SUMMARY
Ordered collections of Arabidopsis thaliana lines containing mapped T-DNA insertions have become an important resource for plant scientists performing genetic studies. Previous reports have indicated that T-DNA insertion lines can have chromosomal translocations associated with the T-DNA insertion site, but the prevalence of these rearrangements has not been well documented. To determine the frequency with which translocations are present in a widely-used collection of T-DNA insertion lines, we analyzed 64 independent lines from the Salk T-DNA mutant collection. Chromosomal translocations were detected in 12 of the 64 lines surveyed (19%). Two assays were used to screen the T-DNA lines for translocations: pollen viability and genome-wide genetic mapping. Although the measurement of pollen viability is an indirect screen for the presence of a translocation, all 11 of the T-DNA lines showing an abnormal pollen phenotype were found to contain a translocation when analyzed using genetic mapping. A normal pollen phenotype does not, however, guarantee the absence of a translocation. We observed one T-DNA line with normal pollen that nevertheless had a translocation based on genetic mapping results. One additional phenomenon that we observed through our genetic mapping experiments was that the T-DNA junctions on the 5′- and 3′-sides of a targeted gene can genetically separate from each other in some cases. Two of the lines in our survey displayed this ‘T-DNA borders separate’ phenomenon. Experimental procedures for efficiently screening T-DNA lines for the presence of chromosomal abnormalities are presented and discussed.
Keywords: T-DNA, translocation, chromosome, Arabidopsis, pollen lethality
INTRODUCTION
T-DNA insertional mutants are a widely used resource in Arabidopsis thaliana research. Currently, there are over 325 000 publicly available Arabidopsis T-DNA insertion lines (O’Malley and Ecker, 2010). Although the precise mechanism of T-DNA integration into the plant genome is not well understood, the site of T-DNA insertion is often associated with local DNA disturbances including small deletions, small duplications, and/or the addition of filler sequence of unknown origin (Gheysen et al., 1991; Mayerhofer et al., 1991; Ohba et al., 1995). In addition to these localized disturbances, T-DNA lines have also been shown to contain chromosomal translocations or paracentric inversions associated with the T-DNA insertion site (Ray et al., 1997; Laufs et al., 1999; Forsbach et al., 2003; Lafleuriel et al., 2004; Yuen et al., 2005; Curtis et al., 2009). In other cases, more complex rearrangements of the genome have also been observed. Nacry et al. (1998) described an Arabidopsis T-DNA line containing a reciprocal translocation between chromosomes II and III, a large inversion on chromosome II, and a short deletion on chromosome III. Tax and Vernon (2001) reported two examples of T-DNA lines with an internal translocation as well as the duplication of the disrupted gene at the T-DNA insertion site. Chromosomal rearrangements associated with T-DNA insertion are not limited to Arabidopsis, as they have also been documented in rice and tobacco (Ohba et al., 1995; Takano et al., 1997).
All of the studies described above have involved documenting the nature of the genomic rearrangements present in one or a few isolated T-DNA lines; the frequency with which translocations occur in collections of T-DNA lines was not addressed in those studies. There have, however, been two studies published that investigated the frequency of translocations in collections of T-DNA lines. Castle et al. (1993) found evidence of chromosomal translocations or inversions in seven of the 56 Arabidopsis lines (12.5%) carrying embryo lethal mutations that they genetically mapped, while Budziszewski et al. (2001) found that 11 of the 32 lines (34%) carrying seedling lethal mutants that they analyzed had potential chromosomal abnormalities such as translocations. Because both of these studies involved estimating the frequency of translocations using T-DNA lines that were pre-selected for the presence of lethal mutations, these samples may have been enriched for plants carrying chromosomal abnormalities. We were therefore interested in measuring the frequency with which chromosomal translocations occur in T-DNA lines using a less biased sample. Accordingly, all of the Salk T-DNA lines (Alonso et al., 2003) used in our study were viable when the T-DNA mutant allele was in the homozygous state. In total, we characterized 64 independent T-DNA insertional lines from the Salk collection and found that 12 of the 64 lines (19%) had chromosomal translocations.
RESULTS
Pollen viability
As first noted by Burnham (1930), plants that are heterozygous for a reciprocal translocation are predicted to create ca. 50% non-viable gametes. By contrast, plants that are homozygous for a reciprocal translocation are genetically balanced and therefore are predicted to produce gametes that are all viable. For a detailed discussion of the consequences of a reciprocal translocation on gametogenesis see Curtis et al. (2009). As a first estimate of the frequency with which translocations are present in T-DNA lines from the Salk collection (Alonso et al., 2003), we examined the viability of pollen collected from plants that were heterozygous for each of the T-DNA insertion alleles. Plants heterozygous for a translocation are expected to produce 50% non-viable pollen. The 64 Salk T-DNA lines used for our study are listed in Table 1. This collection of genes is composed mainly of protein kinase genes that our laboratory is studying. Although this is not a random sample of Salk T-DNA lines, all of these lines are viable when the T-DNA insertion is in the homozygous mutant state (data not shown).
Table 1.
Summary of Salk T-DNA lines tested for chromosomal translocations
| Gene locus ID | Common name |
Salk line number |
Pollen phenotypea |
Genetic mapping |
T-DNA left borderb |
T-DNA right borderc |
5′/ 3′ border separationd |
|---|---|---|---|---|---|---|---|
| At1g59580 | MPK2 | S_047422 | Normal | Translocation | 5′ and 3′ | n.a. | Yes |
| At1g01560 | MPK11 | S_049532 | Abnormal | Translocation | 5′ and 3′ | n.a. | Yes |
| At3g59790 | MPK10 | S_039102 | Abnormal | Translocation | 5′ and 3′ | n.a. | No |
| At3g14720 | MPK19 | S_075214 | Abnormal | Translocation | 5′ and 3′ | n.a. | No |
| At1g73660 | Raf5 | S_001982 | Abnormal | Translocation | 5′ and 3′ | n.a. | No |
| At4g24480 | Raf6 | S_025685 | Abnormal | Translocation | 5′ and 3′ | n.a. | No |
| At3g50730 | Raf45 | S_139852 | Abnormal | Translocation | 5′ and 3′ | n.a. | No |
| At3g58760 | Raf47 | S_033203 | Abnormal | Translocation | 5′ and 3′ | n.a. | No |
