SUMMARY
We set out to review the extent to which molecular karyotyping has overtaken conventional cytogenetics in applications related to epilepsy. Multiplex ligase-dependent probe amplification (MLPA) targeted to predetermined regions such as SCN1A and KCNQ2 has been effectively applied over the past half a decade and oligonucleotide array comparative genome hybridization (array CGH) is now well established for genome wide exploration of microchromosomal variation. Array CGH is applicable to the characterization of lesions present in both sporadic and familial epilepsy, especially where clinical features of affected cases depart from established syndromes. Copy number variants (CNVs) associated with epilepsy and a range of other syndromes and conditions can be recurrent due to non-allelic homologous recombination in regions of segmental duplication. The most common of the recurrent microdeletions associated with generalized epilepsy are typically seen at a frequency of around 1% at 15q13.3, 16p13.11 and 15q11.2, sites that also confer susceptibility for intellectual disability, autism and schizophrenia. Incomplete penetrance and variable expressivity confound the established rules of cytogenetics for determining the pathogenicity for novel CNVs; however, as knowledge is gained for each of the recurrent CNVs, this is translated to genetic counselling. CNVs play a significant role in the susceptibility profile for epilepsies with complex genetics and their comorbidities both from the “hotspots” defined by segmental duplication and elsewhere in the genome where their location and size are often novel.
The road to molecular cytogenetics
Human cytogenetics is the study of chromosomes in relation to inheritance and the origin of abnormal pathology. During the past half century the application of human cytogenetics to the clinical workup of patients has evolved from morphological karyotyping carried out under the light microscope to molecular karyotyping displayed on a computer screen. Molecular karyotyping is now a validated first-tier diagnostic step due to its clinical utility, at least for developmental delay, intellectual disabilities (ID) and congenital abnormalities (Miller et al., 2010, Regier et al., 2010, Xiang et al., 2010, Yu et al., 2009), and we believe for unusual epilepsy cases as well. Implementation of this new technology significantly increases diagnostic yield in the cytogenetics laboratory compared with previous conventional karyotyping. Thus, referring clinicians receive back far more information on many more of their patients than at any time previously.
The foundations for the translation of cytogenetics from insects to medicine were laid down with a series of technical advances in the 1950s. The squash technique was applied to spread the chromosomes into one plane of focus for microscopic examination. Colchicine pretreatment of cells was introduced to arrest and condense chromosomes at metaphase to maximise the number of cells for examination. Hypotonic pretreatment of cells was used to disperse chromosomes and their attached nucleolar material to give a sharp well defined image. Lymphocyte culture was brought in to provide easier access to a human tissue source for routine cytogenetic analysis. Finally, stimulation of cell division by phytohaemagglutinin increased the number of cells arrested at metaphase that were available for observation.
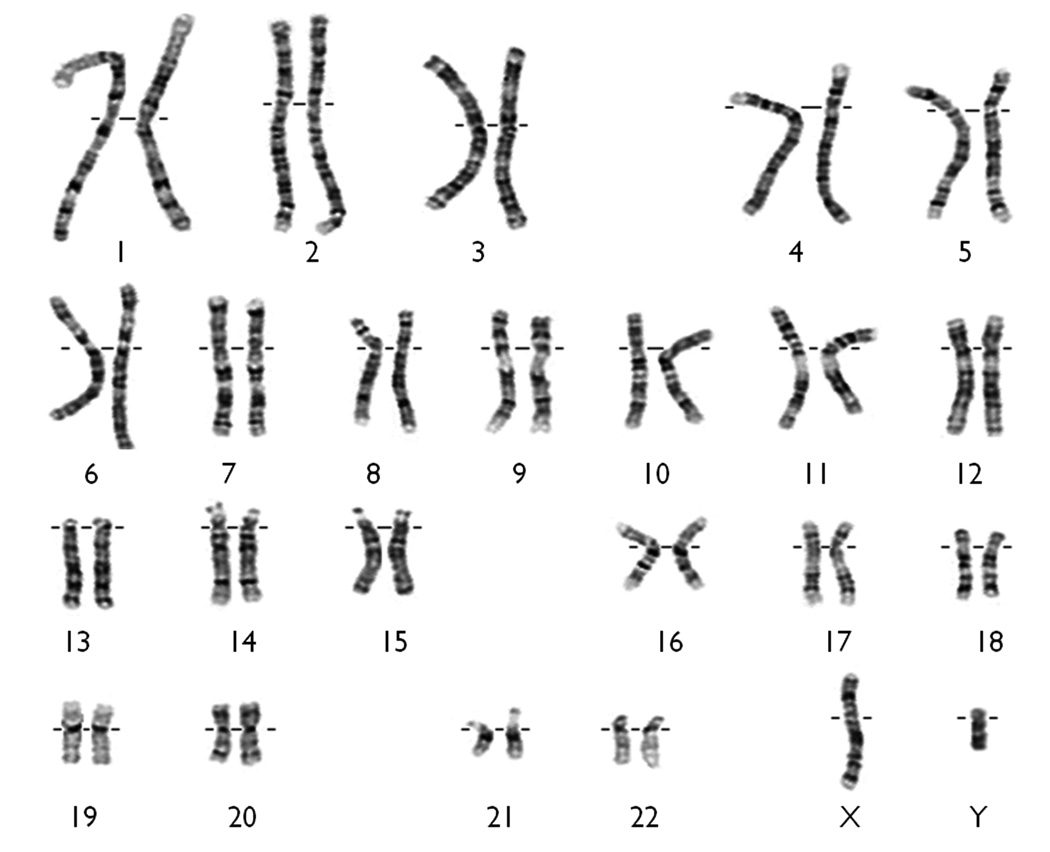
Despite these advances, individual chromosomes did not become physically identifiable until the 1970s. Quinacrine staining in conjunction with fluorescence microscopy, then simplification to permanent G-banding, allowed indefinite storage of slides from which individual chromosomes could be identified by their banding patterns (Fig. 1). This was the foundation for gene mapping allowing genes to be positioned in relation to visible landmarks along the chromosome. Moreover, departures from the unique patterns established for each individual chromosome provided explanations at the gross morphological level for various clinical conditions. Subsequent technical refinement increased the number of detectable bands providing higher resolution on normal and abnormal karyotypes. The association between epileptic seizures and chromosomal abnormalities detected in that way is now well recognised (Battaglia & Guerrini 2005, Singh et al., 2002).
Figure 1.
The conventional G-banded karyotype showing 23 pairs of normal chromosomes as seen under light microscopy.
Fluorescence in situ hybridisation (FISH) during the 1990s ushered cytogenetics into the molecular era. Labelled DNA probes could be hybridised to their corresponding targets on the diploid set of chromosomes spread out on a slide. This greatly improved sensitivity for the detection of small aberrations at predetermined chromosomal regions. Furthermore, researchers cloning genes could send their gene probes to cytogenetics to rapidly position their gene of interest to an identifiable G-banded region anywhere in the genome.
Today, the “molecular karyotype” is rapidly replacing the conventional karyotype in diagnostic and research laboratories. Molecular karyotyping refers to the evaluation of chromosome content and structure using DNA hybridization rather than direct observation of chromosomes under the microscope. Advances in technology over the past several years now allow clinicians and researchers to interrogate the entire genome for copy number variants (CNVs; deletions and duplications) in one experiment. Alternatively, specific genomic regions can be targeted using several different techniques. One important difference from the conventional methods is that molecular karyotypes only detect imbalances caused by deletion or addition of DNA – they cannot detect truly balanced translocations or inversions which still rely on detecting disruption to the normal G-band pattern. Of the apparently “balanced” reciprocal and complex rearrangements as diagnosed by light microscopy about 40% of both de novo and inherited cases show cryptic deletions at the chromosomal breakpoints using the more sensitive molecular approach (De Gregori et al., 2007, Schluth-Bolard et al., 2009). The other important difference from conventional methods is that the physical reference points have shifted from G-bands to genomic coordinates at the basepair level taken from the human DNA reference sequence.
