Abstract
Chromatin regulation provides an important means of controlling cardiac gene expression under different physiological and pathological conditions. Processes that direct the development of normal embryonic hearts and pathology of stressed adult hearts may share general mechanisms that govern cardiac gene expression by chromatin-regulating factors. These common mechanisms may provide a framework for us to investigate the interactions among diverse chromatin remodelers/modifiers and various transcription factors in the fine regulation of gene expression, essential for all aspects of cardiovascular biology. Aberrant cardiac gene expression, triggered by a variety of pathological insults, can cause heart diseases in both animals and humans. The severity of cardiomyopathy and heart failure correlates strongly with abnormal cardiac gene expression. Therefore, controlling cardiac gene expression presents a promising approach to the treatment of human cardiomyopathy. This review focuses on the roles of ATP-dependent chromatin-remodeling factors and chromatin-modifying enzymes in the control of gene expression during cardiovascular development and disease.
Keywords: chromatin, heart development, cardiac hypertrophy, cardiomyopathy, heart failure, differentiation, proliferation
Introduction
Forming a heart that pumps for perhaps 80 years in a human life with near-perfect fluid dynamics is a challenging task. Heart development is thus a precisely regulated process, and the required gene expression is tightly controlled. Many different genes are expressed in fetal and adult hearts to meet the distinct developmental and physiological needs. However, when adult hearts are stressed by various pathological insults such as ischemia, pressure or volume overload, they may express or activate certain genes that are normally only expressed in fetal hearts. This re-expression of “fetal” genes in adult hearts contributes to the disease process that leads to cardiomyopathy and congestive heart failure. Therefore, it is important to understand the mechanisms that control cardiac gene expression under developmental and different pathophysiological conditions.
One important control of gene expression is through chromatin regulation. Human cells have evolved sophisticated packaging methods to compact the 1.7 meter-long DNA into a nucleus that is only about 6 µm in diameter. First, 147 base pairs of DNA is wrapped around an octameric core of histone proteins, composed of two molecules of each of the canonical H2A, H2B, H3 and H4 histones, to form a nucleosome, a basic building block of the chromatin (Figure 1). These repeating units of nucleosomes are then tightly organized into higher-order scaffolds to further condense the DNA. Within each nucleosome, the positively charged residues in the histones contact the negatively charged phosphate backbone of the DNA approximately every 10.4 base pairs, providing 14 weak histone-DNA contacts1. These contacts, in combination, provide positional stability of the nucleosome on the genomic loci. However, such compacted and stable nucleosome structure may preclude regulatory factors from the DNA sequence. To modulate DNA accessibility, many proteins generally known as chromatin “remodelers” or “modifiers” alter the chromatin structure by changing nucleosome positioning and histone-DNA contacts, thereby properly preparing genomic loci for replication, repair, recombination, or transcription.
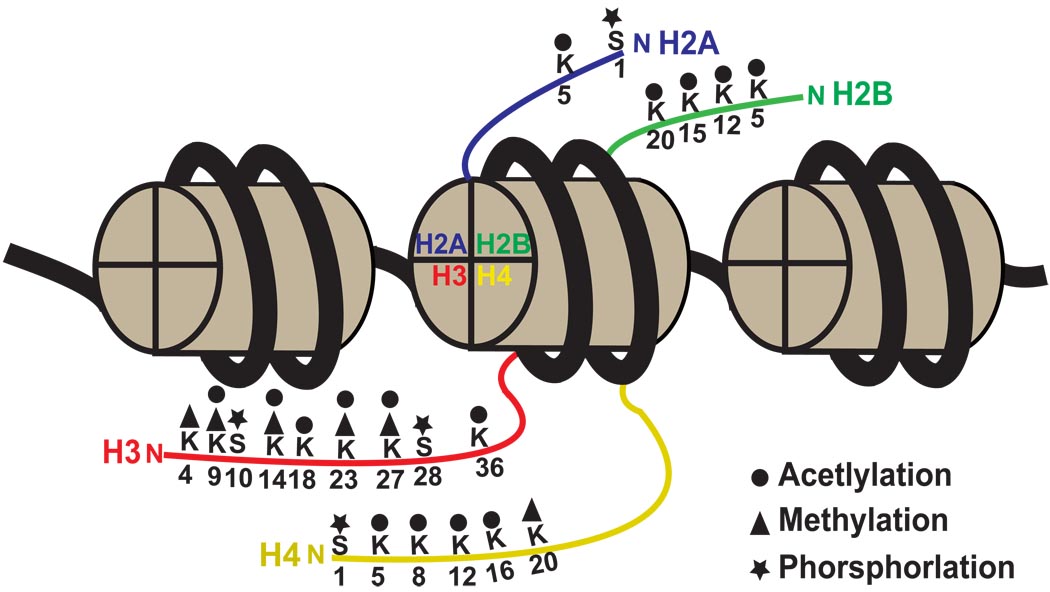
Figure 1. Schematic illustration of the nucleosome structure.
The nucleosome contains 147 base pairs of DNA (bold black line) that encircle an octameric core of histone proteins. Three nucleosomes are illustrated. The histone octamer is composed of two molecules of each of the canonical histone 2A (blue), 2B (green), 3 (red) and 4 (yellow) proteins. The N-terminal tails (colored thin lines) of these histone proteins are subject to a variety of covalent modifications, including acetylation, methylation and phosphorylation. Numbers beneath the colored lines denote the position of amino acid residues of the histones that are covalently modified. Letters above the colored lines indicate the amino acid substrates for these modifications. K: lysine, S: serine.
At least three processes have evolved to control the chromatin structure: DNA methylation, ATP-dependent chromatin remodeling, and covalent histone modifications (Figure 2). Biochemical mechanisms of how these processes regulate chromatin structure are reviewed elsewhere1–3. This review focuses on the developmental and pathophysiological roles of ATP-dependent chromatin remodeling and covalent histone modification. The ATP-dependent chromatin remodeling complexes use energy derived from ATP hydrolysis to move, destabilize, eject, or restructure nucleosomes. They do not perform covalent modification of the DNA or histones. In contrast, other chromatin modifying enzymes, such as histone deacetylases, catalyze the covalent modification of histone proteins to alter the histone-DNA contacts, and thereby “loosen” or “tighten” the chromatin to facilitate or limit genomic loci’s availability to transcriptional or other regulators. Histones can be modified in a variety of ways, including acetylation, methylation, ADP-ribosylation, phosphorylation, ubiquitination, sumoylation, and citrullination. In addition, chromatin remodeling/modifying factors can recruit or be recruited by other chromatin regulators or transcription factors to specific genomic loci, thus contributing to the complexity and specificity of chromatin regulation.
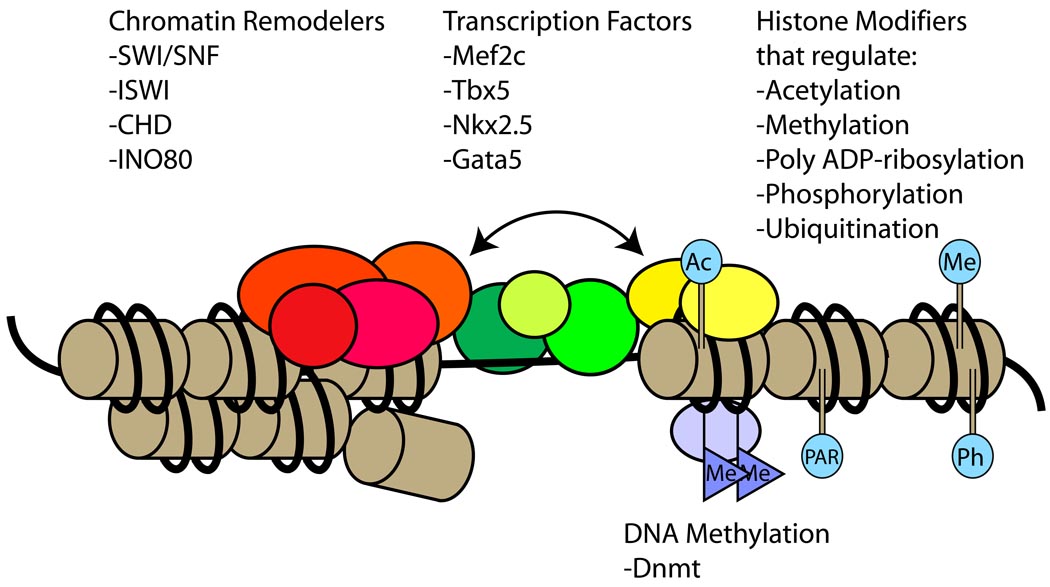
Figure 2. Interactions among chromatin regulators and transcription factors to control gene expression.
Chromatin regulation is an epigenetic mechanism that controls gene expression and function without changes in the DNA sequence. Chromatin remodelers use energy derived from ATP hydrolysis to change chromatin architecture. Histones are covalently modified to modulate access of transcription factors to genomic loci. DNA can be methylated to control transcription. These different forms of chromatin modification often act in concert and are co-regulated to control expression of cardiac genes. Ac: acetylation. Me: methylation. Ph: phosphorylation. PAR: poly ADP-ribosylation. Red/Orange: chromatin remodellers. Yellow: histone modifiers. Purple: DNA methyltransferase (Dnmt). Green/Olive: transcription factors.
Chromatin remodeling and histone modifications play essential roles in heart development and disease by reprogramming gene expression under different pathophysiological conditions. During heart development, chromatin regulators exert their functions in a temporal- and tissue-specific manner to direct the formation of various cardiovascular tissues. In adult hearts, chromatin regulation alters gene expression in response to pathophysiological changes. Pathological stresses trigger individual myocytes to increase in size and the whole heart to enlarge (or hypertrophy). Cardiac hypertrophy, although considered as a compensatory response to workload increased by pathological stresses, leads to a decline of muscle contractility and heart failure. Hypertrophy and heart failure are characterized by transcriptional reprogramming of gene expression and reactivation of fetal genes, many of which are known to be controlled by chromatin regulating factors. This review summarizes the roles of major chromatin remodeling and modifying factors in the control of embryonic heart development and pathogenesis of adult heart diseases.
The ATP-dependent chromatin-remodeling complexes
There are four different families of SWI-like ATP-dependent chromatin remodeling complexes: SWI/SNF (switching defective/sucrose nonfermenting), ISWI (imitation switch), CHD (chromodomain, helicase, DNA binding) and INO80 (inositol requiring 80) complexes (see ref 1, 4 for reviews). All members of the families share an evolutionarily conserved SWI-like ATPase catalytic domain, but each one has its own distinct flanking domains. The ATPase domain serves as a motor to adjust histone-DNA contacts for DNA movement and chromatin restructuring. The other domains may recognize covalently modified histones, modulate the ATPase activity or interact with other chromatin and transcription factors. Therefore, the unique domains and their associated proteins determine the genomic targeting specificity and biological functions of each family of chromatin remodelers.
The SWI/SNF complex in heart development and disease
The SWI/SNF chromatin remodelers, initially purified from Saccharomyces cerevisiae, are composed of 8–14 subunits (see ref 1, 4 for reviews). The vertebrate homologue of the yeast SWI/SNF complex is the BAF (brahma-associated factor) complex. In mammals, the BAF complex contains 12 protein components, including an ATPase subunit encoded by either Brm (brahma) or Brg1 (brahma-related gene 1). These two ATPase subunits, although highly homologous, exhibit non-redundant functions in vivo. Brg1-null mutation is peri-implantation lethal5, while Brm-null mice are viable and develop normally with slight increase in body mass6. Several other subunits of the BAF complex are also encoded by gene families, which give rise to a diverse composition of the BAF complex that have distinct functions in different cell types7–9. Recent studies have demonstrated that BRG1 and individual subunits of the BAF complex are essential in heart development and disease (Table 1).
Table 1.