| At5g55560 | ZIK8 | S_102847 | Abnormal | Translocation | 5′ | 3′ | No |
| At4g26890 | MAPKKK16 | S_003255 | Abnormal | Translocation | 3′ | n.d. | n.a. |
| At1g14000 | Raf17 | S_018487 | Abnormal | Translocation | 3′ | n.d. | n.a. |
| At3g22750 | Raf39 | S_027735 | Abnormal | Translocation | 3′ | n.d. | n.a. |
| At1g10210 | MPK1 | S_063847 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At3g45640 | MPK3 | S_151594 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At1g18150 | MPK8 | S_037501 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At3g18040 | MPK9 | S_064439 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At1g07880 | MPK13 | S_130193 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At4g36450 | MPK14 | S_022928 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At5g19010 | MPK16 | S_059737 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At1g53510 | MPK18 | S_069399 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At4g08500 | MEKK1 | S_052557 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At5g03730 | CTR1 | S_122868 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At4g14780 | Raf26 | S_029496 | Normal | Normal | 5′ and 3′ | n.a. | No |
| At4g01370 | MPK4 | S_056245 | Normal | Normal | 3′ | 5′ | No |
| At1g53165 | MAP4Kα1 | S_033601 | Normal | Normal | 3′ | 5′ | No |
| At2g01450 | MPK17 | S_020801 | Normal | Normal | 5′ | n.d. | n.a. |
| At2g42880 | MPK20 | S_090005 | Normal | Normal | 5′ | n.d. | n.a. |
| At2g43790 | MPK6 | S_073907 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At3g07980 | MPK4Ke2 | S_084747 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At1g09000 | ANP1 | S_033751 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At1g54960 | ANP2 | S_144973 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At3g06030 | ANP3 | S_081990 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At5g55090 | MAPKKK15 | S_047537 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At1g08720 | EDR1 | S_127158 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At1g67890 | Raf11 | S_139302 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At2g31010 | Raf13 | S_137974 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At1g04700 | Raf16 | S_101306 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At1g16270 | Raf18 | S_062362 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At4g38470 | Raf30 | S_112195 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At5g50180 | Raf34 | S_119787 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At3g46930 | Raf43 | S_022462 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At3g50720 | Raf44 | S_103601 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At1g70430 | MAP4K7 | S_108286 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At1g54510 | NEK1 | S_065629 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At3g04810 | NEK2 | S_062288 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At5g28290 | NEK3 | S_123055 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At3g51630 | ZIK1 | S_039004 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At1g64630 | ZIK10 | S_071328 | Normal | n.c. | 5′ and 3′ | n.a. | No |
| At3g06630 | Raf8 | S_120720 | Normal | n.c. | 5′ | 3′ | No |
| At2g24360 | Raf22 | S_120808 | Normal | n.c. | 5′ | n.d. | n.a. |
| At2g35050 | Raf24 | S_107170 | Normal | n.c. | 5′ | n.d. | n.a. |
| AT3G46920 | Raf42 | S_121884 | Normal | n.c. | 5′ | n.d. | n.a. |
| At1g79640 | MAP4K8 | S_067866 | Normal | n.c. | 5′ | n.d. | n.a. |
| At3g22420 | ZIK3 | S_107443 | Normal | n.c. | 5′ | n.d. | n.a. |
| At3g06620 | Raf7 | S_000887 | Normal | n.c. | 3′ | 5′ | No |
| At2g42640 | Raf14 | S_076976 | Normal | n.c. | 3′ | 5′ | No |
| At3g13530 | MPK4Ke1 | S_061724 | Normal | n.c. | 3′ | n.d. | n.a. |
| At4g12020 | MEKK4 | S_097632 | Normal | n.c. | 3′ | n.d. | n.a. |
| At3g06640 | Raf9 | S_022298 | Normal | n.c. | 3′ | n.d. | n.a. |
| At1g79570 | Raf20 | S_143032 | Normal | n.c. | 3′ | n.d. | n.a. |
| At3g59830 | Raf46 | S_130077 | Normal | n.c. | 3′ | n.d. | n.a. |
| At1g69220 | MAP4K3 | S_051369 | Normal | n.c. | 3′ | n.d. | n.a. |
| At3g12200 | NEK7 | S_134382 | Normal | n.c. | 3′ | n.d. | n.a. |
| At3g15220 | MAP4Kα2 | S_116301 | Normal | n.c. | 3′ | n.d. | n.a. |
n.a., not applicable; n.c., not completed; n.d., not detected.
‘Abnormal’ indicates that ca. 50% of the pollen collected from a plant heterozygous for the T-DNA insertion was non-viable and ‘normal’ indicates that a large majority of the pollen was viable.
Presence of the T-DNA left border sequence on either the 5′ and/or 3′ side of the target gene as described in Figure 4 is indicated.
Presence of the T-DNA right border sequence on either the 5′ or 3′ side of the target gene.
‘Yes’ indicates that the 5′- and 3′-T-DNA junctions for the given T-DNA line do not co-segregate 100% of the time.
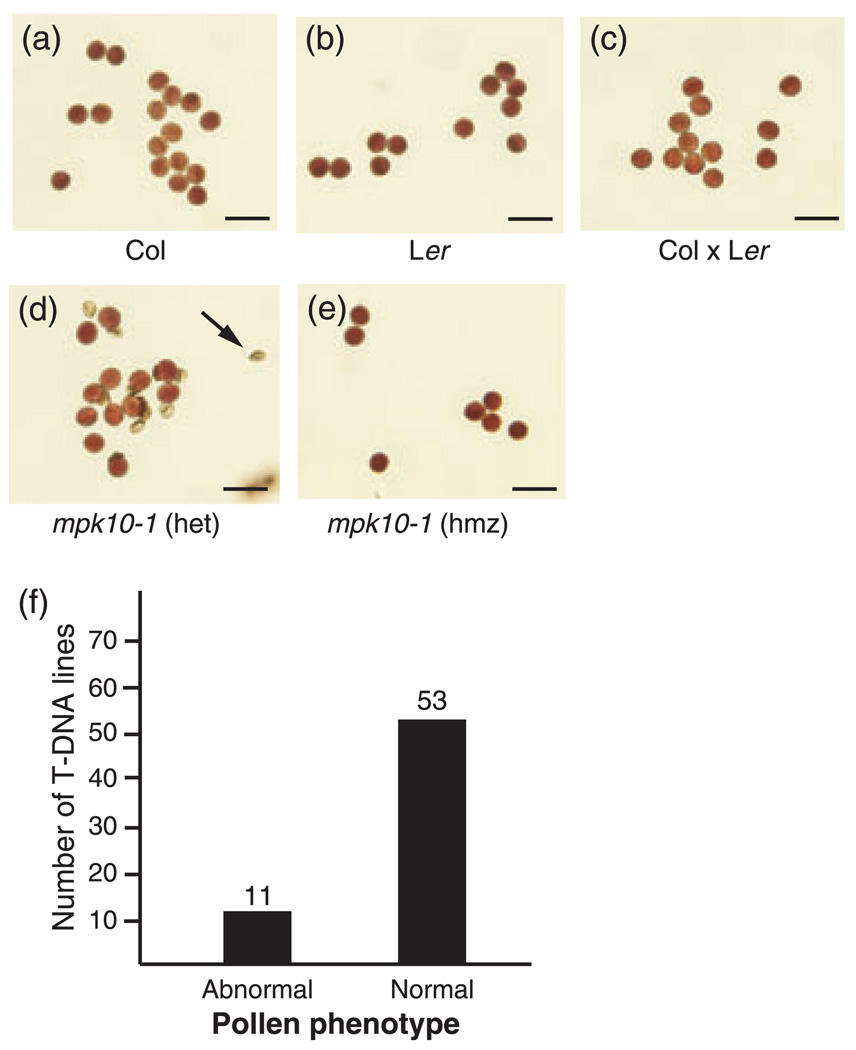
To generate plants that were heterozygous for each of these T-DNA insertions we crossed each T-DNA line to wild-type Landsberg erecta (Ler) ecotype plants. PCR genotyping was then performed using PCR primers that target the T-DNA/plant DNA junctions for each of the T-DNA insertions to confirm that the F1 plants were indeed heterozygous for the given T-DNA allele (data not shown). Pollen was collected from each of the resulting F1 plants and examined for viability using Alexander staining (Alexander, 1969). After staining, viable pollen grains appear round with a green cell wall and darkly stained cytoplasm. Non-viable pollen grains are shriveled with a green cell wall and no apparent cytoplasm (Alexander, 1969). If the majority of the pollen grains appeared viable, we characterized the plants as having a normal pollen phenotype. If ca. 50% of the pollen grains were non-viable, we characterized the plants as having an abnormal pollen phenotype. Of the 64 Salk lines examined, 11 (17%) had an abnormal pollen phenotype, while 53 (83%) were normal (Figure 1f, Table 1).
Figure 1.
Pollen viability screen. (a–e) Alexander stain for pollen viability. Viable pollen grains are round and purple with a green cell wall. Non-viable pollen is green and shriveled, indicated by black arrowhead. The genotypes of the plants that produced the pollen are indicated below each image. Scale bars are 25 µm.
(a) Columbia ecotype (Col).
(b) Landsberg erecta ecotype (Ler).
(c) Columbia × Landsberg erecta F1 heterozygote (Col × Ler).
(d) mpk10-1 heterozygote.
(e) mpk10-1 homozygous mutant.
(f) Pollen phenotype of the 64 Salk T-DNA lines used in this study. ‘Normal’ indicates that the large majority of the pollen produced by a plant heterozygous for the T-DNA insertion was viable. ‘Abnormal’ indicates that ca. 50% of the pollen produced by the heterozygote was non-viable.
