usage of initiation sequences within an origin region 10,42.
Interestingly, the location of most Arabidopsis origins is different from other systems in which a large proportion of highly efficient origins are associated with gene promoters or transcriptional start sites 16,17,37. We have found that the ±0.1kb region around Arabidopsis DNA replication origins possesses a higher than average G+C content and a slight decrease in CG methylation. Consistent with this observation, early-mid replicons in Arabidopsis chromosome 4 have been also found to be depleted of CG methylation 13. One possibility is that in Arabidopsis the relatively high G+C content at origins favors a particular nucleosome organization in these regions. This is reinforced by the colocalization of origins with histone H2A.Z, which affects nucleosome stability 44, and could facilitate pre-RC assembly and/or origin firing. Together, our data show that whereas CG methylation within gene bodies is relevant for gene expression in Arabidopsis 19, it does not seem to be a requirement for origins. Metazoan origins highly correlate with unmethylated CpG islands located at the promoter of active genes or in the proximity to transcriptional start sites 6,42. While CpG islands are not present in the Arabidopsis genome, our results revealed a conserved trend of having relatively lower CG methylation at origins and show a high correlation between origin activity, a local high G+C content and presence of H2A.Z.
Posttranslational histone modifications can also affect origin specification and function. Most Arabidopsis origins tend to be enriched in H3K4me2 and H3K4me3, as well as in H4K5ac, similar to human origins 17,36. Whether all human origins have the same H4ac pattern, as a consequence of HBO1 activity to overcome geminin inhibition 36, and whether all Arabidopsis origins require H4ac for activation remain open questions for the future. However, the H4 acetylation pattern is of particular relevance due to the presence in Arabidopsis of (i) an HBO1-related acetyltransferase 45, (ii) increased tetraH4ac residues around Arabidopsis ORC1-binding sites 26 and (iii) a CDT1-interacting protein, GEM, structurally unrelated to metazoan geminin 46,47. Acetylation in other histone residues may be also relevant for origin function, as suggested by the presence of H3K56ac in early replicons of chromosome 4 (ref. 13).
How replication origins are specified in large eukaryotic genomes has been a long-standing question. The association of early-firing origins with transcribed genomic regions has been reported 48,49. Origins that have been studied in the 0.4–1% of mammalian genomes show a preferential association with active promoters that contain CpG islands 15–17. We have found that origins located in the upstream regions of genes are preferentially associated with highly expressed genes. However, the differences in the genomic distribution of CG methylation pattern in Arabidopsis may contribute to the use of different mechanisms to specify origins. In fact, a higher proportion of origins in Arabidopsis are located in the 5’ half of gene bodies compared to mammalian cells.
Our work has defined a landscape of epigenetic marks associated with a genome-wide set of replication origins in Arabidopsis. The midpoints of most origin regions preferentially colocalize with a significantly higher than average G+C content, but lower CG methylation level, and are enriched in histone H2A.Z, H3K4me2/3 and acetylated H4K5, and depleted in H3K4me1 and H3K9me2. Elucidating how epigenetic mechanisms and gene expression coordinate with DNA replication is of primary importance for understanding these processes in a genomic and developmental context. The Arabidopsis “originome” reported here provides the foundation for future studies to identify the mechanisms of origin specification as well as the regulation and function of DNA replication origins in different eukaryotes.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Martinez-Salas, J.A. Tercero and E. Caro for comments and discussions, and to Sara Diaz-Triviño and P. Hernandez for initial efforts in origin mapping, and to M. Gomez and J. Sequeira-Mendes for advice with the purification and analysis of nascent DNA strands. The technical help of V. Mora-Gil is deeply acknowledged. M.P.S. and C.C. are recipients of JAE-Doc contracts from CSIC. S.F. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. Research has been supported by grants BFU2006-5662, BFU2009-9783 and CSD2007-00057-B (Ministry of Science and Education) and P2006/GEN0191 (Comunidad de Madrid) to C.G, by an institutional grant from Fundación Ramón Areces to CBM, by grant GM60398 (National Institutes of Health) to S.E.J and by grants BIO2004-02502, BIO2007-66935, GEN2003-20218-C02-02 and CSD2007-00057-B (Ministry of Science and Innovation) and GR/SAL/0674/2004 (Comunidad de Madrid) to R.S. S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession codes. The NCBI GEO accession numbers for the datasets generated in this work are GSE21928 (for ORC1 and CDC6 ChIP-chip) and GSE21828 (for BrdU-seq and H4K5ac ChIP-chip).
Supplementary information. Supplementary information is available at the NSMB website.