Roles of ATP-dependent chromatin remodeling factors in cardiovascular development and disease
| Class | Chromatin Remodeling Factor |
Cardiovascular Studies in Animals | Comment | Ref | |
|---|---|---|---|---|---|
| Tissue of Gene Modification |
Phenotype | ||||
| SWI/SNF | BRG1 | Deletion in endothelium and endocardium | Lethality at E10.5–E11.5. Hypotrabeculation, yolk sac vascular defects, anemia. | Brg1 represses endocardial Adamts1 to prevent premature degradation of cardiac jelly, essential for trabeculation. | 10 |
| Deletion in myocardium | Lethality at E11.5. Thin compact myocardium and absent IVS* | Brg1 maintains Bmp10 to promote myocardial proliferation in embryos. Brg1 associates with HDACs and PARPs to repress α-MHC and activate β-MHC in embryonic hearts. | 17 | ||
| Inducible Brg1 deletion in adult myocardium prevents cardiac hypertrophy and fibrosis. | Cardiac stress activates Brg1, which then associates with HDACs and PARPs to control the pathological MHC switch, hypertrophy and fibrosis. | 17 | |||
| Deletion in secondary heart field | Lethality at E10.5. Hypoplastic right ventricle and outflow tract. | Brg1 maintains Bmp10 to promote proliferation of right ventricular and outflow tract myocardium. | 17 | ||
| Heterozygous germline mutation Brg1+/− | Variable heart defects: VSD*, patent foramen ovale, cardiac dilatation, sinus node dysfunction, AV block, conduction abnormalities. | Brg1 interacts dose-dependently with Nkx2.5, Tbx5, and Tbx20 to regulate heart development. | * | ||
| Point mutation; Morpholino knockdown in zebrafish | Mutation of brg1 in zebrafish causes hypoplastic myocardium, abnormal shape and alignment of cardiomyocytes. Knockdown causes cardiac looping defects, chamber narrowing, and contractility reduction. | Brg1 is evolutionarily conserved in zebrafish heart development. | * | ||
| BAF180 | Germline deletion | Lethality at E12.5–15.5. Thin compact myocardium, hypoplastic ventricles, VSD*, coronary defects. | Baf180 interacts with the retinoid acid pathway to regulate cardiac chamber formation. Baf180 is required for expression of genes essential for coronary vessel formation. | 38, 41 | |
| BAF60c | Knockdown in mouse hearts | Lethality at E10.0–11.0. Hypoplastic ventricles, hypotrabeculation, shortened outflow tract, abnormal cardiac looping. | Baf60c regulates heart morphogenesis through Hand2, Bmp10, Irx3 and others. Baf60c controls cardiac looping by activating Notch signaling and Nodal expression. | 42, 43 | |
| Knockdown in zebrafish | Randomized cardiac looping. | Zebrafish and mouse Baf60c share conserved functions for heart looping regulation. | 43 | ||
| BAF45c | Knockdown in zebrafish | Abnormal cardiac looping, poorly defined AV* boundary, reduced cardiac contractility, disarrayed muscle fibers. | Baf45c recognizes acetylated and methylated histones. This recognition may recruit BRG1/BAF to muscle- relevant gene promoters. | 45, 46 | |
| ISWI | SNF2H | Germline deletion | Lethality at E5.5–7.5 due to growth failure of the inner cell mass and trophoblast. | Snf2h may regulate heart development through the Wstf-containing WICH complex. | 51, 59 |
| CHD | CHD7 | Heterozygous germline mutation Chd7+/− | Aortic arch interruptions due to hypoplastic 4th pharyngeal arch artery. | Chd7 interacts with Tbx1 to regulate aortic arch formation in mice. | 61, 62 |
| Knockdown; Expression of dominant-negative Chd7 in frog | Abnormal position of truncus arteriosus and cardiac outflow tract. | CHD7 associates with BRG1/PBAF to control Sox9, Twist, Slug and neural crest cell activation. | 48 | ||
| INO80 | REPTIN PONTIN | Activation of repin or knockdown of pontin in zebrafish | Cardiac muscle hyperplasia. | Pontin and Reptin antagonistically control heart muscle growth, in part, through the β-catenin pathway. | 68 |
B. Bruneau: personal communication. ASD: atrial septal defect. AV: atrioventricular. IVS: interventricular septum. VSD: ventricular septal defect.
BRG1
Brg1 is known to control cardiovascular development in a time- and tissue-specific manner. Ablation of Brg1 in the endothelium causes embryonic lethality around embryonic day 10.5 (E10.5) of mouse development10. The mutant embryos are anemic and display vascular defects in the yolk sac but not in the embryo proper, indicating that endothelial Brg1 is required for primitive erythropoiesis and extraembryonic yolk sac vasculogenesis10, 11.
Within the developing heart, Brg1 acts in the endocardium to control myocardial trabeculation through regulating Adamts1 expression10. Adamts1 is a secreted matrix metalloproteinase whose substrates include versican, a cardiac jelly component required for heart development12–15. Adamts1 is normally repressed by Brg1 from E9.5 to E11.5 to allow the establishment of cardiac jelly for myocardial trabeculation10. Later, from E12.5 to E14.5, Adamts1 is derepressed in the endocardium, releasing the protease into the cardiac jelly to degrade versican and other matrix proteins, thereby terminating myocardial trabeculation at E13.5–E14.5 and preventing excessive trabecular growth10. Mice lacking endocardial Brg1 have Adamts1 prematurely activated in the endocardium at E9.5, resulting in an early degradation of the cardiac jelly and subsequent hypo-trabeculation10. Conversely, mice lacking Adamts1 exhibit hyper-trabeculation in the ventricles10. Therefore, the dynamic control of Adamts1 and cardiac jelly composition by Brg1 provides a developmental mechanism to delimit the extent of myocardial trabeculation. Such regulation is important because inadequate or excessive trabeculation can both cause cardiomyopathy and heart failure16.
Brg1 also functions in the myocardium to control cardiac gene expression, tissue growth and differentiation17. In mouse embryos, Brg1 promotes cardiomyocyte proliferation by maintaining Bmp10 and suppressing p57kip2 expression17. Bmp10 is a key factor required for myocardial proliferation18, while p57kip2 is a cyclin-dependent kinase inhibitor that prevents cell cycle progression18. Mice lacking myocardial Brg1 die around E11.5 because of the thin compact myocardium and absent interventricular septum17. These defects are caused by a failure of myocardial proliferation due to Bmp10 deficiency and ectopic p57kip2 expression. In parallel to proliferation control, Brg1 preserves fetal cardiac differentiation by interacting with histone deacetylases (HDACs) and poly (ADP ribose) polymerases (PARPs) to repress α-myosin heavy chain(α-MHC) and activate β-MHC expression in embryonic hearts17. In mice, α-MHC is the primary MHC isoform expressed in adult hearts, while β-MHC is the major isoform in embryonic hearts. By repressing α-MHC and activating β-MHC, the large Brg1/HDAC/PARP protein complex maintains a fetal state of MHC expression17. Embryonic cardiomyocytes lacking Brg1 switch the expression of MHC from β- to α-MHC. Also, inhibition of HDAC or PARP activity triggers a premature switch from β- to α-MHC expression in embryonic hearts. Therefore, Brg1 commands distinct molecular pathways to maintain cardiomyocytes in a fetal state of proliferation and differentiation.
Brg1, although highly expressed in embryonic hearts, is downregulated in the adult cardiomyocytes17, coinciding with the physiological switch from the fetal β-MHC to adult α-MHC expression. However, when the adult heart is stressed by pressure overload, Brg1 is reactivated in the cardiomyocytes and forms a complex with its embryonic partners, HDACs and PARP1, to repress α-MHC and activate β-MHC expression17. This stress-dependent assembly of fetal Brg1/HDAC/PARP complex triggers the adult heart to return to a fetal state of MHC expression, a critical step in the myopathic process19–27. Preventing such stress-induced Brg1 expression dramatically reduces cardiac hypertrophy, abolishes cardiac fibrosis and reverses the pathological MHC switch in mice17. Interestingly, BRG1 is also upregulated in some patients with hypertrophic cardiomyopathy17. Its level correlates with the disease severity and pathological MHC switch, suggesting that BRG1 may play a role in human hypertrophic heart disease. Since Brg1, HDAC, PARP are three classes of chromatin regulators required for cardiac hypertrophy17, 28–33, the assembly of these chromatin factors on the MHC promoters of stressed hearts suggests that chromatin may ultimately be where all the stress-response signals converge for the regulation of MHC genes.
Brg1 also functions in the secondary heart field to regulate right heart development in mice17. Deletion of Brg1 in the secondary heart field34 results in hypoplastic right ventricle and shortened cardiac outflow tract at E10.5 before embryonic lethality at E11.517. These defects are caused by inadequate proliferation of the right ventricular myocardium due to Bmp10 deficiency and ectopic p57kip2 expression17.
Brg1 is evolutionarily conserved in zebrafish heart development (B. Bruneau, personal communication). Mutation of Brg1 in zebrafish yields hypoplastic myocardium with abnormal shape and alignment of cardiomyocytes. Morpholino knockdown of Brg1 in zebrafish causes cardiac looping defects, chamber narrowing, and contractility reduction.
The gene dosage of Brg1 appears crucial for heart development (B. Bruneau, personal communication). Brg1+/− mice survive at a sub-Mendelian ratio with various degrees of congenital heart anomalies, including ventricular septal defects, patent foramen ovale, and cardiac dilatation. Those Brg1+/− mice that survive to adulthood display abnormal cardiac functions, including sinus node dysfunction, atrioventricular block, conduction abnormalities, and impaired cardiac relaxation. Brg1 genetically interacts with other cardiac transcription factors in a dosage-dependent manner. Compound Brg1 and Tbx5 heterozygous mice exhibit more severe cardiac defects than either mutation alone. Furthermore, when a single Brg1-null allele is introduced alongside with Nkx2.5 or Tbx20 heterozygous mutation that alone causes mild or no cardiac phenotypes35–37, severe cardiac defects with atrial and ventricular septal defects arise, and none of the compound mutants survive beyond E14.5. Transcriptional analyses of E11.5 hearts from single or compound heterozygous mutations for Brg1, Tbx5, or Nkx2.5 demonstrate a high degree of complexity of the genetic program governed by the three genes, with separate and overlapping gene regulation. This complexity appears partly mediated through the dosage of Brg1 and other transcription factors on cardiac gene promoters.
BAF180
Many other subunits of the BAF complex are involved in cardiovascular development. Baf180, also known as polybromo, contains six bromodomains that recognize acetylated histone tails. It is expressed ubiquitously in the developing embryo and is required for heart development. Baf180-null mouse embryos die between E12.5 and E15.538 due to the presence of trophoblast placental defects and severely hypoplastic ventricles with ventricular septal defects. These defects are similar to those observed in embryos lacking RXRα, a component of the retinoic acid (RA) pathway39, 40. It is shown that Baf180 is required for the expression of a subset of RA target genes such as RARβ2 and CRABPII38, suggesting that Baf180 interacts with the RA pathway to regulate cardiac chamber formation. In addition, Baf180 is necessary for the development of coronary vasculature41. Baf180-null epicardium fails to properly undergo epithelial-to-mesenchymal transformation (EMT), and the mice display downregulation of many factors that promote EMT, vasculogenesis, or angiogenesis41. All these defects may result in abnormal coronary development.
BAF60c
Baf60c, a component of the BAF complex, is expressed by the developing heart and skeletal muscle42. Baf60c is required for heart morphogenesis42, 43 and the differentiation of mesodermal cells into cardiomyocytes44. siRNA knockdown of Baf60c in mice causes embryonic lethality around E10.0 to E11.0 with multiple cardiac defects, namely hypoplastic ventricles with reduced trabeculation and shortened outflow tract42. Many genes required for heart morphogenesis are also mis-regulated, including Hand2, Bmp10, Irx3, among others42. These Baf60c knockdown embryos also show randomization of cardiac looping and situs, a function conserved between mouse and zebrafish43. Baf60c stabilizes the interactions between activated Notch and its DNA-binding partner RBP-J to control Nodal expression and thus establishes the left-right asymmetry of embryos and the heart. Also, Baf60c interacts with transcription factors Tbx5, Nkx2.5, and Gata4 to activate cardiac-specific genes for cardiogenesis42. Ectopic expression of Baf60c, Tbx5, and Gata4 in non-cardiogenic mesodermal cells induces these cells to differentiate into contracting cardiomyocytes that express an early cardiac marker Actc144. Therefore, Baf60c is required for early cardiogenesis and subsequent heart morphogenesis.
BAF45c
BAF45c/DPF3 is a subunit of the BAF complex and contains two PHD (plant homeo domain) domains that recognize acetylated and methylated lysine residues of histone H3 and H445, 46. BAF45c/DPF3 is upregulated in the right ventricular myocardium of patients with Tetralogy of Fallot47, and it is expressed by the developing heart and somites in mouse, chicken, and zebrafish embryos45. Morpholino knockdown of dpf3 in zebrafish results in abnormal cardiac looping, poorly defined atrioventricular boundary, reduced cardiac contractility, and disarrayed muscle fibers45. In muscle gene regulation, DPF3 and BRG1 highly overlap in their binding to 21 muscle-relevant gene loci that contain acetylated or methylated H3 and H445, suggesting the DPF3 may recognize those modified histones and help recruit BRG1 there. Indeed, the tandem PHD domains of DPF3- PHD1 and PHD2- act as one functional unit in the sequence-specific recognition of lysine-14-acetylated histone H3 (H3K14ac)46. The binding of DPF3 to histone H3 is promoted by acetylated lysine 14 (H3K14ac) recognized by PHD1 and inhibited by methylated lysine 4 (H3K4me3) recognized by PHD246. This regulation of PHD binding by different histone modifications produces two opposing effects on the transcriptional activation of Dpf3 target genes, such as Pitx2 and Jmjd1c46. Studies support a model in which H3K14 acetylation marks a gene locus for recruiting DPF3/BAF to pre-initiate gene transcription, whereas H3K4 methylation dissociates DPF3/BAF from the locus, thus allowing the entry of transcriptional machinery to initiate and activate gene transcription46. Therefore, the histone H3 interaction modulated by different site-specific modifications may be crucial for DPF3/BAF45c to regulate gene transcription during heart development. Further studies are needed to determine how DPF3/BAF45c recognizes acetylated histone H4 and how such recognition affects interactions with H3 and gene transcription during heart development.
