Abstract
We analyzed brain MRI data from 372 young adult twins to identify cortical regions in which gray matter thickness and volume are influenced by genetics. This was achieved using a A/C/E structural equation model that divides the variance of these traits, at each point on the cortex, into additive genetic (A), shared (C) and unique environmental (E) components. A strong genetic influence was found in frontal and parietal regions. Additionally, we correlated cortical thickness with full-scale IQ for comparison with the A/C/E maps, and several regions where cortical structure was correlated with IQ are under genetic control. These cortical measures may be useful phenotypes to narrow the search for quantitative trait loci influencing brain structure.
Keywords: brain, image analysis, magnetic resonance imaging, cortex, genetics
1 Introduction
Brain structure is influenced by both genetic and environmental factors, and their relative influence varies throughout life, and differs for different brain regions. As the risk for a variety of neurological and psychiatric disorders is partly inherited, one urgent and achievable research goal is to identify aspects of brain structure and function that are under genetic control, prior to searching for specific genes that influence them.
Twin studies have been used extensively to estimate the heritability of many cognitive and behavioral traits, as well as measures derived from brain images. In general, these data sets involve pairs of monozygotic and dizygotic twins; monozygotic twins share 100% of their genes, while dizygotic twins share 50% on average. Various methods have been designed to compare the two groups, yielding estimates of the genetic and environmental contributions to various traits, including brain structure.
Here we studied the influence of genes on cortical thickness and volume; both have been implicated as promising targets for genetic studies. In an early study of 10 monozygotic and 10 dizygotic twin pairs [1], we found regionally-varying genetic influences in 3D maps of the heritability of cortical gray matter density - a measure that is correlated with IQ. Posthuma et al. [2] extended earlier work on gray matter heritability [3,4,5,6]. They found that partially overlapping sets of genes were involved in the control of gray and white matter volumes, and IQ. Recently, Yoon et al. [7] examined the influence of genetic and environmental factors on cortical thickness and volume in 184 eight-year-old twins, in different brain substructures. In [6], the authors showed that there were genetic, shared environmental, and individual-specific environmental influences on the volumes of 96 brain regions of interest (ROIs), in 474 middle-aged male twins from the Vietnam Era Twin Study of Aging.
However, no large-scale study to date has analyzed genetic contributions to cortical volume and thickness in healthy young adult twins. In this paper, we used the A/C/E structural equation model to analyze genetic influences on the brain in a large MRI dataset of twins. We also assessed the relationship between full-scale IQ and cortical thickness at each point on the cortex. This analysis provides a map of significant correlations (p-values) between local cortical thickness and full-scale IQ, and involves using mixed effects regression to account for similarities within families [8].
2 Methods
2.1 Data and Preprocessing
The data consisted of 372 young adult twins of age 21–27 yrs (mean age: 23.7 yrs ± 1.9 yrs, 150 men and 222 women). There were 194 dizygotic and 178 monozygotic twins in the population (37 same-sex male and 60 same-sex female dizygotic pairs, 32 male and 57 female monozygotic pairs). Opposite-sex twin pairs were excluded, to prevent sex differences from inflating estimates of genetic effects. For this study, only complete pairs were used.
All twins were scanned using a 4 T Bruker Medspec whole body scanner at the Center for Magnetic Resonance (University of Queensland, Australia). Three-dimensional T1-weighted images were acquired with a magnetization prepared rapid gradient echo (MP-RAGE) sequence to resolve anatomy at high resolution.
Acquisition pa- rameters were: inversion time (T I) /repetition time (T R) /echo time (T E) = 700 /1500 / 3.35 msec; flip angle = 8°; slice thickness = 0.9 mm with a 256×256×256 acquisition matrix. Each subject was informed of the goals of the study and signed a formal consent. The study was approved by the appropriate Institutional Review and Research Ethics Boards. All subjects underwent physical and psychological screening to exclude cases of pathology known to affect brain structure. No twin subjects reported a history of significant head injury, a neurological or psychiatric illness, substance abuse or dependence, or had a first-degree relative with a psychi- atric disorder. Subjects were all 21–27 years old and were right-handed.
Extracerebral tissues were manually deleted from the MRI images using the Display software from the Montreal Neurological Institute, McGill University, Canada. All scans were then aligned to the ICBM53 template [9] using a 9-parameter registration from the FMRIB's Linear Image Registration Toolbox, FLIRT [10].