The observation that ca. 50% of the pollen collected from an F1 plant is non-viable is consistent with the presence of a translocation in the genome of that plant, but this observation alone does not prove the presence of a translocation. The production of ca. 50% non-viable pollen by a heterozygous plant can be caused by a mutation in a male gametophyte essential gene, a chromosomal inversion, or a chromosomal translocation (Redei and Koncz, 1992). Further genetic characterization is necessary to differentiate between these possibilities. For this reason, we performed genome-wide genetic mapping to more directly test for the presence of translocations.
Genetic mapping
Genetic mapping is used to determine the relative location of a particular mutation in the genome. Genetic mapping is based on the counting of recombination events between the mutation of interest and genetic markers with known chromosomal locations. Normally, a T-DNA insertion should be genetically linked to one location in the genome, i.e. the location of the disrupted gene as indicated by DNA sequencing of the T-DNA/plant DNA junctions at the T-DNA insertion point (Sessions et al., 2002). If there is a translocation associated with a T-DNA insertion, the T-DNA element will be genetically linked to mapping markers on two chromosomes (Burnham, 1930; Castle et al., 1993). Linkage will be observed between the T-DNA and markers near the expected T-DNA insertion site as well as markers located on a second chromosome near the translocation break-point.
T-DNA lines from the Salk collection are derived from the Columbia (Col) ecotype (Alonso et al., 2003). For the pollen viability experiment described above, we created Col × Ler F1 plants for each T-DNA line. In order to generate genetic mapping populations, we next crossed a Col × Ler F1 plant for each T-DNA insertion line chosen for mapping to a wild-type Ler plant (Figure 2a). The resulting F2 progeny were then used for genetic mapping.
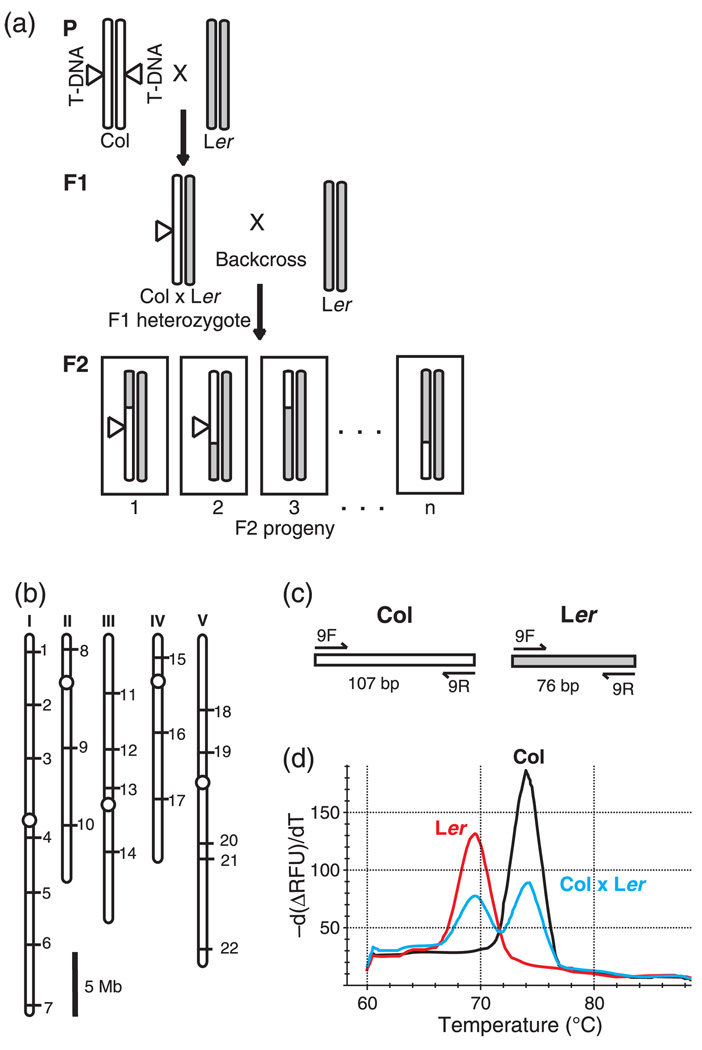
Figure 2.
Genetic mapping strategy.
(a) Creation of the F2 mapping populations. For clarity, only one representative pair of homologous chromosomes is shown. In the parental generation (P), a plant homozygous for a T-DNA insertion (indicated by a triangle) in the Columbia (Col) ecotype is crossed to a wild-type Lansberg erecta (Ler) ecotype plant. The resulting Col × Ler heterozygous plant (F1) is then crossed to another wild-type Ler plant to create a backcrossed mapping population (F2). Example crossover events are indicated in the F2 progeny. Columbia (Col) ecotype chromosomes are white. Landsberg erecta (Ler) ecotype chromosomes are grey.
(b) Locations of the insertion/deletion (INDEL) mapping markers INDEL-1 to INDEL-22 on the five Arabidopsis chromosomes (See also Table S1). Scale bar is 5 Megabases.
(c) PCR product sizes for mapping marker INDEL-9. Primers (indicated by arrows) with predicted PCR product sizes in base pairs (bp) for Columbia (Col) and Landsberg erecta (Ler) templates.
(d) Representative melt-curve genotyping data collected using mapping marker INDEL-9. Melt profiles are shown for INDEL-9 PCR products from plants that are homozygous Columbia (Col), homozygous Landsberg erecta (Ler), and Columbia × Landsberg erecta F1 heterozygote (Col × Ler).
Once a mapping population has been created, the next step in the genetic mapping process is to genotype individual F2 progeny using genetic markers distributed throughout the genome. For our mapping experiments we made use of insertion/deletion (INDEL) polymorphisms between the Col and Ler ecotypes that had been previously characterized (Salathia et al., 2007). The 22 INDEL markers that we chose are spaced across all five chromosomes so that a given gene is no more than ca. 20 centiMorgans from the nearest marker (Figure 2b and Table S1). We designed PCR primers for each marker that are able to amplify products from both Landsberg and Columbia templates (Figure 2c). Differences in the sizes of the PCR products amplified from the two ecotypes allow one to determine if plants are homozygous Col, homozygous Ler, or heterozygous at each of the marker loci (Figure 2d).
We used these 22 INDEL markers to genetically map the locations of the T-DNA insertions in 27 of the Salk T-DNA lines in our survey. We mapped all 11 of the T-DNA lines that produced ca. 50% non-viable pollen, plus an additional 16 lines with a normal pollen phenotype (Table 1). In each case we began by performing PCR reactions to identify F2 individuals containing the T-DNA insertion for each Salk line. For F2 plants carrying the T-DNA insertion, INDEL markers that are not linked to the T-DNA will segregate independently from the T-DNA. In this case the number of F2 plants with the homozygous Ler genotype will be approximately equal to the number with the Col × Ler heterozygous genotype. If, however, an INDEL marker is genetically linked to the T-DNA insertion, then one would expect to see an over-representation of plants with the Col × Ler heterozygous genotype at that INDEL locus. The recombination frequency between the T-DNA insertions and each of the mapping markers was calculated, and the chi-squared test was used to determine if the recombination rate was significantly different from 50% (Figure 3, Figure 4, Figure S1, Figure S2, Figure S3 and Figure S4) (Weigel and Glazebrook, 2002). A recombination rate significantly <50% indicates genetic linkage between the T-DNA insertion and that mapping marker.
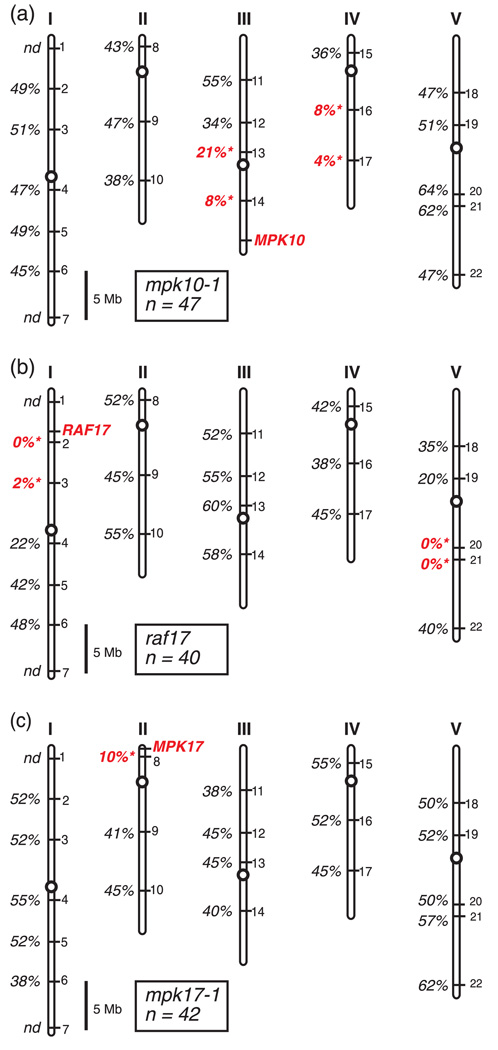
Figure 3.