Implication of BAF in human congenital diseases
BRG1/BRM and BAF proteins have been indirectly implicated in the pathogenesis of CHARGE syndrome48 and Williams syndrome49. CHARGE syndrome will be described below in the section on CHD chromatin remodelers. Williams syndrome is an autosomal dominant disorder characterized by mental and growth deficiency, dysmorphic facial features, aberrant vitamin D metabolism, and cardiovascular abnormalities. Cardiovascular defects linked to the syndrome include supravalvular aortic stenosis, pulmonary arterial stenosis, aortic coarctation, bicuspid aortic valve, and atrioventricular septal defect.
The Williams syndrome transcription factor (WSTF) is among the 28 genes lost by heterozygous deletion of 7q 11.23 in Williams syndrome50. Animal studies suggest that WSTF deficiency may account for some of the cardiac defects observed in Williams syndrome patients51. Wstf-null mice show perinatal lethality with hypo-trabeculation, thin myocardium, atrial septal defect, and ventricular septal defects51. The fourth pharyngeal arch arteries appear hypoplastic, resulting in mild coarctation of aorta between the left common carotid and subclavian arteries. These cardiovascular malformations are also present in Wstf heterozygous mice at a lower frequency, suggesting a partial haploinsufficiency of Wstf and therefore a possible role of WSTF in the pathogenesis of Williams syndrome.
WSTF proteins are present in two different types of chromatin-remodeling complexes: the BRG1/BRM-containing WINAC complex49 and the SNF2H-containing WICH complex 52, 53. In E9.5 mouse hearts, Wstf recruits Brg1, Baf155, and cardiac transcription factors Nkx2.5, Gata4, and Tbx5 to the promoter of Connexin 40 (Cx40 or Gja5)51. Transcriptional studies show that Wstf interacts with Brg1, but not Snf2h, to support the activation of Cx40 promoter51. These ChIP and reporter assays suggest that Wstf regulates heart development through Brg1/WINAC, but not Snf2h/WICH51.
However, the role of SNF2h/WICH in heart development cannot be excluded. WINAC alone does not fully explain the phenotypes of Wstf-null mice. These mice do not have the same cardiovascular defects observed in mice lacking either components of WINAC or its associated transcription factors10, 17, 38, 41, 54–56. Further investigations are necessary to determine the relative contributions of BRG1/WINAC and SNF2h/WICH to the phenotypes of Williams syndrome.
ISWI complex in heart development and disease
Three ISWI complexes, NURF, ACF and CHRAC, were purified from the fruit fly D. melanogaster with subsequent identification of NoRC and WICH in mammals (see ref 4, 57, 58 for reviews). The fly complex has only one single type of core ATPase, while the mammalian complex has two, SNF2H and SNF2L, which are present in different complexes and have non-overlapping expression patterns in mice57. The role of Snf2h in heart development is not defined because of an early lethality of Snf2h-null mouse embryos, which die between E5.5 and E7.5 due to growth failure of inner cell mass and trophoblast59. However, Snf2h may play a role in heart development because it is present in the WICH complex that contains Wstf, whose mutation causes myocardial and septal defects in mice51 (details described above). A tissue-specific deletion of Snf2h in mice will help elucidating the function of Snf2h in cardiovascular development.
CHD in heart development and disease
The CHD family of chromatin remodelers has nine members, CHD1 to CHD94. Haploinsufficiency of CHD7 results in CHARGE syndrome, characterized by coloboma, heart defects, atresia choanae, retarded growth and development, genital hypoplasia and ear abnormalities/deafness60. Patients with CHARGE syndrome may have a variety of heart defects, including conotruncal malformations, atrial/ventricular septal defects, and endocardial cushion anomalies. Chd7+/− mice recapitulate several aspects of the human disease, such as inner-ear vestibular dysfunction61 and heart defects62. These Chd7+/− mice display aortic arch interruption because of hypoplastic pharyngeal arch arteries62. Formation of these arch arteries requires Chd7 and Tbx1 in the pharyngeal ectoderm. Analyses of Tbx1+/−;Chd7+/− double heterozygous mice suggest that Chd7 and Tbx1 synergistically regulate the development of ear, thymus, and the fourth pharyngeal arch.
CHD7 is essential for the development of multipotent migratory neural crest cells, which contribute to the formation of many tissues affected in CHARGE syndrome48. In frog embryos, knockdown of Chd7 or overexpression of dominant-negative form of Chd7 recapitulates major features of CHARGE syndrome: otolith defects, coloboma, craniofacial malformation, and heart defects. The heart defects involve abnormal positioning of the truncus arteriosus and cardiac outflow tract. All these anomalies are consistent with neural crest cell defects. Indeed, Chd7 is required in the frog for the expression of Sox9, Twist and Slug (Snail2), genes essential for early neural crest cell migration and specification. Studies using frog embryos and human neural crest-like cells further suggest that CHD7 associates with BRG1/PBAF to control Sox9, Twist, Slug and neural crest cell activation48. Therefore, CHD7 and BRG1/PBAF interactions may underlie the pathogenesis of CHARGE syndrome.
INO80 in heart development
The INO80 complexes, conserved from the yeast to vertebrate, include the INO80, SRCAP and p400 in humans (see ref 1, 4, 63 for reviews). These INO80 complexes contain DNA helicases: Pontin (alias Rvb1, Tip49, Tip49a) and Reptin (alias Rvb2, Tip48, Tip49b)64. The absence of Pontin or Reptin activity causes disruption of chromatin remodeling activity of the INO80 complexes, suggesting essential roles of those two proteins in chromatin and transcriptional regulation64–67. Interestingly, Pontin and Reptin are known to regulate myocardial growth68. Morpholino knockdown of pontin in zebrafish causes cardiac hyperplasia, while an insertional mutation of reptin that activates the ATPase activity of reptin stimulates cardiomyocyte proliferation in zebrafish68. Studies suggest that Pontin and Reptin antagonistically regulate heart muscle growth, at least in part, through their opposing effects on the β-catenin pathway68. These functions of Pontin and Reptin suggest the involvement of INO80 complexes in heart development. Unfortunately, studies of INO80 subunits in mouse heart development are not yet available. It remains to be investigated how INO80 regulates mammalian heart development.
The Histone Deacetylases
Histone deacetylases (HDACs) are a class of chromatin remodeling factors that usually repress gene expression (see ref 3, 69 for reviews). These enzymes remove acetyl group from conserved lysine residues within the N-terminal tails of histone H3 and H4, thereby increasing histone-histone and histone-DNA contacts and causing condensation of chromatin. Histone deacetylation by HDACs is countered by a class of antagonistic enzymes, histone acetyltransferases (HATs), which add acetyl group to the lysine residues. Thus, dynamic balance in the state of histone acetylation and deacetylation provides flexibility and reversibility in gene expression control.
HDACs are known to function in a genome-wide manner and contribute to an extensive range of biological functions from development to physiology3, 69. The mammalian genome encodes 11 HDAC proteins that contain highly conserved deacetylase domain. The HDACs are classified into four subfamilies (class I, IIa, IIb and IV) based on their protein structure, enzymatic activity, subcellular localization and expression pattern. In addition, the mammalian genome encodes another group of deacetylases (class III HDACs) or sirtuins, which contribute to heterochromatinization though their NAD+-dependent histone deacetylase activity70. Class IV HDAC has only one known member, HDAC1171, whose function in the heart remains elusive. Among HDAC proteins, class I and IIa are the most extensively studied and regarded as classic HDACs. Class I HDACs display high enzymatic activity toward histone substrates, while class IIa HDACs possess only weak enzymatic activity72, 73 due to a tyrosine-to-histidine change in the catalytic pocket of these class IIa proteins74, 75. The intrinsic catalytic activity is not required for class IIa HDACs to repress gene expression. Gene repression by class IIa HDACs is mediated by their interactions with class I HDACs and other transcriptional repressors such as nuclear receptor corepressor (N-CoR) and silencing mediator of retinoic acid and thyroid hormone receptors (SMRT), heterochromatin protein 1 (HP-1), and c-terminal-binding protein (CtBP)72, 76–79. Recent studies of HDAC knockout mice have demonstrated critical functions of HDACs in cardiovascular development and disease (Table 2).
Table 2.
Roles of HDACs in cardiovascular development and disease
| Class | Chromatin Modifying Factor |
Cardiovascular Studies in Animals | Comment | Ref | |
|---|---|---|---|---|---|
| Tissue of Gene Modification |
Phenotype | ||||
| Class I HDAC | HDAC1 | Germline deletion | Lethality before E9.5. General growth retardation. | Hdac1 suppresses Cdk* inhibitor p21 and p27 to promote cell proliferation. | 81 |
| Deletion in myocardium | No apparent cardiac defects. | Hdac1 functions redundantly with Hdac2 in the myocardium. | 81 | ||
| HDAC2 | Germline deletion | Lethality at birth. Thickened myocardium, thickened IVS, hypotrabeculation. | Hdac2-Hop inhibits cardiomyocyte proliferation by suppressing SRF- and Gata4- dependent gene expression. | 31, 81 | |
| Surviving adult mice are resistant to hypertrophy. | Hdac2 suppresses Inppf5, which inhibits the pro-hypertrophic Akt/Gsk3β pathway. | 31, 93 | |||
| Overexpression in myocardium | Cardiac hypertrophy. | Hdac2 overexpression activates the Akt/Gsk3β pathway. | 31 | ||
| Deletion in myocardium | No apparent cardiac defects. | Hdac2 functions redundantly with Hdac1 in the myocardium. | 81 | ||
| Deletion of Hdac1 and Hdac2 in myocardium | Lethality within 2 weeks after birth. Arrhythmias, dilated cardiomyopathy. | Hdac1 and Hdac2 interact with REST to repress fetal genes involved in calcium handling and contractility. | 81 | ||
| HDAC3 | Germline deletion | Lethality at E9.5 due to gastrulation defects. | Hdac3 is required for gastrulation. | 95 | |
| Overexpression | Cardiac hyperplasia without hypertrophy. | Hdac3 suppresses several Cdk* inhibitors to promote cardiomyocyte proliferation. | 94 | ||
| Deletion in myocardium | Lethality at 3–4 months after birth. Cardiac hypertrophy, fibrosis, abnormal fatty acid metabolism, lipid accumulation in heart muscle. | Hdac3 suppresses PPARα activity on the promoters of genes involved in metabolic regulation. | 95 | ||
| Class II HDAC | HDAC5 and HDAC9 | Germline deletion | Single Hdac5- or Hdac9-null mutation causes no apparent cardiac phenotype. Double Hdac5 and Hdac9 mutations cause lethality starting at E15.5, frequent VSD, and occasional thin myocardium. | Hdac5/9 may suppress the transcriptional activity of Mef2, SRF, or myocardin involved in cardiac growth regulation. | 97 |
| Hdac5- or Hdac9-null mice show enhanced hypertrophic response to cardiac stress, and display protection of female hearts from ischemia injury. | Hdac5/9 suppresses the transcriptional activity of pro-hypertrophic Mef2 and CAMTA2. Hdac5/9 suppresses the Mef2-ERα-Vegfa pathway that promotes angiogenesis in ischemic hearts. | 97, 98, 99, 104 | |||
| HDAC7 | Germline deletion | Lethality at E11.5, severe hemorrhage from leaky and dilated blood vessels. | Hdac7 suppresses Mmp10 in the endothelium to maintain the integrity of vessel wall. | 96 | |
| Class III HDAC | SIRT1 | Germline deletion | Lethality at birth, ASD*, VSD*, heart valve defects. | Sirt1 deacetylates p53, preventing p53 from triggering cell apoptosis after DNA damage and stress. | 110, 111, 113 |
| Overexpression in myocardium | Low/moderate Sirt1 overexpression reduces cardiac hypertrophy and apoptosis. High Sirt1 overexpression triggers cardiac hypertrophy and apoptosis. | Sirt1 expression is activated by cardiac stress, and it regulates the stress response in a dose-dependent manner. | 115 | ||
| SIRT3 | Germline deletion | Cardiac hypertrophy and interstitial fibrosis at 2 months of age. | Sirt3 inhibits apoptosis through activating Ku70-Bax interaction and inhibiting Bax’s pro-apoptotic activity. | 118, 120 | |
| Overexpression in myocardium | Resistant to stress-induced cardiac hypertrophy. | Sirt3 activates Foxo3a-dependent antioxidant genes and attenuates the pro-hypertrophic RAS, MAPK/ERK and PI3K/Akt pathways. | 120 | ||
| SIRT7 | Germline deletion | Shortened lifespan, extensive cardiac fibrosis, hypertrophy, and inflammatory cardiomyopathy | Sirt7 deacetylates p53 and protects cardiomyocytes from stress-induced apoptosis. | 122 | |
Cdk: cycline-depedent kinase. ASD: atrial septal defect. VSD: ventricular septal defect.
Class I HDACs in heart development and disease
Class I HDAC proteins, consisting of HDAC1, 2, 3, and 8, are expressed ubiquitously and localized predominantly to the nucleus of the cell. HDAC1 and HDAC2 are generally found together in repressive complexes such as Swi-independent 3 (Sin3), nucleosome remodeling and deacetylase (NuRD), corepressor of RE1-silencing transcription factor (CoREST), and polycomb repressive complex 2 (PRC2) complexes3, 69, 80. HDAC3, on the other hand, associates with two homologous transcriptional corepressors, N-CoR and SMRT, which stimulate its deacetylase activity76. No complex has been described for HDAC869.