We used the cortical reconstruction routine from the Freesurfer software package [11] to segment the pial surface and gray/white matter interfaces, and to generate a tessellation of the resulting surfaces. The software labels a set of 34 cortical subregions per hemisphere, and infers approximate Brodmann areas [12]. We also used Freesurfer to compute the cortical thickness and volume at each vertex over the whole cortex. Thickness was calculated as the average of the distance from the gray matter/cerebrospinal fluid interface to the gray/white matter surface, and vice versa. The volume was defined as the product of the thickness and the area of the surface layer equidistant between the inner and outer cortical surfaces. The area associated with a vertex is defined as the average of the areas of triangles that include that vertex.
Volume values in each individual were filtered using a smoothing kernel of 25 mm FWHM, a value in the range suggested by Lerch et al. [13] to boost the signal to noise ratio. To remove age and gender-related effects from the analysis, we covaried the thickness and volume measures for effects of age and gender by performing linear regression (Male=1, Female=−1). Covarying for age made no significant difference to the analysis, possibly because the age range of our sample is quite small.
2.2 A/C/E model of Variance
We estimated relative contributions of additive genetic (A), shared environmental (C) and unique environmental (E) effects on the variance in cortical thickness and volume across the sample of twins. To do so, we used A/C/E structural equation modeling, as outlined in [14], following the implementation described in [15,8].
The observed variable, Z - here the cortical thickness or volume at each vertex for each member of a twin pair - may be modeled as:
| (1) |
A, C, E are latent variables and a, c, e are the weights of each factor to be determined. The method estimates the vertex-based variance in each of the 3 free model parameters, constrained by the requirement that a2 + c2 + e2 = 1. Measurement and inter-subject registration errors are both classified as part of the E term.
The covariance in the cortical thickness or volume between monozygotic and dizygotic pairs at each vertex was used as an input to the algorithm. The weights were estimated by comparing the covariance matrix implied by the model to the observed sample covariance matrix using maximum-likelihood fitting. The best fitting model was obtained using the Broyden-Fletcher-Goldfarb-Shanno method [16]. We used a permutation distribution to make the results independent of the distribution of the computed statistics [15,8,17].
2.3 Correlation of full scale IQ to thickness
The relationship of full-scale IQ to cortical thickness was assessed at each vertex using a mixed effects regression model to account for similarities within families [8]. Full-scale IQ was added in the model as a fixed effect and a random intercept was included for each family. The analysis was implemented in the R statistical package (version 2.9.2) using the `nlme' library [18]. The nominal p-values represent the significance of the full-scale IQ term.
3 Results
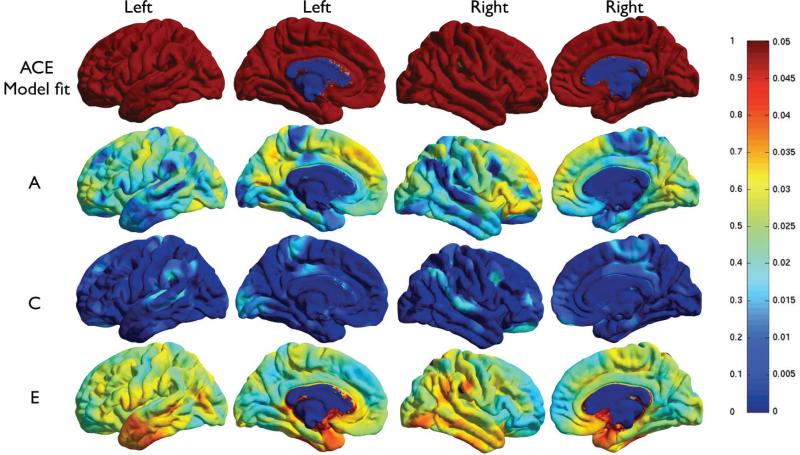
Figure 1 shows detailed cortical surface maps of the additive genetic component (A), common environment (C) and unique environment (E) at each vertex. The p-values for the overall A/C/E model fit are also shown. The A/C/E model fits almost everywhere on the cortex except at the corpus callosum (blue patch on the medial side) where thickness or volume measures are not defined. We also considered the reduced A/E model instead of A/C/E. The common environment (C) term was found to improve the fit of the A/C/E model relative to the reduced A/E model, so we only show results for this model here.
Figure 1.