Genetic mapping data for representative T-DNA lines. Locations of the mapping markers INDEL-1 to INDEL-22 on Arabidopsis chromosomes I–V are indicated by numbers on the right-hand side of each chromosome. The physical location of the wild-type gene disrupted by the T-DNA insertion is shown in red. Percent recombination between the T-DNA insert and each mapping marker is shown. ‘nd’ indicates mapping was not done for that marker. A recombination rate significantly different from 50% is indicated in red (P < 0.001). Scale bar is 5 Megabases.
(a) Mapping data for the mpk10-1 T-DNA line.
(b) Mapping data for the raf17 T-DNA line.
(c) Mapping data for the mpk17-1 T-DNA line.
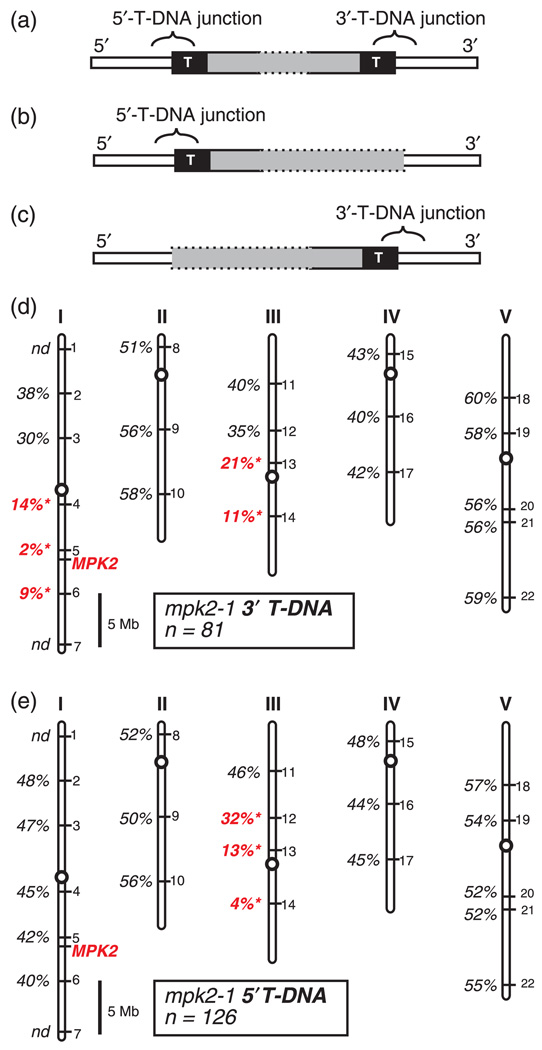
Figure 4.
Genetic mapping the 5′- and 3′-T-DNA junctions of mpk2-1, (a–c) Schematic representation of theoretical T-DNA junction configurations. Generic T-DNA insertion loci are depicted with the T-DNA vector sequence indicated by gray, the Arabidopsis chromosome indicated in white, and the T-DNA border sequences represented as black boxes with the letter ‘T.’
(a) LocuswithdetectableT-DNAjunctionsonboththe5′ and 3′sidesofthe gene.
(b) Locus with a detectable T-DNA junction on only the 5′ side of the gene.
(c) Locus with a detectable T-DNA junction on only the 3′ side of the gene. (d–e) Locations of the mapping markers INDEL-1 to INDEL-22 on Arabidopsis chromosomes I–V are indicated by numbers on the right-hand side of each chromosome. Physical location of the MPK2 gene is shown in red. Percent recombination between each mpk2-1 T-DNA junction and the panel of mapping markers is shown. ‘nd’ indicates mapping not done for that marker. A recombination rate significantly different from 50% is indicated in red (P < 0.001). Scale bar is 5 Megabases.
(d) Mapping data for the mpk2-1 5′-T-DNA junction.
(e) Mapping data for the mpk2-1 3′-T-DNA junction.
Representative mapping data from two T-DNA lines that carry translocations are shown in Figure 3. In the first example, significant genetic linkage is observed between the mapping markers INDEL-13 and INDEL-14 on chromosome III and the T-DNA insertion in MPK10 (Figure 3a). This linkage is expected as INDEL-13 and INDEL-14 are located on the same chromosome as the wild-type MPK10 locus. In addition, however, tight linkage is also observed between the mpk10-1 T-DNA insertion and the markers INDEL-16 and INDEL-17 on the bottom arm of chromosome IV. Genetic linkage between a T-DNA insertion and markers on two chromosomes is indicative of a chromosomal translocation (Burnham, 1930; Castle et al., 2003).
The second example of a translocation shown in Figure 3 involves a T-DNA line carrying an insertion in the RAF17 gene. The raf17 T-DNA insertion shows tight linkage to both chromosomes I and V (Figure 3b), indicative of a translocation. For comparison, mapping data are also shown in Figure 3 for a T-DNA line carrying an insertion in the MPK17 gene. This T-DNA line shows no evidence of a translocation since genetic linkage is observed only to the expected location of the wild-type MPK17 gene and not other chromosomes (Figure 3c). Mapping data for the remaining lines tested are presented in Figure 4, Figure S1, Figure S2, Figure S3 and Figure S4.
Our genetic mapping survey indicated that there were chromosomal translocations in at least 12 of the 64 Salk lines (19%) in our survey (Table 1). All of the 11 Salk lines for which heterozygous plants produced ca. 50% non-viable pollen showed evidence of a translocation in the genetic mapping experiments. For these 11 Salk lines, we observed genetic linkage to the expected site of the T-DNA insertion as well as one or more mapping markers on a different chromosome. With the exception of mpk2-1, no evidence of a translocation was found in the genetic mapping data collected for the 16 Salk lines that produce normal pollen. In the case of mpk2-1, evidence of a translocation was found despite the fact that these plants produce normal pollen.
T-DNA border separation
A canonical T-DNA insertion is characterized by one copy of the T-DNA vector inserted into a single genomic locus. In this situation, the location of the T-DNA insert is defined by the Arabidopsis genomic DNA sequences flanking the insertion site on both the 5′ and 3′ sides of the T-DNA insertion. In order to characterize the structure of the T-DNA inserts present in each of the 64 Salk lines in our study we used PCR primers specific to the T-DNA left border and the T-DNA right border to detect the junctions between each T-DNA insert and the flanking Arabidopsis chromosomal DNA. We call the PCR product amplified using a T-DNA border primer plus a primer from the 5′ side of the target Arabidopsis gene the 5′-T-DNA junction and the product from the 3′ side of the target Arabidopsis gene the 3′-T-DNA junction (Figure 4). It should be noted that the 5′-T-DNA junction can involve either the left border or right border of the T-DNA vector, and likewise for the 3′-T-DNA junction. We were able to detect both 5′- and 3′-T-DNA junctions for 46 of the Salk lines in our study. 40 of these lines had T-DNA left border sequence at both the 5′- and 3′-T-DNA junctions, while six had one left border and one right border. We were able to detect T-DNA junctions on only one side of the predicted T-DNA insertion site for the remaining 18 Salk lines used in our survey (Table 1). It has been documented that T-DNA insertion is often associated with deletion of DNA sequence from the T-DNA border regions of the T-DNA vector (Krysan et al., 2002). It seems likely that those lines for which only one T-DNA junction could be detected may have lost the T-DNA border sequences that are recognized by the T-DNA specific PCR primers used in our study during the T-DNA insertion process.