Molecular platforms for detecting copy number variation
Nowadays the two most commonly used techniques for genome-wide analyses, array comparative genomic hybridization (array-CGH) and single nucleotide polymorphism (SNP) genotyping, use known DNA sequences arranged on a “chip”. Both methods can readily detect microchromosomal aberrations at a much finer resolution than the ~5 Mb limit of the conventional karyotype. The exact resolution depends on the platform that is used. The interpretation of microdeletions is straightforward but microduplications require additional testing by FISH to determine if the duplicated DNA is in tandem or inverted alongside its original location, or inserted elsewhere in the genome.
Array CGH, particularly, allows for flexible, custom designs to interrogate specific regions of interest at very high resolution. Detection of deletions and duplications by array CGH relies on the comparison of two genomes. Fluorescently labelled patient DNA is co-hybridized with DNA from a control individual to an array spotted with either BAC or (more commonly) oligonucleotide probes. Signal intensities for the patient and the control are normalized and reported as a log2 ratio of intensities for each probe. Algorithms to detect CNVs using SNP genotyping arrays involve using both the allele frequency at a given SNP and the total intensity for that SNP. If a SNP is deleted, there will be apparent homozygosity (“AA” which is actually “A-“) and the intensity will be decreased, reflecting DNA dosage.
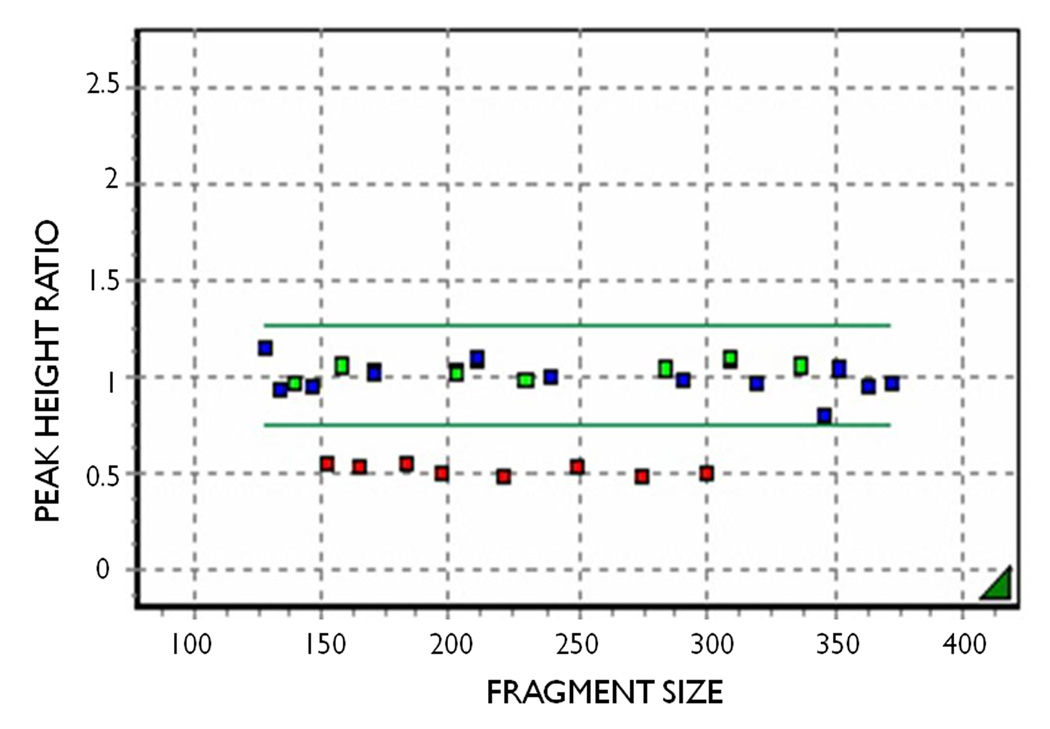
For directed analysis of a specific genomic region, at higher resolution than provided by array technology, one of the most frequently used technologies is multiplex ligation-dependent probe amplification (MLPA) (Fig. 2). MLPA probe kits are available commercially from MRC-Holland, Amsterdam, The Netherlands, for many genes including the epilepsy genes SCN1A and KCNQ2. Duplication and deletion screening is carried out using standard protocols irrespective of the gene. Putative single exon deletions detected by MLPA need to be confirmed by sequencing that exon to exclude allele dropout by mismatch interference between the MLPA probe and the target sequence. Duplication or deletion signal is contiguous where more than one exon is involved, which acts as an internal check for signal validity and sample quality. Confirmation is provided by repeatability in a second run on the same sample. Fragment analysis is carried out by capillary electrophoresis and peak heights for each fragment are converted to a scatterplot using Genemarker software.
Figure 2.
MLPA analysis showing KCNQ2 exons within the normal range (green), control probes from elsewhere in the genome (blue) and deleted exons (red). The display is generated using GeneMarker software. The points represent comparative peak height ratios for each MLPA probe compared with normal samples.
Microchromosomal Mutation in Epilepsy
The following discussion applies to epilepsy presenting primarily as seizures. There are over 200 Mendelian disorders were epilepsy is or can be part of the clinical condition, but not the primary feature (Online Medelian Inheritance in Man, OMIM). Many of these can be associated with DNA sequence based mutations or with various microchromosomal anomalies. Some will be private syndromes and others will be recurrent, with all best detected by array-CGH (Bedoyan et al., 2010, Cardoso et al., 2009, Yeung et al., 2009). Many more novel case reports will follow now that array-CGH is more widely applied as a frontline diagnostic tool. Such cases are not considered as part of this review; however, the value to the clinic and their patients of application of molecular karyotyping on a case by case basis is inestimable.
Soon after discovery of point mutations in the first epilepsy gene, CHRNA4, in association with autosomal dominant nocturnal frontal lobe epilepsy (Steinlein et al., 1995) another CHRNA4 mutation was discovered (Steinlein et al., 1997). This was an insertion of only three nucleotides. Many studies have since shown similar small epilepsy related lesions at the DNA level; for example, many single or few base pair deletions or insertions in SCN1A (Lossin 2009). These also are not considered as part of this review. They are detectable by conventional DNA sequencing, are below the resolution of array-CGH and are not detectable by MLPA unless the site of the lesion within the exon corresponds to part of the probe sequence, causing exon dropout.
In 1998 not long after discovery of CHRNA4 as an epilepsy gene, the second and third epilepsy genes, KCNQ2 and KCNQ3, were discovered in families with benign familial neonatal seizures (Biervert et al., 1998, Charlier et al., 1998, Singh et al., 1998). Discovery of KCNQ2 (Sing, et al. 1998) was based on a microchromosomal deletion that directed the gene search to a defined set of candidate genes known to map to the deleted sequence. These genes were then tested in other families with the same syndrome until mutations were detected which identified the gene. The point is this: microchromosomal copy number variants (CNVs) have been known as causative for epilepsy since the dawn of gene discovery in the idiopathic, or genetic epilepsies, as they are now known (Berg et al., 2010). However, the efficient and routine recognition of microchromosomal lesions ranging in size from lone exons up to multiple genes below the resolution of conventional karyotyping is relatively recent.