A/C/E analysis of cortical thickness. The two leftmost columns show the sagittal and medial surface of the left hemisphere; the two rightmost ones show the right hemi- sphere. The color bar shows the color code used in the bottom three rows a2, c2 and e2 on the left, and associated p-values on the right. Top row: Regions are shown (in red) where the A/C/E model fits. Row 2 to row 4: Maps of the genetic (a2), combined environmental (c2) and unique environmental (e2) contributions to cortical thickness. Figures for cortical volume measures were visually similar and are not shown, but are available upon request.
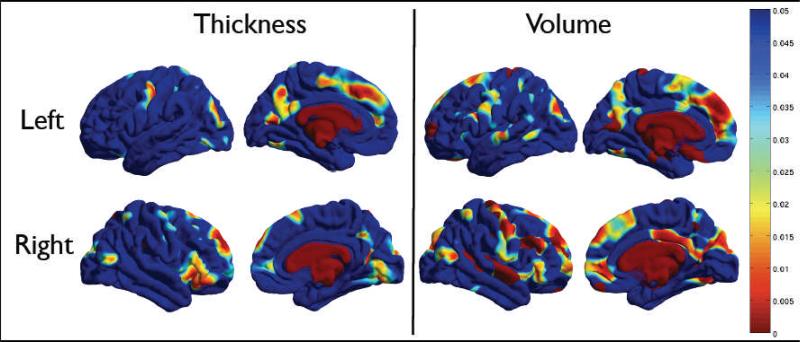
P -values for the A term are shown in Figure 2, for both the thickness and volume measures. Areas in the frontal and parietal lobes, and the primary visual cortices are significantly genetically influenced. Genetic effects on the cingulate and insula were primarily detected on the left, perhaps due to hemispheric differences in sulcal patterning in these regions. Lateral temporal regions showed less evidence for heritability.
Figure 2.
P -value associated with the A term, showing the significance of the genetic contribution to brain structure. Thickness is shown on the left panel, and cortical volume on the right. The top row shows the left hemisphere, and the bottom row shows the right. The color bar displays the p-values.
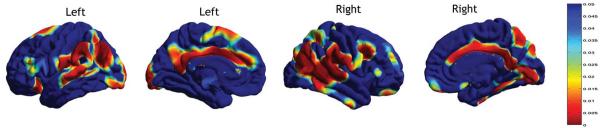
After computing A/C/E analyses of the cortical measures, we also examined correlations between local cortical thickness and full-scale IQ. Genetic contributions to complex traits such as the full-scale IQ may be studied by assessing the heritability of traits that covary with it (thickness, volume; see Fig. 2) and then computing correlations of the full-scale IQ to that trait, as shown in Fig. 3. Cortical thickness in parietal, medial temporal, occipital and cingulate regions was positively correlated with the full-scale IQ. In addition, cortical thickness in Wernicke's area, the region responsible for language comprehension, showed a significant additive genetic (A) component and was correlated with full-scale IQ.
Figure 3.
Correlation of full-scale IQ to cortical gray matter thickness, displayed as a nominal p-value map on the cortex.
4 Discussion
In this paper, we perform a point-wise analysis of cortical measures to determine how they are influenced by genetic versus environmental factors. In prior large-sample studies [19,20], only a regionwise heritability analysis was performed. Here, we took a somewhat different approach by performing the A/C/E analysis at each cortical surface point. Our results generally agree with those of prior pediatric [4,7] and small-sample adult twin studies [1], which reported significant heritability in frontal, temporal and superior parietal areas. However, while [4] found significant heritability in the postcentral and supramarginal gyri, here those results were only found as a trend, perhaps due to age differences. P-values in [19] were generally lower than ours, although statistically significant regions were similar overall.
According to one theory, early-developing regions are more genetically programmed, while late-maturing ones tend to be more environmentally influenced. This theory is not supported here, although tensor-based morphometry and diffusion tensor imaging heritability maps have shown evidence for this in subcortical regions [15].
The maps of full-scale IQ correlation with cortical thickness (Figure 3) and the additive genetic (A) component (Figure 1) shows significant overlap, especially at the Wernicke's areas influencing language.
Our study has some limitations. Although our sample size is large compared to most previous studies, the confidence intervals of additive genetic (A), common environment (C) and unique environment (E) are still wide. This limits the statistical power to identify differences between our results and prior findings. Furthermore, impact of test-retest reliability on these results needs to be evaluated, especially in the temporal lobes where thickness measures are prone to MRI-related distortions and susceptibility artifacts. We will address these issues as our sample size continues to increase in this ongoing study.
Highly heritable cortical measures may be useful phenotypes in the search for trait loci that account for differences in human brain structure (see Enigma project). Identifying heritable aspects of brain morphology is an important step towards understanding specific genes that impact brain structure and function and may help in the search for genetic risk factors for a variety of brain disorders [21].