In an uncorrupted T-DNA insertion, the T-DNA/plant DNA junctions are separated by a few kilobases of T-DNA vector sequence. This small genetic distance and the lack sequence homologous to the T-DNA vector in the Arabidopsis genome makes recombination between the T-DNA vector and the wild-type sister chromosome extremely unlikely. The 5′- and 3′-T-DNA junctions for a given insertion locus are therefore expected to co-segregate 100% of the time. In the case of a reciprocal translocation, the 5′- and 3′-T-DNA junctions may be physically separated on two distinct chromosomes as determined by the translocation break-point. Despite this physical separation, the lethality of unbalanced gametes would preserve complete genetic linkage between the physically separated 5′-T-DNA and the 3′-T-DNA junctions if a reciprocal translocation is present (Curtis et al., 2009). During our characterization of the mpk2-1 T-DNA line, we made the unexpected observation that the 5′- and 3′-T-DNA junctions for this T-DNA insertion are genetically separable from each other.
mpk2-1 homozygous mutant plants have both a 5′-T-DNA junction and a 3′-T-DNA junction that we were able to detect by PCR (Table 1). To analyze the genetic segregation of these two T-DNA junctions we crossed an mpk2-1 homozygous mutant plant to a Ler plant to create an mpk2-1 heterozygous plant. PCR genotyping confirmed that the resulting plant had both the 5′- and 3′-T-DNA junctions. This mpk2-1 heterozygous plant was then backcrossed to a wild-type Ler plant to create F2 progeny. We expected to identify two classes of F2 seedlings: those containing both the 5′- and 3′-T-DNA junctions and those with no T-DNA junctions at all. Instead, progeny were observed that contained all the possible combinations of the two T-DNA junctions. We found 74 plants with both the 5′ and 3′ junctions, seven plants with the 3′ junction only, 52 plants with the 5′ junction only, and 55 plants with no T-DNA junctions. Our observation of F2 plants with only one T-DNA junction indicates that the 5′- and 3′-T-DNA junctions of the mpk2-1 T-DNA insertion are genetically separable from each other.
We next tested whether the 5′- and 3′-T-DNA junctions were able to genetically separate in additional Salk lines in our collection. We genotyped F2 seedlings for all 46 of the T-DNA lines in our study for which we were able to detect by PCR both the 5′- and 3′-T-DNA junctions. If we were able to isolate seedlings with either the 5′-T-DNA junction only or the 3′-T-DNA junction only in F2 individuals we characterized that Salk line as having T-DNA borders that separate. Of the 46 Salk lines tested, two (mpk2-1 and mpk11-1) had 5′- and 3′-T-DNA junctions that were able to genetically separate (Table 1).
Genetic analysis of the mpk2-1 T-DNA line
Analysis of the genetic mapping data obtained for the mpk2-1 Salk line showed evidence of an unusual chromosomal rearrangement. As described above, we observed that the 5′-T-DNA junction and 3′-T-DNA junction of mpk2-1 are genetically separable from each other. Because these two T-DNA junctions do not always co-segregate, we were able to examine the genetic behavior of each of the junctions independently. To begin with, we performed linkage analysis between the mpk2-1 3′-T-DNA junction and our panel of INDEL mapping markers. This analysis showed linkage between the 3′-T-DNA junction and markers on both chromosomes I and III (Figure 4d). Linkage to chromosome I is expected as this is the location of the wild-type MPK2 locus, while linkage to chromosome III is suggestive of a translocation. When a similar analysis was performed using the 5′-T-DNA junction of mpk2-1, however, a different result was observed. The 5′-T-DNA junction was not linked to chromosome I. Linkage was seen only to chromosome III, which does not contain the wild-type MPK2 gene (Figure 4e). These data suggest that the 5′-T-DNA junction is physically located on chromosome III and not on chromosome I.
The standard model for a reciprocal translocation predicts that unbalanced gametes will be lethal due to the absence of entire chromosome arms in those gametes (Burnham, 1930). Our ability to isolate plants in which only one of the mpk2-1 T-DNA junction fragments was present suggested that the rearrangement present in the mpk2-1 T-DNA line may not be a standard reciprocal translocation. In order to further characterize the genetic behavior of the mpk2-1 T-DNA insertion we performed reciprocal crosses between wild-type plants and individuals heterozygous for either the mpk2-1 5′-T-DNA junction alone or the mpk2-1 3′-T-DNA junction alone. We observed that the 5′ T-DNA junction was transmitted to ca. 50% of the progeny through both the male and female gametes in these crosses, indicating that the 5′ T-DNA junction is not linked to any factor that reduces gamete fitness (Table 2). Transmission of the 3′ T-DNA junction also appeared normal through the female gametes, but was significantly reduced through the male (Table 2). This result suggests that the 3′ T-DNA junction of mpk2-1 is associated with some lesion in the genome that reduces pollen fitness. Our observation of normal pollen viability when mpk2-1 was tested using Alexander staining suggests that this reduced pollen fitness is not due to pollen lethality at the stage of development when pollen was collected for viability testing (Table 1).
Table 2.
Transmission of the mpk2-1 5′- and 3′-T-DNA junctions through the male and female gametophyte
| Progeny genotype |
|||
|---|---|---|---|
| Mutant parent | mpk2-1/MPK2 | MPK2/MPK2 | P-value |
| Male 5′-T-DNA | |||
| Observed | 51 | 40 | 0.2489 |
| Expected | 45.5 | 45.5 | |
| Female 5′-T-DNA | |||
| Observed | 45 | 49 | 0.6799 |
| Expected | 47 | 47 | |
| Male 3′-T-DNA | |||
| Observed | 16 | 55 | <0.001 |
| Expected | 35.5 | 35.5 | |
| Female 3′-T-DNA | |||
| Observed | 46 | 44 | 0.8330 |
| Expected | 45 | 45 | |
Mutant parent plants heterozygous for the indicated T-DNA junction were crossed to wild-type Landsberg erecta plants and the resulting progeny were tested by PCR to determine their genotypes. Mutant parent plants were heterozygous for either the mpk2-1 3′-T-DNA junction alone or the mpk2-1 5′-T-DNA junction alone. ‘Observed’ indicates the number of progeny with the indicated genotype. P-value determined using the chi-squared test.
The genetic characterizations described above suggest that the chromosomal rearrangement(s) present in the mpk2-1 T-DNA line do not conform to the model of a simple reciprocal translocation. Examples of complex chromosomal abnormalities associated with T-DNA insertions have been previously documented in the literature (Nacry et al., 1998; Tax and Vernon, 2001). For this reason we did not further pursue mapping of the chromosomal abnormalities present in the mpk2-1 T-DNA line as this information would be of limited value to our overall goal of measuring the rate with which chromosomal rearrangements are present in T-DNA collections. The genetic mapping described above demonstrates that a chromosomal rearrangement is present in the mpk2-1 T-DNA line since the 3′ T-DNA junction of mpk2-1 was found to be genetically linked to two chromosomes. Additional characterization of the precise structure of the genome in these plants would not change the conclusion that there is a chromosomal abnormality present in the mpk2-1 T-DNA line.
DISCUSSION
Through a combination of pollen viability analysis and genetic mapping, we determined that at least 12 of the 64 T-DNA lines (19%) that we tested from the Salk collection (Alonso et al., 2003) display evidence of a chromosomal translocation. We performed pollen viability assays on all 64 of these lines and comprehensive genetic mapping on 27 of them. For the genetic mapping experiments we analyzed all 11 of the lines that displayed an abnormal pollen phenotype, plus an additional 16 lines with normal pollen. We observed that pollen viability screening was an effective tool for predicting the presence of a translocation in a T-DNA line. All 11 of the T-DNA lines with abnormal pollen had evidence of a translocation when tested by genetic mapping, compared with only one of the 16 lines with normal pollen. Because we did not perform genetic mapping on all 64 of the lines in our survey, the fraction of lines carrying translocations may be >19%. Given the low rate with which translocations were observed in the lines that produce normal pollen, however, it seems likely that we have detected the majority of the translocations present in these 64 lines.