HDAC1
Hdac1-null mice die in utero before E9.5 with general growth retardation81. These embryos exhibit reduced cell proliferation and increased expression of cyclin-dependent kinase inhibitor, including p21 and p27. The p21 and p27 promoters of the Hdac1-null embryonic stem cells show modest hyperacetylation of histone 3 and 4, suggesting that Hdac1 may normally deacetylate histones on these promoters to repress p21 and p27 expression, thereby promoting cell proliferation. However, tissue-specific deletion of Hdac1 in the myocardium of mice results in no apparent phenotype81, suggesting that Hdac1 may function redundantly with other Hdac proteins, such as Hdac2, to regulate heart development.
HDAC2
Hdac2 is expressed in the hearts of mouse embryos31. Germline deletion of Hdac2 in mice results in a variety of cardiac defects, and these Hdac2-null mice die within the first 24 hours after birth81. In contrast, another line of Hdac2-deficient mice, generated from a gene-trap embryonic stem cell line, exhibits partial perinatal lethality; the surviving mutant mice, when examined at 2 months of age, appear grossly normal31. Although the exact cause of their difference is not clear, both mouse lines provide insights into the roles of Hdac2 in heart development and disease. Cardiac malformations in these mice include occlusion of the right ventricle81, thickened interventricular septum81, thickened compact myocardium31, and reduced ventricular trabeculation31. These defects are possibly caused by a combination of regional increase of cardiomyocyte proliferation and apoptosis31, 81. Hdac2-mediated cardiomyocyte growth may require the presence of a homeodomain-only protein Hop, a regulator of cardiomyocyte proliferation82, 83. Disruption of Hop in mice results in hyperproliferation of the developing cardiomyocytes82, 83. Hop associates with Hdac2 to inhibit the serum response factor (SRF)-dependent gene expression involved in cardiomyocyte proliferation and differentiation82–84. Also, this Hop-Hdac2 complex deacetylates Gata4, reducing Gata4 transcriptional activity, and downregulates Gata4-controlled cell cycle genes, thereby inhibiting cardiomyocyte proliferation85. Mice with double null mutations of Hop and Hdac2 double mutations exhibit perinatal lethality with hyperacetylation of Gata4, upregulation of Gata4 target genes, and a marked increase in cardiomyocyte proliferation85. These studies support that Hdac2 interacts with Hop to suppress cardiomyocyte growth.
Within the myocardium, Hdac2 functions redundantly with Hdac1 to regulate cardiac gene expression and cardiomyocyte differentiation81. Mice lacking either Hdac1 or Hdac2 in the myocardium have no apparent cardiac defects and survive to adulthood81. However, mice lacking both Hdac genes in the myocardium die within 2 weeks after birth due to cardiac arrhythmias and dilated cardiomyopathy81. These abnormalities are accompanied by an upregulation of genes that encode fetal calcium channels and skeletal muscle-specific contractile proteins81. These fetal genes, including hyperpolarization activated non-selective cation current (If), the T-type Ca2+ current (ICa,T), and α-skeletal actin (α-SA), are transcriptionally repressed in the normal adult myocardium by the RE1-silencing transcription factor (REST, also known as the neuron-restrictive silencer factor, NRSF) through the recruitment of class I and class IIa HDACs86–88. Similar to mice lacking myocardial Hdac1 and Hdac2, mice that overexpress a dominant-negative form of REST develop dilated cardiomyopathy, ventricular arrhythmias, and sudden cardiac death86. Therefore, the combined loss of Hdac1 and Hdac2 may result in the failure of REST to repress fetal genes involved in calcium handling and contractility, thereby causing cardiac arrhythmias and cardiomyopathy.
Those Hdac2-deficient mice that survive to adulthood without cardiomyopathy31 are resistant to hypertrophic stimulation31, 84. Conversely, mice overexpressing Hdac2 display augmented cardiac hypertrophy31. In the mouse heart, Hdac2 suppresses the expression of inositol polyphosphate-5-phosphatase f (Inpp5f), a negative regulator of the pro-hypertrophic PI3 kinase-Akt-Gsk3β pathway31. By reducing phosphorylation of the Akt kinase and glycogen synthase kinase 3β (Gsk3β)31, Inpp5f activates Gsk3β to suppress several pro-hypertrophic pathways31, 89–92, thereby inhibiting cardiac hypertrophy. Without Inpp5f, mice are susceptible to stress-induced cardiac hypertrophy93. Therefore, the resistance of Hdac2-deficient mice to hypertrophy can be explained by an increase of Inpp5f and Gsk3β activity in these mice. Furthermore, unlike Hdac2, overexpression of Hdac1 or Hdac3 in mice do not cause cardiac hypertrophy31, 94, suggesting that among class I proteins, Hdac2 is the major Hdac that modulates Gsk3β activity to regulate cardiac hypertrophy.
HDAC3
Hdac3 overexpression in cardiomyocytes produces cardiac hyperplasia without hypertrophy, resulting in thickening of ventricular myocardium and obstruction of ventricular cavities in newborn mice94. Hdac3 suppresses the expression of several cyclin-dependent kinase (Cdk) inhibitors, such as Cdkn1 and Cdkn2, to promote cardiomyocyte proliferation. Therefore, Hdac3 is a regulator of cardiomyocyte growth during development.
Germline deletion of Hdac3 in mice results in embryonic lethality by E9.5 due to defects in gastrulation95. In contrast, mice with cardiomyocyte-restricted deletion of Hdac3 survive until 3–4 months of age, at which point they develop massive cardiac hypertrophy and fibrosis95. These pathological changes are accompanied by an upregulation of genes that control fatty acid metabolism and by a downregulation of genes that govern glucose utilization. The gene changes result in lipid accumulation in the heart muscle95. ChIP analyses of rat myocytes indicate that Hdac3 and the nuclear hormone receptor PPARα form a protein complex on the promoters of many genes involved in metabolic regulation95. The absence of Hdac3 counterbalance in Hdac3-null hearts may thus cause an excessive PPARα activity on those gene promoters, leading to metabolic derangements and lipid accumulation in the heart. Interestingly, mice with deletion of Hdac1, Hdac2, or other Hdac genes81, 95 do not display such metabolic abnormalities, indicating an unique role of Hdac3 in myocardial energy metabolism.
Class II HDACs in heart development and disease
Class II HDACs are subdivided into class IIa (HDAC4, 5, 7 and 9) and class IIb (HDAC6 and 10) proteins3, 69. Class IIa HDACs contain large N-terminal domains with conserved binding sites for Mef2 (transcription factor) and 14-3-3 (chaperone protein). While most class I HDACs are subunits of multiprotein nuclear complexes crucial for gene repression and epigenetic landscaping, class IIa HDACs regulate cytoplasmic processes or transduce signals between the cytoplasm and the nucleus. Furthermore, unlike class I HDACs, class II HDACs have relatively restricted pattern of expression69. HDAC5 and HDAC9 are highly expressed in muscle, brain and heart. HDAC7 is present in thymocytes and endothelial cells.
HDAC7
Germline deletion of Hdac7 in mice causes embryonic lethality at E11.5, and embryos die from severe hemorrhage due to vascular defects96. The dorsal aortae and cardinal veins of Hdac7-null mice are dilated, leaky and surrounded by fewer smooth muscle cells. Hdac7 normally interacts with Mef2 in the endothelium to repress Mmp10 expression96. When Hdac7 is absent, Mmp10 is de-repressed in the endothelium, releasing this proteinase to damage the blood vessel wall. Concurrent with Mmp10 upregulation, the tissue inhibitor of metalloproteinase 1 (TIMP1) is downregulated in Hdac7-null endothelial cells, further enhancing Mmp10 activity and exacerbating vascular destruction. Therefore, Hdac7 is essential for maintaining the integrity of vessel wall during development.
HDAC5 and HDAC9
Hdac5 and Hdac9 are functionally redundant during heart development. Single Hdac5 or Hdac9 knockout mice survive to adulthood without apparent cardiac defects97. In contrast, double Hdac5 and Hdac9 mutations cause lethality starting at E15.5 with multifocal hemorrhages in embryos. Most of the double mutants exhibit ventricular septal defects; some have thin myocardium. Only few double mutants mice survive to postnatal day 7 or adulthood. These cardiac defects may result from the removal of Hdac’s suppression of Mef2, SRF, myocardin, or CAMTA298 transcriptional activity that regulates cardiac growth.
Hdac5- and Hdac9-null mice display enhanced hypertrophic response to calcineurin activation and pressure overload97, 99. Cardiac stress triggers the phosphorylation of class IIa HDACs by calcium/calmodulin-dependent protein kinase (CaMK) and protein kinase D (PKD). Phosphorylated HDACs bind 14-3-3 and translocate from the nucleus to the cytoplasm100–102. This process dissociates HDACs from Mef2c proteins99, 100, allowing p300 to bind and convert Mef2c to a transcriptional activator69, 103 and thus enabling Mef2c to activate pro-hypertrophic genes. Furthermore, the absence of nuclear HDACs removes the suppression of calmodulin binding transcription activator 2 (CAMTA2), whose hyperactivity triggers cardiac hypertrophy98. Therefore, by removing the suppression of Mef2 and CAMTA2, the loss of Hdac5 or Hdac9 augments the stress-induced cardiac hypertrophy.
Deletion of Hdac5 or Hdac9 reduces the maladaptive remodeling after myocardial infarction in female mice104. This protective effect is attributed in part to an enhanced expression of vascular endothelial growth factor-a (Vegf-a), which promotes neo-angiogenesis with consequent reduction of myocardial infarction. Hdac proteins normally suppress the transcriptional activity of Mef2 and estrogen receptor α (ERα). The loss of Hdac5 or Hdac9 triggers Mef2 to activate its target gene ERα In the presence of estrogen (E2), ERα then activates Vegf-a expression. Therefore, this ERα-dependent transcriptional cascade, released by the loss of Hdac5 or Hdac9, may account for the gender-specific cardiac protection.
HDAC4, HDAC6, and HDAC10
HDAC4 seems unnecessary for cardiovascular development. HDAC4 controls chondrocyte hypertrophy through interacting with Runx2 (Runt related transcription factor 2) and Mef2c during the formation of the skeleton105. No cardiac abnormalities have been reported. HDAC6 also seems dispensable for cardiovascular development as Hdac6-null animals develop normally and grow to adulthood, although with some minor immunological response differences106. Little is known about the functions of HDAC10.
Class III HDACs in heart development and disease
Members of the class III HDACs are homologous to the yeast transcriptional repressor Sir2p and have no sequence homology to class I and II HDACs. Class III HDACs, also called sirtuins, require NAD+ for deacetylation. To date, seven mammalian homologues have been identified, SIRT1–770. Mammalian sirtuins have diverse cellular locations: SIRT1 and SIRT2 are present in both the nucleus and the cytoplasm, while SIRT3, SIRT4, and SIRT5 are localized in the mitochondria, and SIRT6 and SIRT7 are nuclear proteins107–109. A key function of the class III HDACs is their regulation of transcriptional repression70.
SIRT1
Sirt1 knockout mice, when generated in an inbred genetic background, die perinatally with neurological and cardiac malformations110, 111, which include atrial septal, ventricular septal, and heart valve defects. However, in outbred backgrounds, Sirt1 deletion produces viable mice with diverse phenotypes, such as imperfect gametogenesis and autoimmune disease111, 112. Sirt1 is known to deacetylate p53, preventing p53 from triggering cellular senescence and apoptosis in response to DNA damage and stress110, 113. Sirt1-deficient cells exhibit hyperacetylation of p53 and increased cell death after DNA damage110. Furthermore, Sirt1 induction by silibinin protects rat neonatal cardiomyocytes from isoproterenol-induced injury and cell death114. In adult mouse hearts, Sirt1 increases by 4- to 8-fold in response to pressure overload and oxidative stress115. Cardiomyocyte-specific overexpression of Sirt1 in mice shows that a low-to-moderate increase of Sirt1 (2.5-fold to 7.5-fold) protects mice from age-dependent cardiac hypertrophy, apoptosis, and fibrosis115. In contrast, a higher level of Sirt1 (about 13-fold) induces cardiac hypertrophy and apoptosis, ultimately leading to cardiomyopathy115. These studies suggest a narrow window of optimal Sirt1 activity in the regulation of cardiac responses to stress.
SIRT2
Sirt2 is a negative regulator of stress tolerance; knockdown or inhibition of Sirt2 enhances the survival of cells under stress116, 117. Sirt2 knockdown reduces anoxia-reoxygenation injury in H9c2 rat cardiac cells117. Its knockdown induces the expression of a chaperone protein 14-3-3 ζ, which sequesters the pro-apoptotic Bad (Bcl2 antagonist of cell death) protein in the cytoplasm and thus reduces mitochondrial Bad. This prevents Bad proteins from inhibiting the anti-apoptotic activities of Bcl-XL or Bcl-2 in the mitochondria. Therefore, Sirt2 may control 14-3-3 ζ and Bad to regulate cardiac stress response.