5 Conclusion
We performed a genetic analysis of cortical thickness and volume in a large sample of twins, by applying the A/C/E structural equation model. The results showed detailed pointwise genetic and environmental contributions on the whole cortex. Several areas in the parietal and frontal lobes were observed to be genetically influenced.
Acknowledgements
This work was supported by NIH grants R01 HD050735, U54 RR021813 and P41 RR013642a and the National Health and Medical Research Council, Australia grant 496682. The authors thank Stephanie Biglarian and April Ryles for their help in quality control of the processed data.
ABBREVIATIONS
- A, C, E
Additive genetic, common environment and unique environment
- ROI
regions of interest
- MP-RAGE
magnetization prepared rapid gradient echo
- TI, TR, TE
inversion time, repetition time, echo time
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, et al. Genetic Influences on Brain Structure. Nat Neurosci. 2001;4:1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- [2].Posthuma D, De Geus EJC, Baar—e WFC, Hulshoff Pol HE, Kahn RS, Boomsma DI. The association between brain volume and intelligence is of genetic origin. Nat Neurosci. 2002;5:83–84. doi: 10.1038/nn0202-83. [DOI] [PubMed] [Google Scholar]
- [3].Bartley AJ, Jones DW, Weinberger DR. Genetic variability of human brain size and cortical gyral patterns. Brain. 1997;120:257–269. doi: 10.1093/brain/120.2.257. [DOI] [PubMed] [Google Scholar]
- [4].Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, et al. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Hum Brain Mapp. 2009;30:163–174. doi: 10.1002/hbm.20494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sullivan EV, Pfefferbaum A, Swan GE, Carmelli D. Heritability of hippocampal size in elderly twin men: Equivalent influence from genes and environment. Hippocampus. 2001;11:754–762. doi: 10.1002/hipo.1091. [DOI] [PubMed] [Google Scholar]
- [6].Kremen WS, Prom-Wormley E, Panizzon MS, Eyler LT, Fischl B, Neale MC, et al. Genetic and environmental influences on the size of specific brain regions in midlife: The VETSA MRI study. Neuroimage. 2010;49:1213–1223. doi: 10.1016/j.neuroimage.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yoon U, Fahim C, Perusse D, Evans AC. Lateralized genetic and environmental influences on human brain morphology of 8-year-old twins. Neuroimage. 2010;53(3):1117–1125. doi: 10.1016/j.neuroimage.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chiang MC, Barysheva M, Lee AD, Madsen SK, Klunder AD, Toga AW, et al. Genetics of Brain Fiber Architecture and Intelligence. J Neurosci. 2009;29:2212–2224. doi: 10.1523/JNEUROSCI.4184-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- [10].Jenkinson M, Bannister PR, Brady JM, Smith SM. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- [11].Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–94. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- [12].Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–80. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- [13].Lerch JP, Evans AC. Cortical thickness analysis examined through power analysis and a population simulation. Neuroimage. 2005;24:163–173. doi: 10.1016/j.neuroimage.2004.07.045. [DOI] [PubMed] [Google Scholar]
- [14].Neale MC, Maes HM. Methodology for Genetic Studies of Twins and Families. Kluwer Academic Publishers; Dordrecht, Netherlands: 1992. [Google Scholar]
- [15].Lee AD, Lepore N, Brun C, Chou YY, Barysheva M, Chiang MC, et al. Multivariate Statistics in 100 Twins DTI Data. Proceedings, MICCAI. 2009 [Google Scholar]
- [16].Neale MC, Boker SM, Xie G, Maes HH. Mx: Statistical modeling. VCU Department of Psychiatry. 2002 [Google Scholar]
- [17].Nichols TE, Holmes AP. Non-parametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2001;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pinheiro JC, Bates DM. Statistics and computing. Springer; New York: 2000. Mixed-effects models in S and S-PLUS. [Google Scholar]
- [19].Winkler AM, Kochunov P, Blangero J, Almasy A, Zilles K, Fox PT, Duggirala R, Glahn DC. Cortical thickness of gray matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, et al. Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex. 2009;19:2728–2735. doi: 10.1093/cercor/bhp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Thompson PM, Martin NG, Wright MJ. Imaging genomics. Current Opinion in Neurology. 2010;23(4):368–373. doi: 10.1097/WCO.0b013e32833b764c. [DOI] [PMC free article] [PubMed] [Google Scholar]