AUTHORS CONTRIBUTIONS
C.C., M.P.S., Y.Y., S.F., A.B. and I.L.-V. performed experiments. H.S., J.C.O., C.C., M.P.S., X.Z. and R.S. analyzed data, C.G. and S.E.J. prepared the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Huberman JA, Riggs AD. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- 2.DePamphilis ML, et al. Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- 4.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 5.Wyrick JJ, et al. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 6.Antequera F. Genomic specification and epigenetic regulation of eukaryotic DNA replication origins. EMBO J. 2004;23:4365–4370. doi: 10.1038/sj.emboj.7600450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi M, et al. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J. 2007;26:1327–1339. doi: 10.1038/sj.emboj.7601585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert DM. In search of the holy replicator. Nat Rev Mol Cell Biol. 2004;5:848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa S, Blow JJ. The elusive determinants of replication origins. EMBO Rep. 2007;8:332–334. doi: 10.1038/sj.embor.7400954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schepers A, Papior P. Why are we where we are? Understanding replication origins and initiation sites in eukaryotes using ChIP-approaches. Chromosome Res. 2010:63–77. doi: 10.1007/s10577-009-9087-1. [DOI] [PubMed] [Google Scholar]
- 11.Danis E, et al. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- 12.MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes & Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee TJ, et al. Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genet. 2010;6:e1000982. doi: 10.1371/journal.pgen.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucas I, et al. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cadoret JC, et al. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci U S A. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sequeira-Mendes J, et al. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5:e1000446. doi: 10.1371/journal.pgen.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karnani N, Taylor CM, Malhotra A, Dutta A. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell. 2010;21:393–404. doi: 10.1091/mbc.E09-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 19.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns--from conservation to diversity. Trends Plant Sci. 2006;11:199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez M, Caro E, Desvoyes B, Ramirez-Parra E, Gutierrez C. Chromatin dynamics during the plant cell cycle. Semin Cell Dev Biol. 2008;19:537–546. doi: 10.1016/j.semcdb.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Menges M, Murray JA. Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 2002;30:203–212. doi: 10.1046/j.1365-313x.2002.01274.x. [DOI] [PubMed] [Google Scholar]
- 25.Menges M, Murray JA. Synchronization, transformation, and cryopreservation of suspension-cultured cells. Methods Mol Biol. 2006;323:45–61. doi: 10.1385/1-59745-003-0:45. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez MP, Gutierrez C. Arabidopsis ORC1 is a PHD-containing H3K4me3 effector that regulates transcription. Proc Natl Acad Sci U S A. 2009;106:2065–2070. doi: 10.1073/pnas.0811093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C. Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell. 2001;13:2671–2686. doi: 10.1105/tpc.010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prioleau MN, Gendron MC, Hyrien O. Replication of the chicken beta-globin locus: early-firing origins at the 5' HS4 insulator and the rho- and betaA-globin genes show opposite epigenetic modifications. Mol Cell Biol. 2003;23:3536–3549. doi: 10.1128/MCB.23.10.3536-3549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez M, Antequera F. Overreplication of short DNA regions during S phase in human cells. Genes & Dev. 2008;22:375–385. doi: 10.1101/gad.445608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menges M, Hennig L, Gruissem W, Murray JA. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol. 2003;53:423–442. doi: 10.1023/B:PLAN.0000019059.56489.ca. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 34.Hartl T, Boswell C, Orr-Weaver TL, Bosco G. Developmentally regulated histone modifications in Drosophila follicle cells: initiation of gene amplification is associated with histone H3 and H4 hyperacetylation and H1 phosphorylation. Chromosoma. 2007;116:197–214. doi: 10.1007/s00412-006-0092-2. [DOI] [PubMed] [Google Scholar]
- 35.Schwaiger M, et al. Chromatin state marks cell-type- and gender-specific replication the Drosophila genome. Genes & Dev. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miotto B, Struhl K. HBO1 Histone Acetylase Activity Is Essential for DNA Replication Licensing and Inhibited by Geminin. Mol Cell. 2010;37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, Macalpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pak DT, et al. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 39.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- 41.Kim J, Kipreos ET. Control of the Cdc6 replication licensing factor in metazoa: the role of nuclear export and the CUL4 ubiquitin ligase. Cell Cycle. 2008;7:146–150. doi: 10.4161/cc.7.2.5282. [DOI] [PubMed] [Google Scholar]
- 42.Cadoret JC, Prioleau MN. Genome-wide approaches to determining origin distribution. Chromosome Res. 2010:79–89. doi: 10.1007/s10577-009-9094-2. [DOI] [PubMed] [Google Scholar]
- 43.Van't Hof J, Kuniyuki A, Bjerknes CA. The size and number of replicon families of chromosomal DNA of Arabidopsis thaliana. Chromosoma. 1978;68:269–285. [Google Scholar]
- 44.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosomefree regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS. In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J. 2007;52:615–626. doi: 10.1111/j.1365-313X.2007.03264.x. [DOI] [PubMed] [Google Scholar]
- 46.Caro E, Castellano MM, Gutierrez C. A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature. 2007;447:213–217. doi: 10.1038/nature05763. [DOI] [PubMed] [Google Scholar]
- 47.Caro E, Gutierrez C. A green GEM: intriguing analogies with animal geminin. Trends Cell Biol. 2007;17:580–585. doi: 10.1016/j.tcb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Zhou J, et al. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 2005;24:1406–1417. doi: 10.1038/sj.emboj.7600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karnani N, Taylor C, Malhotra A, Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.