SIRT3
Sirt3 is a stress-responsive deacetylase in cardiomyocytes118. In cultured cardiomyocytes, Sirt3 is induced by genotoxic or oxidative stress, and it protects myocytes from cell death caused by such stress118. Induced Sirt3 deacetylates Ku70 and enhances Ku70’s binding to the pro-apoptotic Bax (Bcl2-associated X protein), thereby preventing the entry of Bax proteins into the mitochondria to trigger apoptosis118. Although Sirt3-deficient mice are born grossly normal119, they develop cardiac hypertrophy and interstitial fibrosis at 8 weeks of age120. In addition, when exposed to hypertrophic stimuli, these Sirt3-deficient mice develop severe cardiac hypertrophy; whereas mice overexpressing Sirt3 in the myocardium are resistant to hypertrophy from similar stimuli120. Therefore, Sirt3 possibly protects cardiomyocytes from stress-induced damage by activating the Foxo3a-dependent expression of antioxidant genes and by attenuating the RAS, MAPK/ERK and PI3K/Akt pathways120.
SIRT4, SIRT5, and SIRT6
Sirt4 and Sirt5-deficient mice are born at a Mendelian ratio and appear grossly healthy119. No cardiac defects have been reported. Sirt6-deficient mice appear runted with lymphopenia, loss of subcutaneous fat, lordokyphosis, and severe metabolic defects121. These mice die at about 4 weeks, but no cardiac defects have been reported121.
SIRT7
Sirt7 deletion in mice leads to various pathological changes in adult hearts122. Sirt7-deficient animals have shortened life span and develop cardiac hypertrophy, extensive cardiac fibrosis and inflammatory cardiomyopathy122. Sirt7 interacts with p53 and efficiently deacetylates p53 in neonatal rat cardiomyocytes, and Sirt7-null hearts exhibit p53 hyperacetylation with increased cell apoptosis. Also, Sirt7-deficient primary cardiomyocytes show increased apoptosis after oxidative or genotoxic stress. These results suggest a critical role of Sirt7 in the regulation of cardiac stress responses and cell death.
The Histone Acetyltransferases
Actions by histone deacetylases are counteracted by histone acetyltransferases (HATs), which add acetyl groups to histone tails, resulting in decrease of histone-histone and histone-DNA interaction, thereby loosening the chromatin and increasing transcriptional activation. Both HDACs and HATs are known to regulate heart development and cardiac hypertrophy, suggesting that the balance of acetylation and deacetylation of histones is essential for the heart to respond to developmental and hypertrophic cues (Table 3).
Table 3.
Roles of HAT, histone methylation, and PARP in cardiovascular development and disease
| Class | Chromatin Modifying Factor |
Cardiovascular Studies in Animals | Comment | Ref | |
|---|---|---|---|---|---|
| Tissue of Gene Modification | Phenotype | ||||
| HAT | p300 | Germline deletion | Lethality at E9.5–E11.5. Thin myocardium and diminished trabeculation. | p300-null embryos have generalized reduction in cell proliferation. | 123 |
| Point mutation that ablates p300 acetyltransferase activity | Lethality at E12.5–E15.5. Thin compact myocardium, ASD*, VSD* reduced coronary vessels. | The acetyltransferase activity of p300 is required for heart development. | 124 | ||
| Overexpression in myocardium | Cardiac hypertrophy, dilatation and systolic dysfunction. | p300 is a co-activator of the pro-hypertrophic Gata4. It acetylates Gata4 to enhance Gata4’s DNA binding and transcriptional activity. | 125, 127 | ||
| Histone Methylation | JUMONJI | Germline deletion | Variable defects: VSD, non-compaction, double outlets of right ventricle, trabecular hyperplasia. | Jumonji represses cyclin D1 to inhibit myocardial proliferation. | 132, 133 |
| SMYD1 | Germline deletion | Malformed right ventricle with little trabeculation. | Smyd1 regulates Hand2, which is essential for right ventricle development. | 146, 147 | |
| Knockdown in zebrafish | Disrupted myofibril formation, absent cardiac contraction. | Smyd1 is required for myofibril organization. | 148 | ||
| WHSC1 | Germline deletion | Lethality within 10 days after birth, growth retardation, defective bone formation, ASD, VSD. | Whsc1 associates with Nkx2.5 to repress gene transcription and to regulate cardiac septal formation. | 154 | |
| PARP | PARP-1 and PARP-2 | Germline deletion | Parp-1-null and Parp-2-null mice are viable. Parp-1 and Parp-2 double knockout mice die at E7–E8. | PARP activity is required for Brg1 to repress α-MHC and activate β-MHC in embryonic hearts. | 17, 166 |
| Parp-1-null mutation or inhibition of PARP activity in mice reduces stress-induced cardiac hypertrophy, heart failure, ischemia-reperfusion injury, and infarction. | PARP-1 interacts with HDACs and Brg1 to control cardiac hypertrophy and the pathological MHC switch in hypertrophic hearts. PARP also regulates stress-responsive signaling pathways. | 17, 28, 33, 167–174 | |||
ASD: atrial septal defect. VSD: ventricular septal defect.
p300
One well-defined member of the HATs, p300, is required for heart muscle development and hypertrophy. p300 is broadly expressed by the developing embryo by E7.5, and p300-null embryos die between E9.5 to E11.5 with defects in neural and heart development123. Embryos lacking p300 display poorly vascularized yolk sac and severe pericardial effusion. The mutant heart has thin myocardium and diminished trabeculation at E10.5, and expresses lower levels of MHC and α-actinin. The thin-myocardium phenotype is possibly caused by diminished myocardial cell proliferation because p300-null embryos have generalized reduction in cell proliferation, and p300-null embryonic fibroblasts stop dividing after 3–4 cell generations123. Another mouse model of p300 mutation, in which a single amino acid mutation ablates acetyltransferase activity of p300, shows multiple defects in heart development, although the embryos die a little later between E12.5 and E15.5124. These embryos display a variety of cardiac malformations, including thinning of the compact myocardium, ventricular and atrial septal defects, and reduced coronary vessels124.
In rat neonatal cardiomyocytes, the expression of p300 is induced by phenylephrine, and its acetyltransferase activity is required for the phenylephrine-induced cardiac hypertrophy125, 126. p300 acts as a co-activator of the pro-hypertrophic transcription factor Gata4. It acetylates Gata4 and enhances Gata4’s DNA binding activity125, 126. Transgenic overexpression of p300 in the heart results in acetylation of Gata4 and increased expression of Gata4-dependent cardiac genes such as endothelin-1. These p300-overexpressing mice develop left ventricular (LV) hypertrophy, dilatation and systolic dysfunction125. They also have more severe progression of LV dilation and reduction of systolic function after myocardial infarction in adult mice127. In contrast, mice overexpressing p300 that lacks the acetyltransferase activity do not have such severe progression of LV dysfunction after infarction. Pharmacological inhibition of the acetyltransferase activity of p300/CBP by curcumin also prevents cardiac hypertrophy and LV deterioration in rodent models of hypertrophy and infarction128, 129. In addition, p300 occupancy correlates with the acetylation state of promoters of several cardiac genes that are differentially expressed between the left and right ventricles and also upregulated during hypertrophy, including BNP and ANF130. Therefore, the histone acetyltransferase activity of p300 is required for the activation of Gata4-dependent promoters, LV hypertrophy, and LV dilation, as well as adverse LV remodeling after myocardial infarction.
Histone Methylation and Heart Development
JUMONJI
Methylation of histones at arginine and lysine residues offers another avenue of epigenetic control. Jumonji (jmj, or Jarid2) belongs to a family of histone demethylases containing the conserved Jumonji C (JmjC) domain (see ref 131 for a review). Mice heterozygous of jmj mutation are normal, while homozygous knockouts have various degrees of lethality and embryonic defects according to the genetic background. In one strain, the jmj homozygous mutant animals survive to postnatal day 0 (P0), with ventricular septal defects, ventricular noncompaction, and double outlet of the right ventricle132. On the other hand, in the C3H/He background, embryos die between E11.5 and E12.5 and display defects in neurulation with abnormalities in the right heart and hyperplasia of the trabecular myocardium, resulting in obstruction of the ventricle133. The hyperproliferation is apparently due to overexpression of cyclin D1, which is normally repressed by Jmj134, 135. Although Jmj contains the JmjC domain, it lacks conserved residues for binding iron cofactors, suggesting that Jmj may not have demethylase activity131, 135. Instead, the Jmj complex has methyltransferase activity: it recruits G9a and GLP methyltransferases to methylate H3K9 and repress cyclin D1 promoter135. Also, Jmj recruits polycomb repressive complex (PRC2) to many target genes136–139, where PRC2 represses transcription by methylating histone 3 lysine 27 (H3K27) through the Ezh2 methyltransferase. Besides cyclin D1, Jmj is known to repress several other genes. Jmj associates with Gata4 and Nkx2.5 to repress Nppa (ANF)140, inhibits Mef2a activity to repress α-MHC141, and interacts with retinoblastoma (Rb) proteins to repress gene expression142. It is possible that Jmj also silences these genes by recruiting histone methyltransferases. These studies suggest that Jumonji may associate with other histone modifying factors to regulate gene expression essential for heart development.
SMYD
Smyd proteins belong to a family of histone methyltransferases (HMTs) that contain conserved SET domains necessary for methyltransferase activity143 and MYND domain required for transcriptional repression by interaction with HDAC144. At least two members of the Smyd family are studied in heart development. Smyd1 is expressed in adult cardiac and skeletal muscle145, as well as in embryonic hearts146. Targeted deletion of Smyd1 in mice yields embryonic lethality due to cardiac enlargement146. Smyd1-null animals have hypoplastic right ventricle with little trabeculation. Mutant cardiomyocytes downregulate Hand2, a gene essential for right ventricle development147. In zebrafish, morpholino knockdown of Symd1 produces defects in both cardiac and skeletal muscle function and structure148. In the absence of Smyd1, the fish fail to swim, and their hearts do not contract, consistent with disrupted myofibril formation of Symd1 knockdown myocytes. Another member of the family, Smyd2, is dispensable for normal heart development and function since Symd2 knockout mice have no observable cardiac abnormalities, suggesting redundancy among Smyd family members149.
WHSC1
Patients with Wolf-Hirschhorn Syndrome (WHS) display a range of defects, including mental retardation, epilepsy, craniofacial abnormalities, and cardiac septal defects150, 151. WHS patients have regional deletion of the short arm of chromosome 4 (4p16.3), termed Wolf-Hirschhorn Critical Region (WHSCR)152. Although the extent of deletion is different for many patients, all patients have a specific deletion of Wolf-Hirschhorn Syndrome Candidate (WHSC1) gene, which encodes a histone methyltransferase that tri-methylates lysine 36 of histone 3 (H3K36me3), among other histone targets153. Whsc1 is expressed throughout the embryonic heart except the endocardial cushion154. Most Whsc1 knockout mice die at birth, and none survives past 10 days154. These mice show growth retardation, incomplete bone formation, and defective heart development with atrial and ventricular septal defects. At a low frequency, heterozygous Whsc1 mice display certain features of WHS, such as growth and craniofacial defects, suggesting a partial haploinsufficiency of Whsc1. Interestingly, the Whsc1 haploinsufficiency is enhanced by a concomitant heterozygous mutation of Nkx2.5. One-third of the Whsc1+/−;Nkx2.5+/− double heterozygous mice exhibit atrial and ventricular septal defects, while single Whsc1+/− or Nkx2.5+/− mutants have minimal or no such lesions154. This indicates an interaction between Whsc1 and Nkx2.5 in cardiac septal formation. Indeed, Whsc1 associates with Nkx2.5 and occupies genetic loci to repress transcription of certain genes such as Pdgfra and Isl1, possibly through H3K36me3 modification154. These studies thus suggest that Whsc1 functions with developmental transcription factors to regulate heart development.
The Poly (ADP-ribose) Polymerases
The poly (ADP-ribose) polymerases (PARPs) catalyze the transfer of ADP-ribose units from NAD+ (nicotinamide adenine dinucleotide) to the carboxyl group of glutamic acid, aspartic acid or lysine residues of acceptor proteins (see ref 155 for a review). PARP-1, the most extensively studied of the 17-member PARP family, is a nuclear enzyme known to respond to DNA damage and facilitate DNA repair. Once activated, PARP-1 transfers 50–200 molecules of ADP-ribose to a variety of nuclear proteins, including PARP-1 itself and histone H1 and H2B tails156, 157. This poly ADP-ribosylation (PAR) results in the release of PARP-1 from DNA, modification of histones, and relaxation of the chromatin superstructure158–160. These local chromatin changes facilitate the recruitment and access of repair machinery to the site of DNA lesion. Besides DNA repair, PARP-1 modulates chromatin to control the transcriptional machinery in response to various stimuli. Signals, such as steroids or heat shock, induce PARP-1 activation and the PAR-dependent stripping of histones from chromatin, thereby favoring the opening of chromatin to allow transcriptional regulation160, 161. PARP-1 is present at transcriptionally repressed chromatin regions in the fruit fly161, and biochemical studies have shown that its activation and subsequent dissociation from the chromatin is a prerequisite for transcriptional activity to occur160.