Although pollen viability analysis is an efficient method to screen for the presence of potential translocations in T-DNA lines, it will not detect all types of chromosomal abnormalities. In our survey we found one T-DNA line that produced normal pollen, but nonetheless contained a chromosomal rearrangement associated with the T-DNA insertion. This T-DNA line, which carried an insertion in the MPK2 locus, also displayed the ‘T-DNA borders separate’ phenomena described in the results section. It seems likely that the chromosomal rearrangement(s) present in the mpk2-1 T-DNA line are more complex than that of a simple reciprocal translocation. Our observation that the two T-DNA junctions present in this line display distinct behavior when tested using genetic linkage mapping supports this idea.
Pollen lethality in lines heterozygous for a translocation is due to the production of unbalanced gametes that lack pollen-essential genes. The normal viability of pollen from the mpk2-1 T-DNA line indicates that the gametes produced by this line are not deficient in any pollen-essential genes. One explanation for this result would be that the chromosome segments involved in the translocation do not involve regions of the genome containing pollen-essential genes. Alternatively, the presence of a translocation/duplication rather than a simple translocation could also explain this result. A translocation/duplication would not result in genetically deficient gametes at meiosis, and would therefore not be expected to cause pollen lethality. Further molecular and cytogenetic analyses would be necessary to further explore the precise structure of the mpk2-1 T-DNA line in order to better understand why it does not display pollen lethality. In more practical terms, the mpk2-1 T-DNA line serves as a warning that pollen viability testing alone cannot identify all types of chromosomal rearrangements that can exist in T-DNA lines.
Examples of Arabidopsis T-DNA lines carrying chromosomal rearrangements that are more complex than simple reciprocal translocations have been described. For example, Nacry et al. (1998) reported a T-DNA line carrying a reciprocal translocation that also contained a large inversion within one of the chromosomes involved in the translocation. This rearrangement resulted in the suppression of recombination within the region of the genome involved in the inversion. In addition, Tax and Vernon (2001) characterized two T-DNA lines containing internal translocation/ duplications of genomic DNA associated with the T-DNA insertion. In both of these studies genetic linkage mapping was able to clearly demonstrate the presence of T-DNA-associated chromosomal abnormalities.
In our study, DNA sequencing was used to characterize the Arabidopsis genomic sequences flanking the T-DNA insertion sites. For many of the T-DNA lines carrying reciprocal translocations we were able to generate DNA sequence for both the 5′- and 3′-T-DNA junctions. In all cases these T-DNA flanking sequences corresponded to the target gene of interest. For T-DNA insertions that are associated with chromosomal translocations, one would predict that two additional junction fragments might exist in the genome at the locations of the translocation break-points. It is not, however, straightforward to identify and characterize these additional junction fragments for two reasons. First of all, the structure and orientation of the presumably disrupted T-DNA vector at this location cannot be predicted. Secondly, the precise genomic locations of the translocation breakpoints are not known. Our genetic mapping data allow us to identify a region of the chromosome within which the breakpoint must occur, but the precision of this mapping is much lower than that required to design PCR primers. Although technically challenging, detailed characterization of the structures of the translocation breakpoints could shed more light on the mechanism of T-DNA associated translocation.
Previous studies investigating the frequency of chromosomal translocations in Arabidopsis T-DNA lines have involved collections of lines pre-selected for the presence of lethal mutations (Castle et al., 1993; Budziszewski et al., 2001). All of the T-DNA lines used in our survey were viable when the T-DNA insertion was in the homozygous mutant state. The translocation rate that we observed in our study was similar to that seen by Castle et al. (1993) and Budziszewski et al. (2001), suggesting that T-DNA lines carrying lethal mutations are not enriched for the presence of T-translocations.
The set of T-DNA lines used in our study all carry insertions in genes that encode protein kinases. Although it is formally possible that kinase-encoding genes have a propensity for T-DNA-associated translocations, it is difficult to imagine a mechanism by which such a bias might operate. Comprehensive genetic mapping of a larger set of genes using the methods described in this study would help establish whether certain gene types are more or less prone to T-DNA-associated translocations.
It should be noted that the standard genetic analyses typically performed by researchers studying T-DNA lines will not reveal the presence of a translocation. These standard analyses include performing DNA sequencing to characterize the 5′- and 3′-T-DNA junctions, identifying individuals homozygous for the T-DNA insertion allele, and testing for Mendelian segregation of the T-DNA allele. For the 64 Salk lines used in our survey, we were able to isolate homozygous mutant plants for all of them, including the 12 Salk lines with a chromosomal translocation. We also performed DNA sequencing of the junctions between the T-DNA borders and the plant chromosome in each of these lines. Nine of the Salk lines that were found to contain a chromosomal translocation had both 5′- and 3′-T-DNA junctions that were consistent with insertion of the T-DNA into a single gene. It has previously been documented that plants heterozygous for a reciprocal translocation can exhibit normal Mendelian segregation of the T-DNA insertion associated with the translocation (Castle et al., 1993; Tax and Vernon, 2001; Curtis et al., 2009). Taken together, this information indicates that simply analyzing the genetic behavior of the T-DNA insertion of interest in isolation is not sufficient to detect the presence of a chromosomal translocation.
In order to detect chromosomal translocations associated with a T-DNA insertion, comprehensive genetic mapping must be done to reveal unexpected linkage between the T-DNA insertion and a location on a second chromosome. For this analysis, genetic markers spaced throughout the entire genome must be employed. Although testing the viability of pollen collected from a plant heterozygous for a T-DNA insertion provides a simple, rapid, first-screen for the detection of potential chromosomal translocations, follow-up genetic mapping must be performed before one can conclude that reduced pollen viability is due to a translocation. When performing comprehensive genetic mapping, Ponce et al. (1999) recommend genotyping a minimum of 20 F2 individuals. Our sample size of F2 plants ranged from 21 to 126 plants.
We noted that some of the T-DNA lines carrying a translocation displayed evidence of compression of their genetic map. For example, the raf17 T-DNA insertion shows genetic linkage to mapping markers INDEL-2 (0% recombination) and INDEL-3 (2% recombination) (Figure 3). These markers, located on the same arm of chromosome I, are separated by a physical distance of ca. 5 MB, corresponding to an estimated genetic distance of ca. 20 centiMorgans (Table S1). In addition, the mpk11-1 T-DNA insertion shows significant genetic linkage to all three mapping markers on chromosome IV (Figure S1). One possible explanation for the compression of genetic distances in these lines would be the occurrence of crossover interference. Recombination between homologous chromosomes is based on crossover events during meiosis. Crossover events are not independent from each other, and the occurrence of one crossover event inhibits the formation of a second crossover event in the same region (Jones, 1984). In Arabidopsis, it has been proposed that there are several levels of control of the distribution of crossover events including chromosomal, regional (megabase) and local (kilobase) (Mezard et al., 2006). It is possible that the presence of chromosomal translocations alters the regional or local distribution of crossover events and leads to compression of the genetic map.
Translocations can complicate the genetic analysis of mutant phenotypes (Bonhomme et al., 1998; Guan et al., 2003; Curtis et al., 2009). The mutant phenotype of a plant carrying a chromosomal translocation may be caused by either the loss of the gene disrupted by the T-DNA insertion or by other potential mutations associated with the translocation, such as the disruption of a gene at the translocation breakpoint on the other chromosome. Lafleuriel et al. (2004) noted that the severe developmental delay and sterility phenotypes observed in Arabidopsis plants that were homozygous for a reciprocal translocation were most likely caused by a deletion associated with the translocation and not the loss of function of the gene disrupted by the T-DNA. This example emphasizes the need to confirm any mutant phenotypes observed in T-DNA lines through the use of additional methods such as the identification of an independently generated mutant allele of the gene of interest or the use of genetic rescue with a transgenic, wild-type copy of the gene of interest.