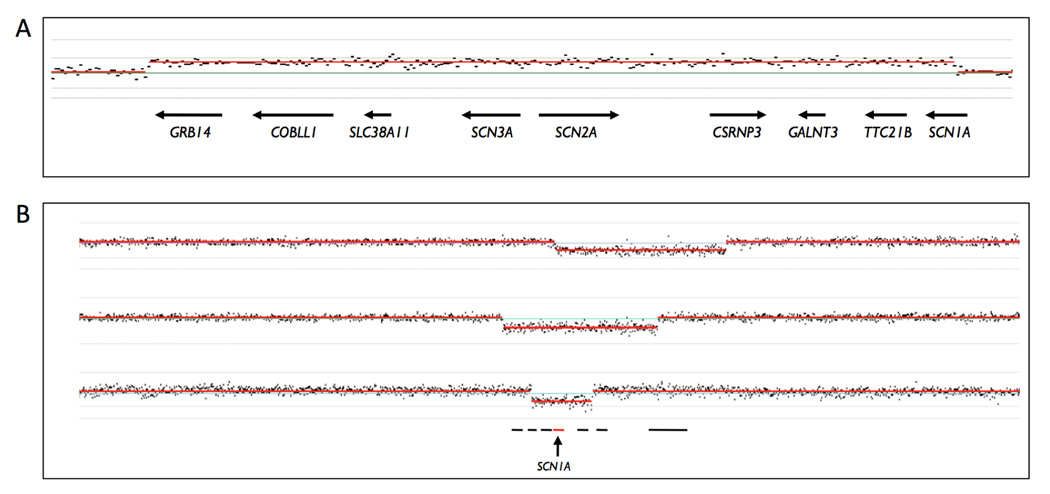
Routine easy application to epilepsy began with implementation of multiplex-ligase dependent probe amplification (MLPA) to the diagnosis of CNVs in the genes SCN1A and KCNQ2 (Mulley et al., 2006; Heron et al., 2007, Marini et al., 2007; 2009). Array-CGH was then used as an adjunct to MLPA and other technologies to molecularly characterise the CNVs for their exact size and gene content (de Kovel et al., 2010, Depienne et al., 2009b, Dibbens et al., 2009, Helbig et al., 2009, Heron et al., 2010, Kurahashi et al., 2009, Marini et al., 2009, Mei et al., 2010, Suls et al., 2010, Wang et al., 2008). Recent advances have come from the application of array-CGH as the frontline tool (Depienne et al., 2009a, Conlin et al., 2010, Heinzen et al., 2010, McMahon et al., 2010, Mefford et al., 2010, Muhle et al., 2010). Figure 3 shows the array-CGH signal for a duplication involving SCN2A, SCN3A and part of SCN1A and three SCN1A deletions of various sizes.
Figure 3.
Signal from Nimblegen chromosome 2-specific oligonucleotide array-CGH. Hybridization data were analysed using SignalMap™ software. A: The familial 1.57 Mb duplication involving 8 contiguous genes including SCN2A, SCN3A and part of SCN1A as reported by Heron et al (2010). B: Three of the Dravet syndrome deletions with excision of SCN1A as reported by Marini et al (2009). Deletion sizes from top to bottom are 1.64 Mb (deleting SCN1A – XIRP2 and all genes between), 1.49 Mb (deleting FAM130A2 – XIRP2 and all genes between) and 588 kb (deleting GALNT3 – SCN9A), respectively.
Sporadic epilepsies
The earliest application of MLPA to epilepsy signalled the potential significance of CNVs in this group of disorders. A substantial number of cases with Dravet syndrome (20–30%) have no DNA sequence based mutations in SCN1A (Harkin et al., 2007). In small pilot studies 11–15% of these unsolved cases had pathogenic CNVs suggesting that this mechanism may represent a significant additional cause (Marini et al., 2007, Mulley et al., 2006). Larger studies confirmed this, with detection rates of 10.1% (Wang et al., 2008), 12.5% (Marini et al., 2009) and 13% (Depienne et al., 2009b) using MLPA and then array-CGH in positives to confirm and refine the size and gene content of the CNVs. The majority were large enough to be detectable by array-CGH alone; however, MLPA has the necessary resolution down to a single exon to detect the smaller CNVs below the resolution of array-CGH. Remarkably, deletions were characterized which removed as much as 9.3Mb and 49 genes without altering the primary SCN1A-related severe Dravet phenotype (Marini et al., 2009). Depienne and colleagues (Depienne et al., 2009a) used genome wide array-CGH to investigate cases of Dravet or Dravet-like syndrome where sequence mutations or CNVs involving SCN1A had been excluded. One case had a deletion in PCDH19 on chromosome X, the gene implicated in the milder familial epilepsy with mental retardation (EFMR) affecting females only (Dibbens et al., 2008). Thus, a new syndrome was born similar in severity to Dravet syndrome but with mutations in PCDH19 rather than SCN1A (Depienne, et al. 2009a). Other case reports utilizing array-CGH relate to a late onset Lennox-Gastaut syndrome with 15q11.2-q13.1 microduplication (Orrico et al., 2009), epilepsy with infantile spasms with 19p13.13 microdeletion (Auvin et al., 2009) and a myoclonic epilepsy with 15q26 microdeletion (Veredice et al., 2009). Such reports point to genomic locations harbouring putative epilepsy genes.
Recently, twenty cases of treatment refractory epilepsy were analysed by whole genome array-CGH in a search for CNVs to test a hypothesis that such lesions may contain epilepsy related genes not responsive to antiepileptic drugs (McMahon et al., 2010). Surprisingly, one of the 20 cases tested was found to have a 15q13.3 microdeletion. Individuals with the 15q13.3 microdeletion usually respond to medication; hence, it was likely a major risk factor for the patient’s epilepsy but not likely the reason for non-response to medication. A second case within the same small cohort carried a novel microduplication at 10q21.2 present in the unaffected mother, suggesting an interpretation equivalent of that for the 15q13.3 CNV. Although a small sample of just 20 cases, these data suggest that the two CNVs detected might be largely responsible for the seizures, but they cannot be blamed for the refractory nature of the seizures. In contrast, Heinzen et al. (Heinzen et al., 2010) detected 23 cases of the 16p13.11 microdeletion, 16 of which (70%) were refractory to treatment, much higher than the 30% generally regarded as refractory to treatment (Kwan & Brodie 2000). Interestingly, the detection rate for CNVs responsible for seizures in the small study (McMahon, et al. 2010) was consistent with the firmer rate of about 10% from the two much larger studies (Heinen, et al. 2010; Mefford, et al. 2010).
Familial epilepsies
The first observation of familial epilepsy with a CNV was made in 1998 (Singh et al., 1998) after microsatellite maker results led to inference of the deletion. Nearly a decade later Heron and colleagues (Heron et al., 2007) carried out a directed search for familial CNVs using MLPA to interrogate the potassium channel gene KCNQ2. Forty four per cent of cases for whom DNA sequence based mutation had been excluded were found to have a microdeletion or microduplication, establishing this as a necessary second tier test strategy for KCNQ2 related disorders. Kurahashi et al (Kurahashi et al., 2009) also used MLPA to ascertain CNVs in KCNQ2, followed by array-CGH to determine the size of their deletions, and found a frequency of 4/22 (18%). Remarkably, two of their deletions confirmed by array-CGH extended beyond KCNQ2 to include the CHRNA4 gene, also an established epilepsy gene. Symptoms associated with these larger deletions were consistent only with benign familial neonatal seizures associated with KCNQ2 loss of function, or haploinsufficiency. Autosomal dominant nocturnal frontal lobe epilepsy associated with specific CHRNA4 missense mutations is triggered by change of function mutations, rather than loss of function, so haploinsufficiency of CHRNA4 had no effect over and above what was expected for KCNQ2 associated loss of function mutations.
Familial epilepsies with intellectual disability not fitting known syndromes are prime candidates for rare and unique pathogenic CNVs. An unusual multigenerational family segregating seizures and ID was detected by multiplex amplicon quantification (Suls et al., 2010). Complete microdeletion of SCN1A is not generally compatible with reproduction and not generally associated with such a mild case of Dravet syndrome. Another family with seizures and ID segregated an associated and previously undescribed microduplication inclusive of the known epilepsy associated gene SCN2A (Heron et al., 2010). It was initially detected by accident when microsatellite markers displayed three alleles, confirmed as a microduplication by array-CGH and its location and orientation shown by FISH to be partially inverted and inserted near SCN1A. Another family with early onset absence epilepsy segregated a previously undescribed microduplication inclusive of the known epilepsy gene CHRNB2 (Muhle et al., 2010). Both cases with the inherited microduplications were confounded with biparental inheritance and one of them with intellectual disability as well. Thus, array CGH can assist with teasing out some of the factors where complexity exists and has efficacy for familial epilepsies as well as the sporadic cases. It should be considered first for situations where the phenotypes do not fit within conventional syndrome designations, whether sporadic or familial.