PARP1 and PARP2
Although PARP activity is required for the maintenance of genome integrity162–165, Parp-1-null mice are viable without apparent cardiovascular defects; Parp-1 and Parp-2 double knockout mice die at early embryogenesis before E7–E8166 (Table 3). These phenotypes suggest redundant functions of Parp-1 and Parp-2 in early mouse embryogenesis166. A recent study shows that PARP activity is essential for maintaining fetal MHC expression in embryonic hearts17. Mouse embryos treated with a chemical inhibitor that blocks PARP activity exhibit increased α-MHC and reduced α-MHC expression. In embryos, Parp-1 associates with Brg1 and/or Hdac proteins on the cardiac MHC promoters to repress α-MHC and activate α-MHC expression17. This fetal mechanism of MHC control, although silenced in normal adult hearts, is reactivated by cardiac stress to trigger the pathological switch from adult α-MHC to fetal α-MHC, a crucial step in the myopathic process of stressed adult hearts17, 22.
PARP-1 is activated in cardiac hypertrophy, and its activity increases further in the failing hearts of mice and humans33, 167, 168. Null mutation of Parp-1 in mice or pharmacological inhibition of PARP activity reduces cardiac hypertrophy stimulated by angiotensin II33 or pressure overload167, 169, delays the progression from hypertensive cardiomyopathy to heart failure28, diminishes myocardial ischemia-reperfusion injury170, 171, and decreases cell death and heart failure after myocardial infarction172, 173.
Besides MHC regulation, PARP-1 contributes to cardiac hypertrophy and failure in a NAD+- and Sirt1-dependent manner33, 169. The PARP-mediated cell death in stressed hearts appears to be caspase 3-independent168, and caused at least in part by a depletion of NAD+169 and mitochondria-to-nucleus translocation of an apoptosis-inducing factor (AIF)167, 173, 174. The PARP-induced depletion of NAD+ and accumulation of nicotinamide reduce Sirt1 deacetylase activity169, 175, which may lead to hyperacetylation and activation of the pro-apoptotic factor p53110, 113, and cell death. Also, the nuclearly translocated AIF fragments the chromatin and condenses the nucleus, resulting in cell death. Experiments using chemical inhibitors of PARP suggest that PARP regulates several stress-responsive signaling pathways, including ERK1/2, p38 MAP kinase, JNK and PI3 kinase-Akt-Gsk3β pathways, to control cardiac responses to hypertrophic stimuli, ischemia-reperfusion, and infarction28, 172, 176, 177. Furthermore, through its interaction with TEF1, Brg1, and HDACs17, 178, 179, PARP-1 controls the expression of cardiac specific genes, including troponin T and MHC, thereby contributing to pathological gene expression in stressed hearts. Interestingly, under inflammatory stimuli, PARP-1 proteins are acetylated and deacetylated by p300/CBP and class-I HDACs180, respectively, raising the possibility that p300/CBP and HDACs may also contribute to the PARP-1-mediated cardiac hypertrophy and failure.
Discussion
Target Specificities of Chromatin Regulating Factors
This review summarizes the cardiovascular functions of BRG1/BAF, ISWI, CHD, INO80, HDAC, SIRT, HAT, HMT, and PARP. These families of chromatin regulators can form large protein complexes with one another to modify chromatin structure and interact with various transcription factors to control gene expression (Figure 2). For example, BRG1/BAF forms a complex with HDAC and PARP proteins to regulate MHC in both developing and hypertrophic hearts. BRG1 associates with CHD7 to regulate Twist and Slug in neural crest cells, essential for cardiac outflow tract development. Given the large families of BAF, HDAC, PARP, and CHD, a combinatorial assembly of epigenetic complexes may provide a unique protein composition that enables specific targeting of cardiac gene loci under different pathophysiological conditions. Furthermore, the stoichiometry of chromatin regulators may modulate transcriptional activities, adding another layer of gene expression control. Brg1 interacts dose-dependently with Nkx2.5, Tbx5 and Tbx20 to control heart development. The gene dosage of Chd7 and Tbx1 are critical during aortic arch formation. Whsc1 synergizes with Nkx2.5 to direct cardiac septal formation. Therefore, both the quantity and composition of chromatin regulating complexes are critical for cardiac gene expression.
How the genomic loci are marked and present themselves to chromatin regulators is not fully understood. Histone modifications may represent one means to mark the epigenetic landscape and form “histone codes” on the genome (see ref 181 for a review). A large number of enzymes can covalently add or remove chemical groups at specific residues of histone tails. These marks can act as beacons or docking sites recognized by proteins such as transcription factors or other chromatin remodelers, thereby introducing target gene specificity and additional regulatory control. Modification of specific residues of a histone tail can change histone-DNA contacts, modulate modification of other residues on the same or neighboring histone tail, or alter the binding affinity of other modified histone residues to chromatin remodelers46, 182, 183. These combined effects may create a specific epigenetic scene or histone code for chromatin remodelers to recognize. On the other hand, chromatin remodelers may change the conformation and position of nucleosomes to produce an epigenetic architecture for histone modifying enzymes to identify and modify specific histone residues. Further investigations are needed to fully define how chromatin regulators interact with each other and with transcription factors to target to specific genomic loci.
Chromatin Regulation and Extracellular Matrix Control
Besides cell-autonomously regulating cardiac growth and differentiation, chromatin regulators can indirectly control cardiovascular development by modulating the extracellular matrix environment. Within the blood vessel, endothelial Hdac7 suppresses a matrix metalloproteinase Mmp10 to maintain the matrix environment and integrity of subjacent vessel wall, thereby preventing aneurysmal dilation and leakage of developing vessels. In the heart, endocardial Brg1 represses Adamts1, which encodes a secreted metalloproteinase that degrades cardiac jelly matrix. This repression of Adamts1 allows the establishment of cardiac jelly that promotes myocardial trabeculation. Therefore, chromatin regulators can coordinate the development of adjacent cardiac tissues through extracellular matrix. Whether chromatin regulation can also modulate extracellular matrix in adult hearts to synchronize the responses of different cardiac tissues to pathological stress remains to be elucidated.
Non-chromatin Functions of Histone Modifying Enzymes
Histone modifiers may control cardiac pathophysiology through functions not directly related to their chromatin activities. For instance, by acetylating the transcription factor Gata4, p300 enhances DNA-binding affinity of Gata4 and promotes Gata4-dependent pro-hypertrophic cardiac gene expression. This aspect of p300’s contribution to cardiac hypertrophy does not require histone modification by p300. On the other hand, Hdac2 deacetylates Gata4 and downregulates Gata4-activated cell cycle genes, thereby inhibiting cardiomyocyte proliferation. This function of Hdac2 requires its interaction with Hop, but not its chromatin activities. Furthermore, Sirt1 and Sirt7 deacetylate p53 to suppress apoptosis and prevent cardiomyopathy. It remains to be determined how such non-chromatin functions interplay with chromatin activities of these histone regulators to control cardiac pathophysiology.
Chromatin Link between Fetal Heart Development and Adult Heart Disease
The epigenetic landscape of cardiomyocytes and other cell types of the heart undergoes profound changes as the heart goes through development, maturation, and disease. Chromatin regulation provides a mechanism underlying the transcriptional reprogramming and fetal gene activation in diseased hearts. Studies have suggested that many chromatin regulators are required to suppress fetal gene expression in postnatal hearts. For instance, Hdac1 and Hdac2 suppress fetal calcium channel and contractile protein gene expression as cardiomyocytes mature in neonatal mice. Loss of Hdac1 and Hdac2 causes an ectopic expression of those fetal genes, leading to severe cardiac arrhythmias and dilated cardiomyopathy. On the other hand, chromatin remodelers can be induced by cardiac stress and promote reactivation of fetal genes in an diseased adult heart. For example, stressed adult hearts reactivate the expression of a fetal gene Brg1, which then associates with its embryonic partners, HDAC and PARP, to form an epigenetic complex that triggers fetal MHC expression, cardiac hypertrophy, and fibrosis. Prevention of such stress-induced Brg1 activation reverses pathological MHC expression, diminishes hypertrophy, and abolishes cardiac fibrosis. These studies thus suggest a link at the chromatin level between fetal hearts and myopathic hearts. Therefore, targeting chromatin regulation may provide a promising approach to prevent and possibly reverse the myopathic process. To realize that goal, further studies are necessary to determine the dynamic changes of chromatin architecture, genomic specificities of chromatin regulators, and interactions among chromatin regulators under different pathophysiological conditions of the heart.
Acknowledgments
Sources of Funding
C.-P.C. was supported by funds from the National Institute of Health (NIH)(HL085345), American Heart Association (AHA), Baxter Foundation, Children’s Heart Foundation, March of Dimes Foundation, Office of the University of California (TRDRP), California Institute of Regenerative Medicine, Stanford Cardiovascular Institute, and Oak foundation. C.T.H was support by an NIH training fellowship (5T32 CA09302) and an AHA Predoctoral Fellowship.
Abbreviation
- ACF
ATP-utilizing Chromatin Assembly and Remodeling Factor
- BAF
Brahma-Associated Factor
- BRG1
Brahma-Related Gene1
- BRM
Brahma
- CHD
Cromodomain, Helicase, DNA binding
- CHRAC
Chromatin Accessibility Complex
- HAT
Histone Acetyltransferase
- HDAC
Histone Deacetylases
- HMT
Histone Methyltransferase
- INO80
Inositol requiring 80
- ISWI
Imitation Switch
- JMJ
Jumonji
- NoRC
Nucleolar Remodeling Complex
- NuRD
Nucleosome Remodeling and Deacetylase
- NURF
Nucleosome Remodeling Factor
- PAR
Poly ADP-ribosylation
- PARP
Poly (ADP-ribose) Polymerase
- REST/ NRSF
RE1-silencing Transcription Factor/ Neuron-Restrictive Silencer Factor
- CoREST
Corepressor of RE1-silencing Transcription Factor
- SIRT
Sirtuin
- SMYD1
SET, MYND domain containing 1
- SRCAP
SNF2-related CREB-activator Protein
- SWI/SNF
Switching defective/Sucrose Nonfermenting
- WICH
WSTF-ISWI Chromatin Remodeling Complex
- WINAC
WSTF Including Nucleosome Assembly Complex
- WHSCR
Wolf-Hirschhorn Critical Region
- WHSC1
Wolf-Hirschhorn Syndrome Candidate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
C.T.H. and P.H. contributed equally; the order of authorship does not reflect their relative contributions.
Disclosures
None.