Previous studies of T-DNA-associated chromosomal translocations used T-DNA lines from a variety of collections, including the Salk collection (Curtis et al., 2009), a collection created by Feldmann and Marks (1987) (Castle et al., 1993; Ray et al., 1997), plants from T-DNA lines created in the investigators’ own laboratories (Nacry et al., 1998; Budziszewski et al., 2001; Lafleuriel et al., 2004) and other T-DNA mutant collections (Tax and Vernon, 2001). Based on this information, it appears that the presence of translocations in T-DNA lines is a general phenomenon and is not limited to particular T-DNA mutant collections. Xing and Zachgo (2007) noted that at least 10% of the heterozygous transgenic Arabidopsis plants that they analyzed produced 20–50% abnormal pollen, regardless of the T-DNA vector sequence. The authors concluded that pollen lethality in heterozygous plants is a common phenomenon in transgenic plant populations and may be caused by chromosomal rearrangements. When using T-DNA insertional lines from any source, one should expect that a significant fraction of these lines may contain chromosomal translocations.
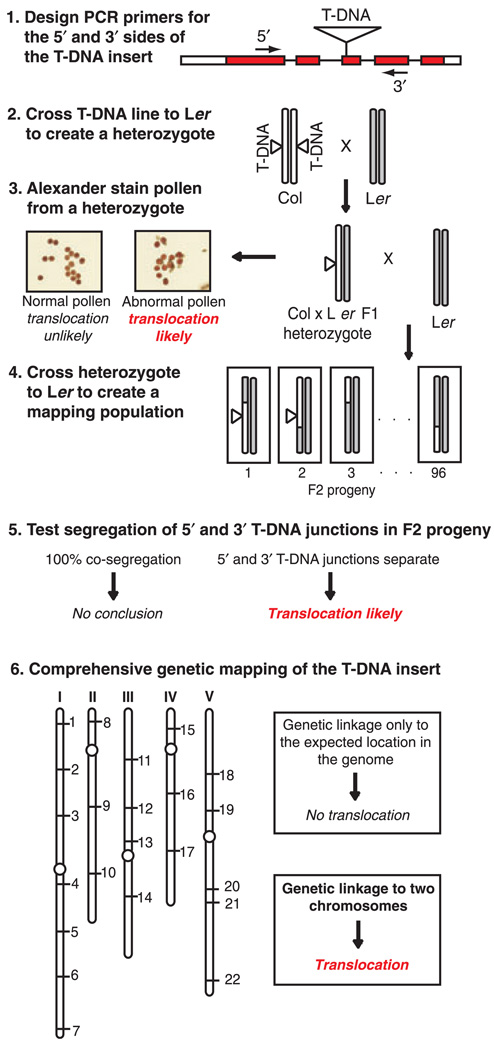
We have developed an efficient procedure for screening T-DNA insertional lines for the presence of chromosomal translocations (Figure 5). The first step in the process is to design PCR genotyping primers that flank the predicted T-DNA insertion site and use these primers to determine if both the 5′- and 3′-T-DNA junctions can be detected. Assuming that the T-DNA insertion is in the Col ecotype, the next step is to cross the T-DNA line to a wild-type Ler plant to create a heterozygous plant. This cross represents the first step in the process of creating a mapping population for the T-DNA line, and also provides a heterozygous plant that can be used to screen for pollen viability using Alexander staining (Alexander, 1969). To examine pollen viability, it is not necessary to use a Col × Ler heterozygous plant. A heterozygous plant with any genetic background may be used. If the heterozygous plant produces all viable pollen, it is unlikely that the T-DNA line has a chromosomal translocation. However, as demonstrated by the mpk2-1 T-DNA line characterized in this study, a plant with a chromosomal translocation may be able to produce apparently normal pollen. If the heterozygous plant produces ca. 50% non-viable pollen, it is likely that the T-DNA line has a chromosomal translocation. It is essential that the plant used for pollen viability screening is heterozygous for the T-DNA insertion since plants that are homozygous for a translocation are genetically balanced and will produce all viable pollen (Burnham, 1930).
Figure 5.
Flow chart outlining a strategy for screening T-DNA lines for the presence of chromosomal translocations. In this example the T-DNA insertion is present in the Columbia ecotype (Col). ‘Abnormal pollen’ refers to the situation where a heterozygous plant produces ca. 50% non-viable pollen. Co-segregation of the 5′ and 3′ T-DNA junctions can only be performed when both junctions are detectable by PCR, which may not be the case with all T-DNA lines.
The final test for the presence of a translocation is to perform comprehensive genetic mapping. To create the necessary mapping population, one should cross the Col × Ler heterozygous plant described above to a wild-type Ler plant to create an F2 mapping population. Ideally one should perform enough genetic crosses to produce a few hundred F2 progeny. If the T-DNA line has both 5′- and 3′-T-DNA junctions, one should genotype F2 seedlings to determine if the junctions co-segregate 100% of the time. If the 5′- and 3′-T-DNA junctions are genetically separable, that indicates that the T-DNA line most likely has a chromosomal translocation. Compete co-segregation of the 5′- and 3′-T-DNA junctions does not indicate the absence of a chromosomal translocation, however (Table 1).
The final step in the process is to perform genetic mapping using a panel of marker loci distributed throughout the genome, such as the INDEL markers described in this study. If the T-DNA insertion is genetically linked to one location in the genome, there is most likely no translocation associated with the T-DNA insertion. Genetic linkage of the T-DNA insertion to two chromosomes indicates the presence of a translocation associated with the T-DNA insertion.
The underlying cause of chromosomal translocations in T-DNA transformed lines is not known. Improving our understanding of the mechanism of T-DNA insertion into the plant genome may help us to understand why these translocations occur with such a high frequency in T-DNA lines. The mechanism of T-DNA insertion is believed to occur via a mechanism of illegitimate recombination that begins at the site of a double-strand break in the plant chromosome (Gelvin, 2010). It is possible that cells that are amendable to T-DNA insertion have an enhanced potential for the generation of translocations due to the required presence of double-strand breaks in those cells. Future studies investigating the frequency of chromosomal translocations in non-mutagenized Arabidopsis populations could be helpful for further investigating this possibility. Regardless of the mechanism responsible for the generation of T-DNA associated translocations, it is important for plant scientists to recognize that these genomic rearrangements are common phenomena in the public collections of T-DNA lines that are widely used for studying gene function in Arabidopsis.
EXPERIMENTAL PROCEDURES
Seedling growth and DNA preparation
Arabidopsis plants were grown in soil under constant light at 20–24°C. To collect tissue from F2 seedlings for the genetic mapping experiments, seedlings were grown in 96-well plates using the Ice-Cap method as described (Clark and Krysan, 2007). DNA extraction from root tissue harvested using Ice-Cap was performed as described (Clark and Krysan, 2007) with the following modifications: 12.5 µl of a solution of 500 mM Tris pH 8.0, 50 mmol EDTA pH 8.0 was added to each well of the 96-well plate after root tissue had been collected. Following centrifugation of the disrupted root tissue, 50 µl of the resulting cleared supernatant was then diluted 1:10 (v/v) with deionized water; 2 µl of this DNA extract was used as the template for the PCR genotyping reactions.
PCR genotyping
PCR reactions were set up in 96-well full-skirted hard shell PCR plates (Eppendorf, catalog number 951020401, http://www.eppendorf.com). PCR reactions with a final volume of 20 µl contained 0.2 µm of each PCR primer, 0.2× EvaGreen fluorescent dye (Biotium Inc., catalog number 31000, http://www.biotium.com), 0.2 mm of each dNTP (Promega Corporation, http://www.promega.com), 75 mm Tris–HCl pH 9, 20 mm (NH4)2SO4, 3 mm MgCl2, 0.01% Tween 20 (v/v), 2 µl of DNA extract, and 0.5 µl of Taq polymerase. 25 µl of Mineral Seal Oil (CQ Concepts, catalog no. 64742-46-7, http://www.cqconcepts.com) was added to each well to prevent evaporation during thermal cycling. PCR plates were sealed with Temp-Plate Sealing film plate seals (USA Scientific, catalog no. 2921-0000, http://www.usascientific.com). PCR was performed using 40 cycles of the following thermal cycling profile: 96°C for 1 min, 94°C for 4 sec, annealing temperature as indicated in Table S1 for 30 sec, 72°C for 15 sec. Insertion/deletion INDEL PCR products were analyzed using the Bio-Rad CFX96 Detection System (Bio-Rad Laboratories, USA, http://www.biorad.com). The SYBR/FAM emission/ excitation detection channel was used to detect the EvaGreen fluorescent dye present in the reactions. Melt-curve data was collected using the following thermal profile: 65°C for 30 sec followed by a ramp from 65 to 95°C at a rate of 0.5°C sec−1. Fluorescence data was captured once every second.