Genesis of Recurrent Deletions
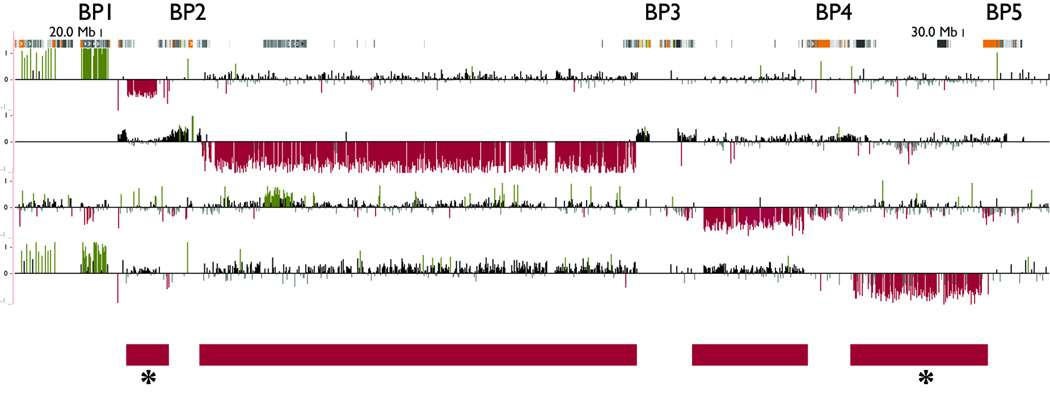
Many regions of the human genome are susceptible to recurrent rearrangement due to the local genomic architecture, including the regions on proximal 15q and 16p that have recently been associated with epilepsy (see below) (Bailey et al., 2002, Makoff & Flomen 2007, Zody et al., 2006). For each of these regions, breakpoint (BP) hotspots correspond to blocks of segmental duplications that flank the deleted or duplicated sequence, prompting suggestion of non-allelic homologous recombination as the likely mechanism (Lupski & Stankiewicz 2005, Pujana et al., 2002). On proximal 15q, for example, deletions and duplications involving various combinations of five BPs are associated with Prader-Willi and Angleman syndromes (BP1-BP3 or BP2-BP3 deletions), autism (BP2-BP3 duplications), and most relevant to this review, genetic generalized epilepsy (BP4-BP5 deletions; Fig. 4). The BP4-BP5 deletions associated with epilepsy contain CHRNA7 and another six genes (Sharp et al., 2008).
Figure 4.
Microdeletions associated with breakpoint (BP) “hotspots” in the 15q11-q13 region associated with Prader-Willi and Angelman syndromes, autism and genetic generalized epilepsy as described in the text. Blocks of semental duplications, in which the breakpoints lie, are represented at the top by orange/yellow/grey blocks. Red bars represent the unique sequence deleted (or duplicated) between blocks of segmental duplications. Deletions between BP1-BP2 and BP4-BP5, marked with an asterisk, have been associated with generalized epilepsy. The clinical significance of deletions between BP3-BP4 is unknown.
Mefford et al (Mefford et al., 2010) used a custom designed array targeting 107 BP rearrangement hotspots throughout the genome (Bailey et al., 2002) together with dense coverage of the remainder of the genome. Overall, 9% of patients harboured rare gene-containing CNVs ranging in size from 13 kb to 15.9 Mb, averaging 1.2 Mb – 4% were recurrent rearrangements at hotspot regions while 5% were rare, non-recurrent events. The three “common” recurrent deletions comprising 3% of patients were at 15q13.3 (1.5 Mb), 16p13.11 (900 kb) and 15q11.2 (600 kb). Collectively an additional 1% of patients had rearrangements at other “hotspot” regions. Some patients had more than one putative pathogenic CNV thus providing insights into a possible origin of genetic modifiers and a mechanism for phenotypic variability associated with mutations in the monogenic epilepsies as well as causes for the syndrome constellations associated with the 15q13.3, 16p13.11 and 15q11.2 CNVs (Mefford & Mulley 2010). Some of the phenotypic variation might arise from the presence of more than one pathogenic CNV in a patient, as in severe developmental delay associated with the 16p12 CNV where the second hit leads to greater phenotypic severity (Girirajan et al., 2010). Overall, 4% of the cohort had CNVs at a BP hotspot, emphasising the need for whole genome coverage in addition to coverage of the BP hotspots for thorough analysis of DNA for CNVs from any patient to detect the remaining 5%.
New Microdeletion Syndromes
The role of the 15q13.3 CNV in cohorts where genetic (idiopathic) generalised epilepsy was the main feature was established by Helbig et al (Helbig et al., 2009) as an extension of the original observation of seizures associated with intellectual disability (Sharp et al., 2008). The role of the 15q13.3 microdeletion in generalized epilepsy was quickly confirmed (Dibbens et al., 2009, Mefford et al., 2010). The phenotypic variability associated with this lesion was subsequently extended in another study (van Bon et al., 2009). Interestingly, the 15q13.3 deletion was absent from a cohort of nearly 3000 patients with partial epilepsies, suggesting that the deletion may confer a risk specifically of generalized epilepsy (Heinzen et al., 2010). Most recurrent 15q13.3 deletions are approximately equivalent in genomic length with similar breakpoints within large blocks of segmental duplications flanking the deleted region. The deletion and the reciprocal duplication are thought to arise due to nonallelic homologous recombination mediated by the segmental duplications, thus leading to recurrence of the same lesion in unrelated individuals.
The 15q13.3 microdeletion is associated with ID, autism, schizophrenia or epilepsy, sometimes with more than one of these in the same affected individual, and sometimes with none of them when it is completely nonpenetrant. There is significant phenotypic variability associated with the 15q13.3 microdeletion with a remarkable range of syndromes seen between and within families. Several mechanisms have been proposed to explain this phenomenon, which would apply across the board to all deletion CNVs (Heinzen et al., 2010, Mulley & Dibbens 2009, Sharp 2009), but none of the mechanisms have been validated. Haploinsufficiency of CHRNA7 is postulated as the most likely component responsible for the pathology associated with the 15q13.3 deletion; however, neighbouring genes within the deletion could potentially be involved (Mulley & Dibbens 2009). Apart from CHRNA7, these include six other genes ARHGAP11B, MTMR10, MTMR15, TRPM1, KLF13 and OTUD7A.
Recently, Shinawi et al (Shinawi et al., 2009) narrowed down the likely pathogenic portion of the CNV to CHRNA7 and/or OTUD7A through observation of a smaller recurrent microdeletion in 10 cases affecting expression of both genes. This smaller and rarer 680 kb CNV is entirely within the boundaries of the more common 1.5 Mb 15q13.3 microdeletion. The 680 kb CNV probably forms by nonallelic homologous recombination in individuals who are heterozygous for a naturally occurring inversion polymorphism within 15q13.3 (Makoff & Flomen 2007).
The recurrent presence of the 15q13.3 CNV provides a finite probability for its homozygosity. Such a case has been described with visual impairment, hypotonia, profound ID and refractory epilepsy (Lepichon et al., 2010). The complex clinical picture reflected the complete loss of function of the genes in the region due to homozygosity for the microdeletion. Absence of CHRNA7 was postulated as the cause of refractory seizures.
The perceived dilemma relating to the 15q13.3 deletion for disease aetiology revolves around uncertainty of phenotypic prediction, since it has been associated with such a broad range and severity of phenotypes. Risks associated with these CNVs appear to be much higher than SNP variants; however, the penetrance of effects modulated by the CNV are less than seen in the monogenic epilepsies, otherwise more large families would be detected segregating with CNVs. These insights come from estimates of penetrance for a number of schizophrenia related CNVs (Vassos et al. 2010) and for 15q13.3 in epilepsy (Dibbens et al. 2009).