References
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 3.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9(3):206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6(6):1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 6.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) Embo J. 1998;17(23):6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55(2):201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56(1):94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106(13):5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, Chang CP. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14(2):298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135(3):493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386(Pt 1):15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kern CB, Twal WO, Mjaatvedt CH, Fairey SE, Toole BP, Iruela-Arispe ML, Argraves WS. Proteolytic cleavage of versican during cardiac cushion morphogenesis. Dev Dyn. 2006;235(8):2238–2247. doi: 10.1002/dvdy.20838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wirrig EE, Snarr BS, Chintalapudi MR, O'Neal JL, Phelps AL, Barth JL, Fresco VM, Kern CB, Mjaatvedt CH, Toole BP, Hoffman S, Trusk TC, Argraves WS, Wessels A. Cartilage link protein 1 (Crtl1), an extracellular matrix component playing an important role in heart development. Dev Biol. 2007;310(2):291–303. doi: 10.1016/j.ydbio.2007.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kern CB, Norris RA, Thompson RP, Argraves WS, Fairey SE, Reyes L, Hoffman S, Markwald RR, Mjaatvedt CH. Versican proteolysis mediates myocardial regression during outflow tract development. Dev Dyn. 2007;236(3):671–683. doi: 10.1002/dvdy.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenni R, Rojas J, Oechslin E. Isolated noncompaction of the myocardium. N Engl J Med. 1999;340(12):966–967. doi: 10.1056/NEJM199903253401215. [DOI] [PubMed] [Google Scholar]
- 17.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, Chen Z, Yang Z, Schneider MD, Chien KR, Conway SJ, Yoder MC, Haneline LS, Franco D, Shou W. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131(9):2219–2231. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herron TJ, McDonald KS. Small amounts of alpha-myosin heavy chain isoform expression significantly increase power output of rat cardiac myocyte fragments. Circ Res. 2002;90(11):1150–1152. doi: 10.1161/01.res.0000022879.57270.11. [DOI] [PubMed] [Google Scholar]
- 20.Krenz M, Robbins J. Impact of beta-myosin heavy chain expression on cardiac function during stress. J Am Coll Cardiol. 2004;44(12):2390–2397. doi: 10.1016/j.jacc.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 21.James J, Martin L, Krenz M, Quatman C, Jones F, Klevitsky R, Gulick J, Robbins J. Forced expression of alpha-myosin heavy chain in the rabbit ventricle results in cardioprotection under cardiomyopathic conditions. Circulation. 2005;111(18):2339–2346. doi: 10.1161/01.CIR.0000164233.09448.B1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lompre AM, Schwartz K, d'Albis A, Lacombe G, Van Thiem N, Swynghedauw B. Myosin isoenzyme redistribution in chronic heart overload. Nature. 1979;282(5734):105–107. doi: 10.1038/282105a0. [DOI] [PubMed] [Google Scholar]
- 23.Swynghedauw B. Molecular mechanisms of myocardial remodeling. Physiol Rev. 1999;79(1):215–262. doi: 10.1152/physrev.1999.79.1.215. [DOI] [PubMed] [Google Scholar]
- 24.Miyata S, Minobe W, Bristow MR, Leinwand LA. Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ Res. 2000;86(4):386–390. doi: 10.1161/01.res.86.4.386. [DOI] [PubMed] [Google Scholar]
- 25.Abraham WT, Gilbert EM, Lowes BD, Minobe WA, Larrabee P, Roden RL, Dutcher D, Sederberg J, Lindenfeld JA, Wolfel EE, Shakar SF, Ferguson D, Volkman K, Linseman JV, Quaife RA, Robertson AD, Bristow MR. Coordinate changes in Myosin heavy chain isoform gene expression are selectively associated with alterations in dilated cardiomyopathy phenotype. Mol Med. 2002;8(11):750–760. [PMC free article] [PubMed] [Google Scholar]
- 26.Lowes BD, Gilbert EM, Abraham WT, Minobe WA, Larrabee P, Ferguson D, Wolfel EE, Lindenfeld J, Tsvetkova T, Robertson AD, Quaife RA, Bristow MR. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346(18):1357–1365. doi: 10.1056/NEJMoa012630. [DOI] [PubMed] [Google Scholar]
- 27.Blaxall BC, Tschannen-Moran BM, Milano CA, Koch WJ. Differential gene expression and genomic patient stratification following left ventricular assist device support. J Am Coll Cardiol. 2003;41(7):1096–1106. doi: 10.1016/s0735-1097(03)00043-3. [DOI] [PubMed] [Google Scholar]
- 28.Bartha E, Solti I, Kereskai L, Lantos J, Plozer E, Magyar K, Szabados E, Kalai T, Hideg K, Halmosi R, Sumegi B, Toth K. PARP inhibition delays transition of hypertensive cardiopathy to heart failure in spontaneously hypertensive rats. Cardiovasc Res. 2009;83(3):501–510. doi: 10.1093/cvr/cvp144. [DOI] [PubMed] [Google Scholar]
- 29.Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113(22):2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antos CL, McKinsey TA, Dreitz M, Hollingsworth LM, Zhang CL, Schreiber K, Rindt H, Gorczynski RJ, Olson EN. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278(31):28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 31.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13(3):324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 32.Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, Ahn Y, Jeong MH, Bang YJ, Kim N, Kim JK, Kim KK, Epstein JA, Kook H. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113(1):51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 33.Pillai JB, Gupta M, Rajamohan SB, Lang R, Raman J, Gupta MP. Poly(ADP-ribose) polymerase-1-deficient mice are protected from angiotensin II-induced cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 2006;291(4):H1545–H1553. doi: 10.1152/ajpheart.01124.2005. [DOI] [PubMed] [Google Scholar]
- 34.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287(1):134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 35.Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Koentgen F, Robb L, Feneley M, Harvey RP. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ Res. 2000;87(10):888–895. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
- 36.Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, Kupershmidt S, Roden DM, Schultheiss TM, O'Brien TX, Gourdie RG, Berul CI, Izumo S. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113(8):1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, Elliott DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132(10):2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang C, Skarnes WC, Tjian R. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18(24):3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kastner P, Grondona JM, Mark M, Gansmuller A, LeMeur M, Decimo D, Vonesch JL, Dolle P, Chambon P. Genetic analysis of RXR alpha developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994;78(6):987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 40.Sucov HM, Dyson E, Gumeringer CL, Price J, Chien KR, Evans RM. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994;8(9):1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- 41.Huang X, Gao X, Diaz-Trelles R, Ruiz-Lozano P, Wang Z. Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev Biol. 2008;319(2):258–266. doi: 10.1016/j.ydbio.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 42.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432(7013):107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG. Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc Natl Acad Sci U S A. 2007;104(3):846–851. doi: 10.1073/pnas.0608118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459(7247):708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lange M, Kaynak B, Forster UB, Tonjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, Mebus S, Lehrach H, Lurz R, Gobom J, Rottbauer W, Abdelilah-Seyfried S, Sperling S. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22(17):2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466(7303):258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaynak B, von Heydebreck A, Mebus S, Seelow D, Hennig S, Vogel J, Sperling HP, Pregla R, Alexi-Meskishvili V, Hetzer R, Lange PE, Vingron M, Lehrach H, Sperling S. Genome-wide array analysis of normal and malformed human hearts. Circulation. 2003;107(19):2467–2474. doi: 10.1161/01.CIR.0000066694.21510.E2. [DOI] [PubMed] [Google Scholar]
- 48.Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463(7283):958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitagawa H, Fujiki R, Yoshimura K, Mezaki Y, Uematsu Y, Matsui D, Ogawa S, Unno K, Okubo M, Tokita A, Nakagawa T, Ito T, Ishimi Y, Nagasawa H, Matsumoto T, Yanagisawa J, Kato S. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113(7):905–917. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 50.Lu X, Meng X, Morris CA, Keating MT. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics. 1998;54(2):241–249. doi: 10.1006/geno.1998.5578. [DOI] [PubMed] [Google Scholar]
- 51.Yoshimura K, Kitagawa H, Fujiki R, Tanabe M, Takezawa S, Takada I, Yamaoka I, Yonezawa M, Kondo T, Furutani Y, Yagi H, Yoshinaga S, Masuda T, Fukuda T, Yamamoto Y, Ebihara K, Li DY, Matsuoka R, Takeuchi JK, Matsumoto T, Kato S. Distinct function of 2 chromatin remodeling complexes that share a common subunit, Williams syndrome transcription factor (WSTF) Proc Natl Acad Sci U S A. 2009;106(23):9280–9285. doi: 10.1073/pnas.0901184106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Poot RA, Bozhenok L, van den Berg DL, Steffensen S, Ferreira F, Grimaldi M, Gilbert N, Ferreira J, Varga-Weisz PD. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol. 2004;6(12):1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- 53.Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. Embo J. 2002;21(9):2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9(13):1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 55.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106(6):709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 56.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11(8):1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 57.Dirscherl SS, Krebs JE. Functional diversity of ISWI complexes. Biochem Cell Biol. 2004;82(4):482–489. doi: 10.1139/o04-044. [DOI] [PubMed] [Google Scholar]
- 58.Corona DF, Tamkun JW. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta. 2004;1677(1–3):113–119. doi: 10.1016/j.bbaexp.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 59.Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci U S A. 2003;100(24):14097–14102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36(9):955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 61.Hurd EA, Capers PL, Blauwkamp MN, Adams ME, Raphael Y, Poucher HK, Martin DM. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome. 2007;18(2):94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- 62.Randall V, McCue K, Roberts C, Kyriakopoulou V, Beddow S, Barrett AN, Vitelli F, Prescott K, Shaw-Smith C, Devriendt K, Bosman E, Steffes G, Steel KP, Simrick S, Basson MA, Illingworth E, Scambler PJ. Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J Clin Invest. 2009;119(11):3301–3310. doi: 10.1172/JCI37561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bao Y, Shen X. INO80 subfamily of chromatin remodeling complexes. Mutat Res. 2007;618(1–2):18–29. doi: 10.1016/j.mrfmmm.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gallant P. Control of transcription by Pontin and Reptin. Trends Cell Biol. 2007;17(4):187–192. doi: 10.1016/j.tcb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 65.Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16(3):465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 66.Jonsson ZO, Dhar SK, Narlikar GJ, Auty R, Wagle N, Pellman D, Pratt RE, Kingston R, Dutta A. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J Biol Chem. 2001;276(19):16279–16288. doi: 10.1074/jbc.M011523200. [DOI] [PubMed] [Google Scholar]
- 67.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406(6795):541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 68.Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, Wo ZG, Kemler R, Kingston R, Wu C, Fishman M. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell. 2002;111(5):661–672. doi: 10.1016/s0092-8674(02)01112-1. [DOI] [PubMed] [Google Scholar]
- 69.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome biology. 2004;5(5):224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277(28):25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 72.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9(1):45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 73.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, Koch U, De Francesco R, Steinkuhler C, Gallinari P. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104(44):17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones P, Altamura S, De Francesco R, Gallinari P, Lahm A, Neddermann P, Rowley M, Serafini S, Steinkuhler C. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett. 2008;18(6):1814–1819. doi: 10.1016/j.bmcl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 75.Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, Mazitschek R, Kwiatkowski NP, Lewis TA, Maglathin RL, McLean TH, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J Biol Chem. 2008;283(17):11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21(18):6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang CL, McKinsey TA, Lu JR, Olson EN. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J Biol Chem. 2001;276(1):35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 78.Dressel U, Bailey PJ, Wang SC, Downes M, Evans RM, Muscat GE. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J Biol Chem. 2001;276(20):17007–17013. doi: 10.1074/jbc.M101508200. [DOI] [PubMed] [Google Scholar]
- 79.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22(20):7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.rOoi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8(7):544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 81.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21(14):1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, Nazarian R, Schnepp R, Jen K, Biben C, Runke G, Mackay JP, Novotny J, Schwartz RJ, Harvey RP, Mullins MC, Epstein JA. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110(6):713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 83.Shin CH, Liu ZP, Passier R, Zhang CL, Wang DZ, Harris TM, Yamagishi H, Richardson JA, Childs G, Olson EN. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110(6):725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 84.Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man Yung W, Mackay J, Zhou R, Ferrari V, Gruber P, Epstein JA. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest. 2003;112(6):863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ, Li L, Hannenhalli S, Epstein JA. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell. 2010;19(3):450–459. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuwahara K, Saito Y, Takano M, Arai Y, Yasuno S, Nakagawa Y, Takahashi N, Adachi Y, Takemura G, Horie M, Miyamoto Y, Morisaki T, Kuratomi S, Noma A, Fujiwara H, Yoshimasa Y, Kinoshita H, Kawakami R, Kishimoto I, Nakanishi M, Usami S, Saito Y, Harada M, Nakao K. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. Embo J. 2003;22(23):6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuwahara K, Saito Y, Ogawa E, Takahashi N, Nakagawa Y, Naruse Y, Harada M, Hamanaka I, Izumi T, Miyamoto Y, Kishimoto I, Kawakami R, Nakanishi M, Mori N, Nakao K. The neuron-restrictive silencer element-neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol Cell Biol. 2001;21(6):2085–2097. doi: 10.1128/MCB.21.6.2085-2097.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakagawa Y, Kuwahara K, Harada M, Takahashi N, Yasuno S, Adachi Y, Kawakami R, Nakanishi M, Tanimoto K, Usami S, Kinoshita H, Saito Y, Nakao K. Class II HDACs mediate CaMK-dependent signaling to NRSF in ventricular myocytes. J Mol Cell Cardiol. 2006;41(6):1010–1022. doi: 10.1016/j.yjmcc.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 89.Haq S, Choukroun G, Kang ZB, Ranu H, Matsui T, Rosenzweig A, Molkentin JD, Alessandrini A, Woodgett J, Hajjar R, Michael A, Force T. Glycogen synthase kinase-3beta is a negative regulator of cardiomyocyte hypertrophy. J Cell Biol. 2000;151(1):117–130. doi: 10.1083/jcb.151.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Antos CL, McKinsey TA, Frey N, Kutschke W, McAnally J, Shelton JM, Richardson JA, Hill JA, Olson EN. Activated glycogen synthase-3 beta suppresses cardiac hypertrophy in vivo. Proc Natl Acad Sci U S A. 2002;99(2):907–912. doi: 10.1073/pnas.231619298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morisco C, Zebrowski D, Condorelli G, Tsichlis P, Vatner SF, Sadoshima J. The Akt-glycogen synthase kinase 3beta pathway regulates transcription of atrial natriuretic factor induced by beta-adrenergic receptor stimulation in cardiac myocytes. J Biol Chem. 2000;275(19):14466–14475. doi: 10.1074/jbc.275.19.14466. [DOI] [PubMed] [Google Scholar]