The following PCR primers were used for PCR reactions involving the T-DNA border. T-DNA left border: MLB1 5′-GTGGACTCTTGTT CCAAACTG-3′ or p745 5′-AACGTCCGCAATGTGTTATTAAGTTGTC-3′. T-DNA right border: JMRB2 5′-TGATAGTGACCTTAGGCGACTT TTGAACGC-3′.
Pollen viability assay
We determined pollen viability using Alexander’s stain (Alexander, 1969) with the following modifications: we omitted the choral hydrate and diluted the stain 1:10 (v/v) in dH2O to create a working concentration. Dehiscent anthers were removed from flowers, placed in the diluted Alexander stain on a microscope slide, covered with a glass cover slip and examined using a light microscope.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Susan Bush for assistance with the development of the melt-curve genotyping assay used in this study and for helpful discussions. This work was supported by a grant from The National Science Foundation (grant number MCB-0447750).
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Figure S1. Genetic mapping data for mpk11-1, mpk19-1, raf5, raf45, and raf47.
Figure S2. Genetic mapping data for zik8, mapkkk16, raf39, mpk1, mpk3, and mpk8.
Figure S3. Genetic mapping data for mpk9-1, mpk13-1, mpk14-1, mpk16-1, and mpk18-1, mekk1.
Figure S4. Genetic mapping data for ctr1, raf26, mpk4, map4kα1, and mpk20-1.
Table S1. PCR primers for amplifying the INDEL mapping markers.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
REFERENCES
- Alexander MP. Differential staining of aborted and nonaborted pollen. Stain Technol. 1969;3:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Bonhomme S, Horlow C, Vezon D, de Laissardière S, Guyon A, Férault M, Marchand M, Bechtold N, Pelletier G. T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distortion alone. Mol. Gen. Genet. 1998;260:444–452. doi: 10.1007/s004380050915. [DOI] [PubMed] [Google Scholar]
- Budziszewski GJ, Lewis SP, Glover LW, et al. Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics. 2001;159:1765–1778. doi: 10.1093/genetics/159.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham CR. Genetical and cytological studies of semisterility and related phenomena in maize. Proc. Natl Acad. Sci. USA. 1930;16:269–277. doi: 10.1073/pnas.16.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol. Gen. Genet. 1993;241:504–514. doi: 10.1007/BF00279892. [DOI] [PubMed] [Google Scholar]
- Clark KA, Krysan PJ. Protocol: an improved high-throughput method for generating tissue samples in 96-well format for plant genotyping (Ice-Cap 2.0) Plant Methods. 2007;12:3–8. doi: 10.1186/1746-4811-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Belcream K, Boilmann SR, Timoney CM, Hoffman PD, Mercier R, Hays JB. Reciprocal chromosomal translocation associated with TDNA-insertion mutation in Arabidopsis: genetic and cytological analysis of consequences for gametophyte development and for construction of doubly mutant lines. Planta. 2009;229:731–745. doi: 10.1007/s00425-008-0868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann KA, Marks MD. Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: a non-tissue culture approach. Mol. Gen. Genet. 1987;208:1–9. [Google Scholar]
- Forsbach A, Schubert D, Lechtenberg B, Gils M, Schmidt R. A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol. Biol. 2003;52:161–176. doi: 10.1023/a:1023929630687. [DOI] [PubMed] [Google Scholar]
- Gelvin SB. Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu. Rev. Phytopathol. 2010;48:45–68. doi: 10.1146/annurev-phyto-080508-081852. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Villarroel R, Van Montagu M. Illegitimate recombination in plants: a model for T-DNA integration. Genes Dev. 1991;5:287–297. doi: 10.1101/gad.5.2.287. [DOI] [PubMed] [Google Scholar]
- Guan C, Rosen ES, Boonsirichai K, Poff KL, Masson PH. The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway. Plant Physiol. 2003;133:100–112. doi: 10.1104/pp.103.023358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GH. The control of chiasma distribution. Symp. Soc. Exp. Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Jester PJ, Monson S, Copenhaver G, Preuss D, Sussman MR. Characterization of T-DNA insertion sites in Arabidopsis thaliana and the implications for saturation mutagenesis. OMICS. 2002;6:163–174. doi: 10.1089/153623102760092760. [DOI] [PubMed] [Google Scholar]
- Lafleuriel J, Degroote F, Depeiges A, Picard G. A reciprocal translocation, induced by a canonical integration of a single T-DNA, interrupts the HMG-I/Y Arabidopsis thaliana gene. Plant Physiol. Biochem. 2004;42:171–179. doi: 10.1016/j.plaphy.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Laufs P, Autran D, Traas J. A chromosomal paracentric inversion associated with T-DNA integration in Arabidopsis. Plant J. 1999;18:131–139. doi: 10.1046/j.1365-313x.1999.00436.x. [DOI] [PubMed] [Google Scholar]
- Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C. T-DNA integration: a mode of illegitimate recombination in plants. EMBO J. 1991;10:697–704. doi: 10.1002/j.1460-2075.1991.tb07999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezard C, Vignard J, Drouaud J, Mercier R. The road to crossovers: plants have their say. Trends Genet. 2006;23:91–99. doi: 10.1016/j.tig.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Nacry P, Camillleri C, Coutial B, Caboche M, Bouchez D. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics. 1998;149:641–650. doi: 10.1093/genetics/149.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Yoshioka Y, Machida C, Machida Y. DNA rearrangement associated with the integration of T-DNA in tobacco: an example for multiple duplications of DNA around the integration target. Plant J. 1995;7:157–164. doi: 10.1046/j.1365-313x.1995.07010157.x. [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Ecker JR. Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 2010;61:928–940. doi: 10.1111/j.1365-313X.2010.04119.x. [DOI] [PubMed] [Google Scholar]
- Ponce MR, Robles P, Micol JL. High-throughput genetic mapping in Arabidopsis thaliana. Mol. Gen. Genet. 1999;261:408–415. doi: 10.1007/s004380050982. [DOI] [PubMed] [Google Scholar]
- Ray S, Park S-S, Ray A. Pollen tube guidance by the female gametophyte. Development. 1997;124:2489–2498. doi: 10.1242/dev.124.12.2489. [DOI] [PubMed] [Google Scholar]
- Redei GP, Koncz C. Classical mutagenesis. In: Koncz C, Chau N-H, Schell J, editors. Methods in Ara-bidopsis Research. River Edge: World Scientific; 1992. pp. 16–82. [Google Scholar]
- Salathia N, Lee HN, Sangster TA, et al. Indel arrays: an affordable alternative for genotyping. Plant J. 2007;51:727–737. doi: 10.1111/j.1365-313X.2007.03194.x. [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, et al. A high-throughput Arabidopsis reverse genetic system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Egawa H, Ikeda J-H, Wakasa K. The structures of integration sites in transgenic rice. Plant J. 1997;11:353–361. doi: 10.1046/j.1365-313x.1997.11030353.x. [DOI] [PubMed] [Google Scholar]
- Tax FE, Vernon DM. T-DNA-associated duplication/translocations in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiol. 2001;126:1527–1538. doi: 10.1104/pp.126.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Xing S, Zachgo S. Pollen lethality: a phenomenon in Arabidopsis RNA interference plants. Plant Physiol. 2007;145:330–333. doi: 10.1104/pp.107.103127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen CY, Sedbrook JC, Perrin RM, Carroll KL, Masson PH. Loss- of-function mutations of ROOT HAIR DEFECTIVE3 suppress root waving, skewing, and epidermal cell file rotation in Arabidopsis. Plant Physiol. 2005;138:701–714. doi: 10.1104/pp.105.059774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.