Where passed through several generations the question is whether a given CNV “breeds true”. If so, cis-acting genetic modifiers would need to be involved, and they would need to be linked in close physical proximity to the lesion to maintain phenotypic stability. Alternatively, trans-acting modifiers on the chromosome 15 homologue or any other unlinked modifier on any other chromosome would promote extensive clinical variability within families expressed as ID, autism, schizophrenia and/or epilepsy. Highly variable intra- and inter-familial phenotypes (van Bon et al., 2009) argue in favour of trans-acting elements, although in their study few cases were associated with seizures. Familial epilepsy when inherited in association with the 15q13.3 CNV appears more homogeneous than one might expect if expressivity was governed solely by unlinked modifiers (Dibbens et al., 2009). This would suggest involvement of at least some cis-acting effects, but more observations are needed.
Like any autosomal dominant monogenic disorder with incomplete penetrance and variable expressivity, the phenotype displayed by the 15q13.3 microdeletion is just as unpredictable. That can be seen by observation of the extent of its associated phenotypic variability in families, which include normal unaffected carriers where the effects of the lesion appear to be non-penetrant. The likely reason is that the 15q13.3 CNV is just one part of the polygenic profile contributing to the risk of presenting with seizures in any given subject. Standard cytogenetics generally adheres to the principle that de novo microchromosomal lesions in an affected proband are most likely causative. Alternatively, when the same lesion is present in a parent who is phenotypically normal, the CNV is considered benign, with affection in the proband likely due to some other cause. However, these guidelines are beginning to shift as experience with CNVs is gained. The 15q13.3 CNV can commonly be carried by an unaffected family member like a parent or grandparent, as observed in family follow up, despite its known pathogenicity in another family member (van Bon et al., 2009). Ledbetter (Ledbetter 2009) argues correctly that such recurrent lesions are exceptional, and once knowledge and experience is gained, for each of them, the clinical geneticist recognises these situations and can counsel accordingly. This does not necessarily cast doubt on the well-established principles that will always apply to the inheritance of the vast majority of CNVs that are non-recurrent. For the less common of the recurrent CNVs there remains insufficient knowledge and experience at this time to make a judgement on their pathogenicity and penetrance if pathogenic.
The same detection protocols relate to the other recurrent microdeletions commonly associated with epilepsy, at 15q11.2 and 16p13.11 (de Kovel et al., 2010, Heinzen et al., 2010, Mefford et al., 2010). The combined haploinsufficiency of the gene content within the 15q13.3 (7 genes), 16p13.11 (12 genes) and 15q11.2 (8 genes) CNVs represents a major susceptibility determinant in ~3% of genetic generalised epilepsies (Mefford et al., 2010), and all three loci have been associated to varying degrees with a range of neurocognitive and neuropsychiatric disorders. The epilepsy so far associated with deletions of 15q13.3 and 15q11.2 is primarily generalized epilepsy, while deletions of 16p13.11 are seen in both generalized and localization-related epilepsies. Prenatal diagnosis would be extremely problematic for any of these recurrent CNVs due to the inability to accurately predict the associated phenotype or to be certain of a phenotype at all, other than normal. Accurate prediction for a genetically complex disorder would require information on all of its susceptibility genes, not just the one (albeit perhaps the most significant one, when present). The less frequent recurrent CNVs associated with epilepsy are at 1q21.1, 16p12 and two regions within 16p11.2 (de Kovel et al., 2010, Mefford et al., 2010) and until more is known about these they need to be managed the same way as the three more common of the recurrent CNVs. That is, their carriers are at greatly increased risk of the morbidities characterized for each of the lesions. Nothing more can be definitively stated.
The management of rare or novel non-recurrent variants on the other hand will likely remain under “the well established principles that will always apply to the vast majority of inherited CNVs”. That is, de novo lesions are usually pathogenic and familial ones inherited from a phenotypically normal parent are benign. These rare variants are collectively common at around 9% in the genetic generalised and focal epilepsies combined (Mefford et al., 2010) so will need to be routinely tested for, detected and counselled. Many of these, particularly the de novo ones, are likely to confer a relatively large pathogenic risk in the absence of any other carriers in the family displaying evidence to the contrary. Rare but inherited events will remain a diagnostic dilemma for some time, but educated inferences on likely pathogenicity may be gleaned from in silico analysis of the CNV gene content.
Detecting a CNV by array-CGH: What does it mean?
The three most common recurrent CNVs at 15q13.3, 16p13.11 and 15q11.2 have been intensively studied so we now have some knowledge of their significance in relation to epilepsy. In the absence of sufficient knowledge, the rarer recurrent CNVs at 1q21.1, 16p12 and the two regions within 16p11.2 need to be regarded similarly to the three most common CNVs. All of these recurrent CNVs most likely are part of the polygenic architecture underlying susceptibility to epilepsy, but for any of their carriers, these lesions are not the complete picture. Other susceptibility variants at other loci are not yet identifiable but are part of the profile for each affected individual for each of the conditions associated with these deletions.
In the clinical setting, genetic counselling with respect to CNVs remains a challenge. In the case where a patient with epilepsy carries one of the common recurrent CNVs discussed above, it is reasonable to conclude that the CNV is a major genetic factor contributing to the patient’s disease. This is true even when there are unaffected family members with the same CNV, as it has been shown that the penetrance is incomplete. On the other hand, the broad phenotypic spectrum associated with 15q13.3, 16p13.11 and other recurrent CNVs makes predictive counselling more problematic. There is simply not enough known about the additional factors – genetic, epigenetic or environmental – that influence the final phenotypic outcome.
Rare, novel or non-recurrent CNVs on the other hand require serious interpretation to determine their likely pathogenicity or normality. If these non-recurrent CNVs are de novo, then the chance they are pathogenic is high, especially if supported by in silico analysis of genes within the microdeletion suggesting potential relevance to the condition. If familial, then the phenotype of the carrier parent provides the best guide. Interpreting the pathogenicity of a rare, inherited CNV can be complicated (Mencarelli et al., 2008). It requires searching the literature and patient databases for reports of similar or overlapping CNVs in affected individuals. Just as important is the evaluation of control databases and published studies (Itsara et al., 2009, Shaikh et al., 2009) to determine the frequency of overlapping CNVs in unaffected individuals. Finally, the expression pattern, function and disease association (if applicable) of each gene affected by the CNV must be considered. Some general interpretative guidelines for results from array-CGH analysis that can be translated to epilepsy are presented by Sharp (Sharp 2009), Paciorkowski and Fang (Paciorkowski & Fang 2009), Miller et al (Miller et al., 2010) and Xiang et al (Xiang et al., 2010).
Summary and conclusions
Molecular karyotyping has revolutionized the discovery of rare and common copy number changes associated with various diseases – epilepsy is no exception. Large, recurrent microdeletions at 15q13.3, 16p13.11 and 15q11.2 have been established as substantial risk factors for epilepsy. With the continued application of genome-wide approaches to large cohorts of individuals with epilepsy syndromes, additional recurrent and non-recurrent CNVs will be added to this list, facilitating the discovery of novel genes and genomic regions important to the genetic etiology of this complex and common neurological disorder.