- 92.Hardt SE, Sadoshima J. Glycogen synthase kinase-3beta: a novel regulator of cardiac hypertrophy and development. Circ Res. 2002;90(10):1055–1063. doi: 10.1161/01.res.0000018952.70505.f1. [DOI] [PubMed] [Google Scholar]
- 93.Zhu W, Trivedi CM, Zhou D, Yuan L, Lu MM, Epstein JA. Inpp5f is a polyphosphoinositide phosphatase that regulates cardiac hypertrophic responsiveness. Circ Res. 2009;105(12):1240–1247. doi: 10.1161/CIRCRESAHA.109.208785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trivedi CM, Lu MM, Wang Q, Epstein JA. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem. 2008;283(39):26484–26489. doi: 10.1074/jbc.M803686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118(11):3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126(2):321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 97.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24(19):8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Song K, Backs J, McAnally J, Qi X, Gerard RD, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125(3):453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 99.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110(4):479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408(6808):106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24(19):8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Passier R, Zeng H, Frey N, Naya FJ, Nicol RL, McKinsey TA, Overbeek P, Richardson JA, Grant SR, Olson EN. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J Clin Invest. 2000;105(10):1395–1406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97(8):4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van Rooij E, Fielitz J, Sutherland LB, Thijssen VL, Crijns HJ, Dimaio MJ, Shelton J, De Windt LJ, Hill JA, Olson EN. Myocyte Enhancer Factor 2 and Class II Histone Deacetylases Control a Gender-Specific Pathway of Cardioprotection Mediated by the Estrogen Receptor. Circ Res. 2009 doi: 10.1161/CIRCRESAHA.109.207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119(4):555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 106.Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, Lombard D, Mizeracki A, Matthias G, Alt FW, Khochbin S, Matthias P. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28(5):1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 108.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158(4):647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Perrod S, Cockell MM, Laroche T, Renauld H, Ducrest AL, Bonnard C, Gasser SM. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. Embo J. 2001;20(1–2):197–209. doi: 10.1093/emboj/20.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100(19):10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6(6):759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 112.Coussens M, Maresh JG, Yanagimachi R, Maeda G, Allsopp R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One. 2008;3(2):e1571. doi: 10.1371/journal.pone.0001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smith J. Human Sir2 and the 'silencing' of p53 activity. Trends Cell Biol. 2002;12(9):404–406. doi: 10.1016/s0962-8924(02)02342-5. [DOI] [PubMed] [Google Scholar]
- 114.Zhou B, Wu LJ, Li LH, Tashiro S, Onodera S, Uchiumi F, Ikejima T. Silibinin protects against isoproterenol-induced rat cardiac myocyte injury through mitochondrial pathway after up-regulation of SIRT1. J Pharmacol Sci. 2006;102(4):387–395. doi: 10.1254/jphs.fpj06005x. [DOI] [PubMed] [Google Scholar]
- 115.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circulation research. 2007;100(10):1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 116.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317(5837):516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 117.Lynn EG, McLeod CJ, Gordon JP, Bao J, Sack MN. SIRT2 is a negative regulator of anoxia-reoxygenation tolerance via regulation of 14-3-3 zeta and BAD in H9c2 cells. FEBS Lett. 2008;582(19):2857–2862. doi: 10.1016/j.febslet.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28(20):6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27(24):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119(9):2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 122.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102(6):703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 123.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93(3):361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 124.Shikama N, Lutz W, Kretzschmar R, Sauter N, Roth JF, Marino S, Wittwer J, Scheidweiler A, Eckner R. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. Embo J. 2003;22(19):5175–5185. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yanazume T, Hasegawa K, Morimoto T, Kawamura T, Wada H, Matsumori A, Kawase Y, Hirai M, Kita T. Cardiac p300 is involved in myocyte growth with decompensated heart failure. Mol Cell Biol. 2003;23(10):3593–3606. doi: 10.1128/MCB.23.10.3593-3606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gusterson RJ, Jazrawi E, Adcock IM, Latchman DS. The transcriptional co-activators CREB-binding protein (CBP) and p300 play a critical role in cardiac hypertrophy that is dependent on their histone acetyltransferase activity. J Biol Chem. 2003;278(9):6838–6847. doi: 10.1074/jbc.M211762200. [DOI] [PubMed] [Google Scholar]
- 127.Miyamoto S, Kawamura T, Morimoto T, Ono K, Wada H, Kawase Y, Matsumori A, Nishio R, Kita T, Hasegawa K. Histone acetyltransferase activity of p300 is required for the promotion of left ventricular remodeling after myocardial infarction in adult mice in vivo. Circulation. 2006;113(5):679–690. doi: 10.1161/CIRCULATIONAHA.105.585182. [DOI] [PubMed] [Google Scholar]
- 128.Morimoto T, Sunagawa Y, Kawamura T, Takaya T, Wada H, Nagasawa A, Komeda M, Fujita M, Shimatsu A, Kita T, Hasegawa K. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J Clin Invest. 2008;118(3):868–878. doi: 10.1172/JCI33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li HL, Liu C, de Couto G, Ouzounian M, Sun M, Wang AB, Huang Y, He CW, Shi Y, Chen X, Nghiem MP, Liu Y, Chen M, Dawood F, Fukuoka M, Maekawa Y, Zhang L, Leask A, Ghosh AK, Kirshenbaum LA, Liu PP. Curcumin prevents and reverses murine cardiac hypertrophy. J Clin Invest. 2008;118(3):879–893. doi: 10.1172/JCI32865. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 130.Mathiyalagan P, Chang L, Du XJ, El-Osta A. Cardiac ventricular chambers are epigenetically distinguishable. Cell Cycle. 2010;9(3):612–617. doi: 10.4161/cc.9.3.10612. [DOI] [PubMed] [Google Scholar]
- 131.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7(9):715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 132.Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res. 2000;86(9):932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- 133.Takeuchi T, Kojima M, Nakajima K, Kondo S. jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech Dev. 1999;86(1–2):29–38. doi: 10.1016/s0925-4773(99)00100-8. [DOI] [PubMed] [Google Scholar]
- 134.Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev Cell. 2003;5(1):85–97. doi: 10.1016/s1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- 135.Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y, Takeuchi T. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem. 2009;284(2):733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- 136.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24(4):368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 138.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim TG, Chen J, Sadoshima J, Lee Y. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol Cell Biol. 2004;24(23):10151–10160. doi: 10.1128/MCB.24.23.10151-10160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim TG, Jung J, Mysliwiec MR, Kang S, Lee Y. Jumonji represses alpha-cardiac myosin heavy chain expression via inhibiting MEF2 activity. Biochem Biophys Res Commun. 2005;329(2):544–553. doi: 10.1016/j.bbrc.2005.01.154. [DOI] [PubMed] [Google Scholar]
- 142.Jung J, Kim TG, Lyons GE, Kim HR, Lee Y. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J Biol Chem. 2005;280(35):30916–30923. doi: 10.1074/jbc.M414482200. [DOI] [PubMed] [Google Scholar]
- 143.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 144.Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18(12):7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hwang I, Gottlieb PD. The Bop gene adjacent to the mouse CD8b gene encodes distinct zinc-finger proteins expressed in CTLs and in muscle. J Immunol. 1997;158(3):1165–1174. [PubMed] [Google Scholar]
- 146.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, Nakagawa O, Srivastava D. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31(1):25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 147.Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270(5244):1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- 148.Tan X, Rotllant J, Li H, De Deyne P, Du SJ. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci U S A. 2006;103(8):2713–2718. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Diehl F, Brown MA, van Amerongen MJ, Novoyatleva T, Wietelmann A, Harriss J, Ferrazzi F, Bottger T, Harvey RP, Tucker PW, Engel FB. Cardiac deletion of Smyd2 is dispensable for mouse heart development. PLoS One. 2010;5(3):e9748. doi: 10.1371/journal.pone.0009748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet. 2005;21(3):188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 151.Hirschhorn K, Cooper HL, Firschein IL. Deletion of short arms of chromosome 4–5 in a child with defects of midline fusion. Humangenetik. 1965;1(5):479–482. doi: 10.1007/BF00279124. [DOI] [PubMed] [Google Scholar]
- 152.Wright TJ, Ricke DO, Denison K, Abmayr S, Cotter PD, Hirschhorn K, Keinanen M, McDonald-McGinn D, Somer M, Spinner N, Yang-Feng T, Zackai E, Altherr MR. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum Mol Genet. 1997;6(2):317–324. doi: 10.1093/hmg/6.2.317. [DOI] [PubMed] [Google Scholar]
- 153.Marango J, Shimoyama M, Nishio H, Meyer JA, Min DJ, Sirulnik A, Martinez-Martinez Y, Chesi M, Bergsagel PL, Zhou MM, Waxman S, Leibovitch BA, Walsh MJ, Licht JD. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111(6):3145–3154. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460(7252):287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 155.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7(7):517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 156.Ogata N, Ueda K, Hayaishi O. ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J Biol Chem. 1980;255(16):7610–7615. [PubMed] [Google Scholar]
- 157.Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem. 1980;255(16):7616–7620. [PubMed] [Google Scholar]
- 158.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A. 1982;79(11):3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Realini CA, Althaus FR. Histone shuttling by poly(ADP-ribosylation) J Biol Chem. 1992;267(26):18858–18865. [PubMed] [Google Scholar]
- 160.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119(6):803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 161.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299(5606):560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 162.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94(14):7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11(18):2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Masutani M, Nozaki T, Nishiyama E, Shimokawa T, Tachi Y, Suzuki H, Nakagama H, Wakabayashi K, Sugimura T. Function of poly(ADP-ribose) polymerase in response to DNA damage: gene-disruption study in mice. Mol Cell Biochem. 1999;193(1–2):149–152. [PubMed] [Google Scholar]
- 165.Masutani M, Suzuki H, Kamada N, Watanabe M, Ueda O, Nozaki T, Jishage K, Watanabe T, Sugimoto T, Nakagama H, Ochiya T, Sugimura T. Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proc Natl Acad Sci U S A. 1999;96(5):2301–2304. doi: 10.1073/pnas.96.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo J. 2003;22(9):2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Xiao CY, Chen M, Zsengeller Z, Li H, Kiss L, Kollai M, Szabo C. Poly(ADP-Ribose) polymerase promotes cardiac remodeling, contractile failure, and translocation of apoptosis-inducing factor in a murine experimental model of aortic banding and heart failure. J Pharmacol Exp Ther. 2005;312(3):891–898. doi: 10.1124/jpet.104.077164. [DOI] [PubMed] [Google Scholar]
- 168.Pillai JB, Russell HM, Raman J, Jeevanandam V, Gupta MP. Increased expression of poly(ADP-ribose) polymerase-1 contributes to caspase-independent myocyte cell death during heart failure. Am J Physiol Heart Circ Physiol. 2005;288(2):H486–H496. doi: 10.1152/ajpheart.00437.2004. [DOI] [PubMed] [Google Scholar]
- 169.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280(52):43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 170.Zhou HZ, Swanson RA, Simonis U, Ma X, Cecchini G, Gray MO. Poly(ADP-ribose) polymerase-1 hyperactivation and impairment of mitochondrial respiratory chain complex I function in reperfused mouse hearts. Am J Physiol Heart Circ Physiol. 2006;291(2):H714–H723. doi: 10.1152/ajpheart.00823.2005. [DOI] [PubMed] [Google Scholar]
- 171.Szabo G, Bahrle S, Stumpf N, Sonnenberg K, Szabo EE, Pacher P, Csont T, Schulz R, Dengler TJ, Liaudet L, Jagtap PG, Southan GJ, Vahl CF, Hagl S, Szabo C. Poly(ADP-Ribose) polymerase inhibition reduces reperfusion injury after heart transplantation. Circ Res. 2002;90(1):100–106. doi: 10.1161/hh0102.102657. [DOI] [PubMed] [Google Scholar]
- 172.Palfi A, Toth A, Hanto K, Deres P, Szabados E, Szereday Z, Kulcsar G, Kalai T, Hideg K, Gallyas F, Jr, Sumegi B, Toth K, Halmosi R. PARP inhibition prevents postinfarction myocardial remodeling and heart failure via the protein kinase C/glycogen synthase kinase-3beta pathway. J Mol Cell Cardiol. 2006;41(1):149–159. doi: 10.1016/j.yjmcc.2006.03.427. [DOI] [PubMed] [Google Scholar]
- 173.Xiao CY, Chen M, Zsengeller Z, Szabo C. Poly(ADP-ribose) polymerase contributes to the development of myocardial infarction in diabetic rats and regulates the nuclear translocation of apoptosis-inducing factor. J Pharmacol Exp Ther. 2004;310(2):498–504. doi: 10.1124/jpet.104.066803. [DOI] [PubMed] [Google Scholar]
- 174.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 175.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305(5686):1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 176.Kovacs K, Toth A, Deres P, Kalai T, Hideg K, Sumegi B. Myocardial protection by selective poly(ADP-ribose) polymerase inhibitors. Exp Clin Cardiol. 2004;9(1):17–20. [PMC free article] [PubMed] [Google Scholar]
- 177.Palfi A, Toth A, Kulcsar G, Hanto K, Deres P, Bartha E, Halmosi R, Szabados E, Czopf L, Kalai T, Hideg K, Sumegi B, Toth K. The role of Akt and mitogen-activated protein kinase systems in the protective effect of poly(ADP-ribose) polymerase inhibition in Langendorff perfused and in isoproterenol-damaged rat hearts. J Pharmacol Exp Ther. 2005;315(1):273–282. doi: 10.1124/jpet.105.088336. [DOI] [PubMed] [Google Scholar]
- 178.Butler AJ, Ordahl CP. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol Cell Biol. 1999;19(1):296–306. doi: 10.1128/mcb.19.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Vyas DR, McCarthy JJ, Tsika GL, Tsika RW. Multiprotein complex formation at the beta myosin heavy chain distal muscle CAT element correlates with slow muscle expression but not mechanical overload responsiveness. J Biol Chem. 2001;276(2):1173–1184. doi: 10.1074/jbc.M007750200. [DOI] [PubMed] [Google Scholar]
- 180.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280(49):40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 181.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 182.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5(6):905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 183.Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138(6):1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]