Acknowledgement
JM is supported by SA Pathology within the South Australian Department of Health. HCM is supported by the National Institutes of Health (National Institute of Neurologic Disease and Stroke) and is a recipient of the Career Award for Medical Scientists from the Burroughs Wellcome Fund
Footnotes
Disclosure: Neither author has a conflict of interest to disclose. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- Auvin S, Holder-Espinasse M, Lamblin MD, Andrieux J. Array-CGH detection of a de novo 0.7-Mb deletion in 19p13.13 including CACNA1A associated with mental retardation and epilepsy with infantile spasms. Epilepsia. 2009;50:2501–2503. doi: 10.1111/j.1528-1167.2009.02189.x. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Guerrini R. Chromosomal disorders associated with epilepsy. Epileptic Disord. 2005;7:181–192. [PubMed] [Google Scholar]
- Bedoyan JK, Kumar RA, Sudi J, Silverstein F, Ackley T, Iyer RK, Christian SL, Martin DM. Duplication 16p11.2 in a child with infantile seizure disorder. Am J Med Genet A. 2010;152A:1567–1574. doi: 10.1002/ajmg.a.33415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, van Emde Boas W, Engel J, French J, Glauser TA, Mathern GW, Moshe SL, Nordli D, Plouin P, Scheffer IE. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2010;51:676–685. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- Biervert C, Schroeder BC, Kubisch C, Berkovic SF, Propping P, Jentsch TJ, Steinlein OK. A potassium channel mutation in neonatal human epilepsy. Science. 1998;279:403–406. doi: 10.1126/science.279.5349.403. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Boys A, Parrini E, Mignon-Ravix C, McMahon JM, Khantane S, Bertini E, Pallesi E, Missirian C, Zuffardi O, Novara F, Villard L, Giglio S, Chabrol B, Slater HR, Moncla A, Scheffer IE, Guerrini R. Periventricular heterotopia, mental retardation, and epilepsy associated with 5q14.3-q15 deletion. Neurology. 2009;72:784–792. doi: 10.1212/01.wnl.0000336339.08878.2d. [DOI] [PubMed] [Google Scholar]
- Charlier C, Singh NA, Ryan SG, Lewis TB, Reus BE, Leach RJ, Leppert M. A pore mutation in a novel KQT-like potassium channel gene in an idiopathic epilepsy family. Nat Genet. 1998;18:53–55. doi: 10.1038/ng0198-53. [DOI] [PubMed] [Google Scholar]
- Conlin LK, Kramer W, Hutchinson AL, Li X, Riethman H, Hakonarson H, Mulley JC, Scheffer IE, Berkovic SF, Hosain SA, Spinner NB. Molecular analysis of ring chromosome 20 syndrome reveals two distinct groups of patients. J Med Med. 2010 doi: 10.1136/jmg.2010.080382. (in press) [DOI] [PubMed] [Google Scholar]
- De Gregori M, Ciccone R, Magini P, Pramparo T, Gimelli S, Messa J, Novara F, Vetro A, Rossi E, Maraschio P, Bonaglia MC, Anichini C, Ferrero GB, Silengo M, Fazzi E, Zatterale A, Fischetto R, Previdere C, Belli S, Turci A, Calabrese G, Bernardi F, Meneghelli E, Riegel M, Rocchi M, Guerneri S, Lalatta F, Zelante L, Romano C, Fichera M, Mattina T, Arrigo G, Zollino M, Giglio S, Lonardo F, Bonfante A, Ferlini A, Cifuentes F, Van Esch H, Backx L, Schinzel A, Vermeesch JR, Zuffardi O. Cryptic deletions are a common finding in "balanced" reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet. 2007;44:750–762. doi: 10.1136/jmg.2007.052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, Kluck C, Muhle H, von Spiczak S, Ostertag P, Obermeier T, Kleefuss-Lie AA, Hallmann K, Steffens M, Gaus V, Klein KM, Hamer HM, Rosenow F, Brilstra EH, Kasteleijn-Nolst Trenite D, Swinkels ME, Weber YG, Unterberger I, Zimprich F, Urak L, Feucht M, Fuchs K, Moller RS, Hjalgrim H, De Jonghe P, Suls A, Ruckert IM, Wichmann HE, Franke A, Schreiber S, Nurnberg P, Elger CE, Lerche H, Stephani U, Koeleman BP, Lindhout D, Eichler EE, Sander T. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–33. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Bouteiller D, Keren B, Cheuret E, Poirier K, Trouillard O, Benyahia B, Quelin C, Carpentier W, Julia S, Afenjar A, Gautier A, Rivier F, Meyer S, Berquin P, Helias M, Py I, Rivera S, Bahi-Buisson N, Gourfinkel-An I, Cazeneuve C, Ruberg M, Brice A, Nabbout R, Leguern E. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 2009a;5:e1000381. doi: 10.1371/journal.pgen.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Trouillard O, Saint-Martin C, Gourfinkel-An I, Bouteiller D, Carpentier W, Keren B, Abert B, Gautier A, Baulac S, Arzimanoglou A, Cazeneuve C, Nabbout R, LeGuern E. Spectrum of SCN1A gene mutations associated with Dravet syndrome: analysis of 333 patients. J Med Genet. 2009b;46:183–191. doi: 10.1136/jmg.2008.062323. [DOI] [PubMed] [Google Scholar]
- Dibbens LM, Mullen S, Helbig I, Mefford HC, Bayly MA, Bellows S, Leu C, Trucks H, Obermeier T, Wittig M, Franke A, Caglayan H, Yapici Z, Sander T, Eichler EE, Scheffer IE, Mulley JC, Berkovic SF. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum Mol Genet. 2009;18:3626–3631. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, Tarpey PS, Hynes K, Bayly MA, Scheffer IE, Smith R, Bomar J, Sutton E, Vandeleur L, Shoubridge C, Edkins S, Turner SJ, Stevens C, O'Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Jones D, Lee R, Madison M, Mironenko T, Varian J, West S, Widaa S, Wray P, Teague J, Dicks E, Butler A, Menzies A, Jenkinson A, Sheperd R, Gusella JF, Afawi Z, Mazarib A, Neufeld MY, Kivity S, Lev D, Lerman-Sagie T, Korczyn AD, Derry CP, Sutherland GR, Friend K, Shaw M, Corbett M, Kim HG, Geschwind DH, Thomas P, Haan E, Ryan S, McKee S, Berkovic SF, Futreal PA, Stratton MA, Mulley JC, Gecz J. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776–781. doi: 10.1038/ng.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W, McDonald MT, Hoo JJ, French BN, Hudson C, Johnson JP, Ozmore JR, Moeschler JB, Surti U, Escobar LF, El-Khechen D, Gorski JL, Kussmann J, Salbert B, Lacassie Y, Biser A, McDonald-McGinn DM, Zackai EH, Deardorff MA, Shaikh TH, Haan E, Friend KL, Fichera M, Romano C, Gecz J, DeLisi LE, Sebat J, King MC, Shaffer LG, Eichler EE. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin LA, McMahon JM, Iona X, Dibbens L, Pelekanos JT, Zuberi SM, Sadleir LG, Andermann E, Gill D, Farrell K, Connolly M, Stanley T, Harbord M, Andermann F, Wang J, Batish SD, Jones JG, Seltzer WK, Gardner A, Sutherland G, Berkovic SF, Mulley JC, Scheffer IE. The spectrum of SCN1A-related infantile epileptic encephalopathies. Brain. 2007;130:843–852. doi: 10.1093/brain/awm002. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Radtke RA, Urban TJ, Cavalleri GL, Depondt C, Need AC, Walley NM, Nicoletti P, Ge D, Catarino CB, Duncan JS, Kasperaviciute D, Tate SK, Caboclo LO, Sander JW, Clayton L, Linney KN, Shianna KV, Gumbs CE, Smith J, Cronin KD, Maia JM, Doherty CP, Pandolfo M, Leppert D, Middleton LT, Gibson RA, Johnson MR, Matthews PM, Hosford D, Kalviainen R, Eriksson K, Kantanen AM, Dorn T, Hansen J, Kramer G, Steinhoff BJ, Wieser HG, Zumsteg D, Ortega M, Wood NW, Huxley-Jones J, Mikati M, Gallentine WB, Husain AM, Buckley PG, Stallings RL, Podgoreanu MV, Delanty N, Sisodiya SM, Goldstein DB. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet. 2010;86:707–718. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, de Kovel C, Baker C, von Spiczak S, Kron KL, Steinich I, Kleefuss-Lie AA, Leu C, Gaus V, Schmitz B, Klein KM, Reif PS, Rosenow F, Weber Y, Lerche H, Zimprich F, Urak L, Fuchs K, Feucht M, Genton P, Thomas P, Visscher F, de Haan GJ, Moller RS, Hjalgrim H, Luciano D, Wittig M, Nothnagel M, Elger CE, Nurnberg P, Romano C, Malafosse A, Koeleman BP, Lindhout D, Stephani U, Schreiber S, Eichler EE, Sander T. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron SE, Cox K, Grinton BE, Zuberi SM, Kivity S, Afawi Z, Straussberg R, Berkovic SF, Scheffer IE, Mulley JC. Deletions or duplications in KCNQ2 can cause benign familial neonatal seizures. J Med Genet. 2007;44:791–796. doi: 10.1136/jmg.2007.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron SE, Scheffer IE, Grinton BE, Eyre H, Oliver KL, Bain S, Berkovic SF, Mulley JC. Familial neonatal seizures with intellectual disability caused by microduplication of chromosome 2q24.3. Epilepsia. 2010;51:1865–1869. doi: 10.1111/j.1528-1167.2010.02558.x. [DOI] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, Mefford H, Ying P, Nickerson DA, Eichler EE. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Wang JW, Ishii A, Kojima T, Wakai S, Kizawa T, Fujimoto Y, Kikkawa K, Yoshimura K, Inoue T, Yasumoto S, Ogawa A, Kaneko S, Hirose S. Deletions involving both KCNQ2 and CHRNA4 present with benign familial neonatal seizures. Neurology. 2009;73:1214–1217. doi: 10.1212/WNL.0b013e3181bc0158. [DOI] [PubMed] [Google Scholar]
- Kwan P, Brodie MJ. Early identification of refractory epilepsy. N Engl J Med. 2000;342:314–319. doi: 10.1056/NEJM200002033420503. [DOI] [PubMed] [Google Scholar]
- Ledbetter DH. New microdeletion syndromes: complex, but no new paradigms. J Med Genet. 2009;46:576. doi: 10.1136/jmg.2009.068916. [DOI] [PubMed] [Google Scholar]
- Lepichon JB, Bittel DC, Graf WD, Yu S. A 15q13.3 homozygous microdeletion associated with a severe neurodevelopmental disorder suggests putative functions of the TRPM1, CHRNA7, and other homozygously deleted genes. Am J Med Genet A. 2010;152A:1300–1304. doi: 10.1002/ajmg.a.33374. [DOI] [PubMed] [Google Scholar]
- Lossin C. A catalog of SCN1A variants. Brain Dev. 2009;31:114–130. doi: 10.1016/j.braindev.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makoff AJ, Flomen RH. Detailed analysis of 15q11-q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader-Willi, Angelman, and inv dup(15) syndromes. Genome Biol. 2007;8:R114. doi: 10.1186/gb-2007-8-6-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini C, Mei D, Temudo T, Ferrari AR, Buti D, Dravet C, Dias AI, Moreira A, Calado E, Seri S, Neville B, Narbona J, Reid E, Michelucci R, Sicca F, Cross HJ, Guerrini R. Idiopathic epilepsies with seizures precipitated by fever and SCN1A abnormalities. Epilepsia. 2007;48:1678–1685. doi: 10.1111/j.1528-1167.2007.01122.x. [DOI] [PubMed] [Google Scholar]
- Marini C, Scheffer IE, Nabbout R, Mei D, Cox K, Dibbens LM, McMahon JM, Iona X, Carpintero RS, Elia M, Cilio MR, Specchio N, Giordano L, Striano P, Gennaro E, Cross JH, Kivity S, Neufeld MY, Afawi Z, Andermann E, Keene D, Dulac O, Zara F, Berkovic SF, Guerrini R, Mulley JC. SCN1A duplications and deletions detected in Dravet syndrome: Implications for molecular diagnosis. Epilepsia. 2009;50:1670–1678. doi: 10.1111/j.1528-1167.2009.02013.x. [DOI] [PubMed] [Google Scholar]
- McMahon JM, Scheffer IE, Nicholl JK, Waters W, Eyre H, Hinton L, Nelson P, Yu S, Dibbens LM, Berkovic SF, Mulley JC. Detection of microchromosomal aberrations in refractory epilepsy: a pilot study. Epileptic Disord. 2010;12:192–198. doi: 10.1684/epd.2010.0326. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, Franke A, Malafosse A, Genton P, Thomas P, Gurnett CA, Schreiber S, Bassuk AG, Guipponi M, Stephani U, Helbig I, Eichler EE. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genetics. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Mulley JC. Genetically complex epilepsies, copy number variations and syndrome constellations. Genome Med. 2010;2:71. doi: 10.1186/gm192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei D, Marini C, Novara F, Bernardina BD, Granata T, Fontana E, Parrini E, Ferrari AR, Murgia A, Zuffardi O, Guerrini R. Xp22.3 genomic deletions involving the CDKL5 gene in girls with early onset epileptic encephalopathy. Epilepsia. 2010;51:647–654. doi: 10.1111/j.1528-1167.2009.02308.x. [DOI] [PubMed] [Google Scholar]
- Mencarelli MA, Katzaki E, Papa FT, Sampieri K, Caselli R, Uliana V, Pollazzon M, Canitano R, Mostardini R, Grosso S, Longo I, Ariani F, Meloni I, Hayek J, Balestri P, Mari F, Renieri A. Private inherited microdeletion/microduplications: implications in clinical practice. Eur J Med Genet. 2008;51:409–416. doi: 10.1016/j.ejmg.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhle H, Steinich I, von Spiczak S, Franke A, Weber Y, Lerche H, Wittig M, Heidemann S, Suls A, de Jonghe P, Marini C, Guerrini R, Scheffer IE, Berkovic SF, Stephani U, Siebert R, Sander T, Helbig I, Tonnies H. A duplication in 1q21.3 in a family with early onset and childhood epilepsy. Epilepsia. 2010 doi: 10.1111/j.1528-1167.2010.02712.x. (in press) [DOI] [PubMed] [Google Scholar]
- Mulley JC, Dibbens LM. Chipping away at the common epilepsies with complex genetics: the 15q13.3 microdeletion shows the way. Genome Med. 2009;1:33. doi: 10.1186/gm33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulley JC, Nelson P, Guerrero S, Dibbens L, Iona X, McMahon JM, Harkin L, Schouten J, Yu S, Berkovic SF, Scheffer IE. A new molecular mechanism for severe myoclonic epilepsy of infancy: exonic deletions in SCN1A. Neurology. 2006;67:1094–1095. doi: 10.1212/01.wnl.0000237322.04338.2b. [DOI] [PubMed] [Google Scholar]
- Orrico A, Zollino M, Galli L, Buoni S, Marangi G, Sorrentino V. Late-onset Lennox- Gastaut syndrome in a patient with 15q11.2-q13.1 duplication. Am J Med Genet A. 2009;149A:1033–1035. doi: 10.1002/ajmg.a.32785. [DOI] [PubMed] [Google Scholar]
- Paciorkowski AR, Fang M. Chromosomal microarray interpretation: what is a child neurologist to do? Pediatr Neurol. 2009;41:391–398. doi: 10.1016/j.pediatrneurol.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Guitart M, Armengol L, Gratacos M, Estivill X. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur J Hum Genet. 2002;10:26–35. doi: 10.1038/sj.ejhg.5200760. [DOI] [PubMed] [Google Scholar]
- Regier DA, Friedman JM, Marra CA. Value for money? Array genomic hybridization for diagnostic testing for genetic causes of intellectual disability. Am J Hum Genet. 2010;86:765–772. doi: 10.1016/j.ajhg.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluth-Bolard C, Delobel B, Sanlaville D, Boute O, Cuisset JM, Sukno S, Labalme A, Duban-Bedu B, Plessis G, Jaillard S, Dubourg C, Henry C, Lucas J, Odent S, Pasquier L, Copin H, Latour P, Cordier MP, Nadeau G, Till M, Edery P, Andrieux J. Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: array CGH study of 47 unrelated cases. Eur J Med Genet. 2009;52:291–296. doi: 10.1016/j.ejmg.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O'Hara R, Casalunovo T, Conlin LK, D'Arcy M, Frackelton EC, Geiger EA, Haldeman-Englert C, Imielinski M, Kim CE, Medne L, Annaiah K, Bradfield JP, Dabaghyan E, Eckert A, Onyiah CC, Ostapenko S, Otieno FG, Santa E, Shaner JL, Skraban R, Smith RM, Elia J, Goldmuntz E, Spinner NB, Zackai EH, Chiavacci RM, Grundmeier R, Rappaport EF, Grant SF, White PS, Hakonarson H. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ. Emerging themes and new challenges in defining the role of structural variation in human disease. Hum Mutat. 2009;30:135–144. doi: 10.1002/humu.20843. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De Gregori M, Ciccone R, Broomer A, Casuga I, Wang Y, Xiao C, Barbacioru C, Gimelli G, Bernardina BD, Torniero C, Giorda R, Regan R, Murday V, Mansour S, Fichera M, Castiglia L, Failla P, Ventura M, Jiang Z, Cooper GM, Knight SJ, Romano C, Zuffardi O, Chen C, Schwartz CE, Eichler EE. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinawi M, Schaaf CP, Bhatt SS, Xia Z, Patel A, Cheung SW, Lanpher B, Nagl S, Herding HS, Nevinny-Stickel C, Immken LL, Patel GS, German JR, Beaudet AL, Stankiewicz P. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet. 2009;41:1269–1271. doi: 10.1038/ng.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NA, Charlier C, Stauffer D, DuPont BR, Leach RJ, Melis R, Ronen GM, Bjerre I, Quattlebaum T, Murphy JV, McHarg ML, Gagnon D, Rosales TO, Peiffer A, Anderson VE, Leppert M. A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat Genet. 1998;18:25–29. doi: 10.1038/ng0198-25. [DOI] [PubMed] [Google Scholar]
- Singh R, Gardner RJ, Crossland KM, Scheffer IE, Berkovic SF. Chromosomal abnormalities and epilepsy: a review for clinicians and gene hunters. Epilepsia. 2002;43:127–140. doi: 10.1046/j.1528-1157.2002.19498.x. [DOI] [PubMed] [Google Scholar]
- Steinlein OK, Magnusson A, Stoodt J, Bertrand S, Weiland S, Berkovic SF, Nakken KO, Propping P, Bertrand D. An insertion mutation of the CHRNA4 gene in a family with autosomal dominant nocturnal frontal lobe epilepsy. Hum Mol Genet. 1997;6:943–947. doi: 10.1093/hmg/6.6.943. [DOI] [PubMed] [Google Scholar]
- Steinlein OK, Mulley JC, Propping P, Wallace RH, Phillips HA, Sutherland GR, Scheffer IE, Berkovic SF. A missense mutation in the neuronal nicotinic acetylcholine receptor alpha 4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 1995;11:201–203. doi: 10.1038/ng1095-201. [DOI] [PubMed] [Google Scholar]
- Suls A, Velizarova R, Yordanova I, Deprez L, Van Dyck T, Wauters J, Guergueltcheva V, Claes LR, Kremensky I, Jordanova A, De Jonghe P. Four generations of epilepsy caused by an inherited microdeletion of the SCN1A gene. Neurology. 2010;75:72–76. doi: 10.1212/WNL.0b013e3181e62088. [DOI] [PubMed] [Google Scholar]
- van Bon BW, Mefford HC, Menten B, Koolen DA, Sharp AJ, Nillesen WM, Innis JW, de Ravel TJ, Mercer CL, Fichera M, Stewart H, Connell LE, Ounap K, Lachlan K, Castle B, Van der Aa N, van Ravenswaaij C, Nobrega MA, Serra-Juhe C, Simonic I, de Leeuw N, Pfundt R, Bongers EM, Baker C, Finnemore P, Huang S, Maloney VK, Crolla JA, van Kalmthout M, Elia M, Vandeweyer G, Fryns JP, Janssens S, Foulds N, Reitano S, Smith K, Parkel S, Loeys B, Woods CG, Oostra A, Speleman F, Pereira AC, Kurg A, Willatt L, Knight SJ, Vermeesch JR, Romano C, Barber JC, Mortier G, Perez-Jurado LA, Kooy F, Brunner HG, Eichler EE, Kleefstra T, de Vries BB. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46:511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassos E, Collier DA, Holden S, Patch C, Rujescu D, St Clair D, Lewis CM. Penetrance for copy number variants associated with schizophrenia. Hum Mol Genet. 2010;19:3477–3481. doi: 10.1093/hmg/ddq259. [DOI] [PubMed] [Google Scholar]
- Veredice C, Bianco F, Contaldo I, Orteschi D, Stefanini MC, Battaglia D, Lettori D, Guzzetta F, Zollino M. Early onset myoclonic epilepsy and 15q26 microdeletion: Observation of the first case. Epilepsia. 2009;50:1810–1815. doi: 10.1111/j.1528-1167.2009.02078.x. [DOI] [PubMed] [Google Scholar]
- Wang JW, Kurahashi H, Ishii A, Kojima T, Ohfu M, Inoue T, Ogawa A, Yasumoto S, Oguni H, Kure S, Fujii T, Ito M, Okuno T, Shirasaka Y, Natsume J, Hasegawa A, Konagaya A, Kaneko S, Hirose S. Microchromosomal deletions involving SCN1A and adjacent genes in severe myoclonic epilepsy in infancy. Epilepsia. 2008;49:1528–1534. doi: 10.1111/j.1528-1167.2008.01609.x. [DOI] [PubMed] [Google Scholar]
- Xiang B, Zhu H, Shen Y, Miller DT, Lu K, Hu X, Andersson HC, Narumanchi TM, Wang Y, Martinez JE, Wu BL, Li P, Li MM, Chen TJ, Fan YS. Genome-wide oligonucleotide array comparative genomic hybridization for etiological diagnosis of mental retardation: a multicenter experience of 1499 clinical cases. J Mol Diagn. 2010;12:204–212. doi: 10.2353/jmoldx.2010.090115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung A, Bruno D, Scheffer IE, Carranza D, Burgess T, Slater HR, Amor DJ. 4.45 Mb microduplication in chromosome band 14q12 including FOXG1 in a girl with refractory epilepsy and intellectual impairment. Eur J Med Genet. 2009;52:440–442. doi: 10.1016/j.ejmg.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Yu S, Bittel DC, Kibiryeva N, Zwick DL, Cooley LD. Validation of the Agilent 244K oligonucleotide array-based comparative genomic hybridization platform for clinical cytogenetic diagnosis. Am J Clin Pathol. 2009;132:349–360. doi: 10.1309/AJCP1BOUTWF6ERYS. [DOI] [PubMed] [Google Scholar]
- Zody MC, Garber M, Sharpe T, Young SK, Rowen L, O'Neill K, Whittaker CA, Kamal M, Chang JL, Cuomo CA, Dewar K, FitzGerald MG, Kodira CD, Madan A, Qin S, Yang X, Abbasi N, Abouelleil A, Arachchi HM, Baradarani L, Birditt B, Bloom S, Bloom T, Borowsky ML, Burke J, Butler J, Cook A, DeArellano K, DeCaprio D, Dorris L, 3rd, Dors M, Eichler EE, Engels R, Fahey J, Fleetwood P, Friedman C, Gearin G, Hall JL, Hensley G, Johnson E, Jones C, Kamat A, Kaur A, Locke DP, Madan A, Munson G, Jaffe DB, Lui A, Macdonald P, Mauceli E, Naylor JW, Nesbitt R, Nicol R, O'Leary SB, Ratcliffe A, Rounsley S, She X, Sneddon KM, Stewart S, Sougnez C, Stone SM, Topham K, Vincent D, Wang S, Zimmer AR, Birren BW, Hood L, Lander ES, Nusbaum C. Analysis of the DNA sequence and duplication history of human chromosome 15. Nature. 2006;440:671–675. doi: 10.1038/nature04601. [DOI] [PubMed] [Google Scholar]