Synopsis
The Phosphoinositide-3-kinase (PI3K) pathway regulates cell proliferation, survival and migration and is consequently of great interest for targeted cancer therapy. Using a panel of small molecule PI3K isoform-selective inhibitors in a diverse set of breast cancer cell lines, we demonstrate that the biochemical and biological responses were highly variable and dependent on the genetic alterations present. p110α inhibitors were generally effective in inhibiting the phosphorylation of Akt and S6, two downstream components of PI3K signaling, in most cell lines examined. In contrast, 110β selective inhibitors only reduced Akt phosphorylation in PTEN mutant cell lines, and was associated with a lesser decrease in S6 phosphorylation. PI3K inhibitors reduced cell viability by causing a cell cycle arrest in the G1 phase of the cell cycle, with multi-targeted inhibitors causing the most potent effects. Cells expressing mutant Ras were resistant to the cell cycle effects of PI3K inhibition, which could be reversed using inhibitors of Ras signaling pathways. Taken together our data indicates that these compounds, alone or in suitable combinations, may be useful as breast cancer therapeutics, when used in appropriate genetic contexts.
Keywords: PI3K inhibitors, PKB/Akt, S6, phosphorylation, breast cancer, genetic alterations
Introduction
The phosphoinositide-3-kinase (PI3K) pathway has crucial roles in many cellular processes including cell survival, proliferation and migration, driven by many downstream signaling pathways [1]. PI3K consists of a catalytic subunit (termed p110) and a regulatory subunit (p85, p101, p55 depending on the enzyme) that acts to integrate stimulatory signals and direct the catalytic subunit to appropriate cellular locations. Four distinct class I p110 isoforms (α, β, γ and δ) generate phosphatidylinositol (3,4,5) trisphosphate (PtdIns(3,4,5)P3), which recruits downstream effectors to the sites of generation. Key to this process is the recruitment of 3-phosphoinositide dependent kinase 1 (PDK1) which plays a critical role in mediating the activation of AGC kinases including protein kinase B (PKB/Akt) and p70 ribosomal protein S6 kinase (p70S6K). In turn these kinases are known to play essential roles in cell proliferation, survival and growth [2].
Breast tumors can be defined by their morphological features as well as their distinct patterns of underlying genetic alterations [3]. In particular, the involvement of the PI3K pathway in breast cancer is implicated from a variety of genetic changes which include EGFR amplification, HER2/neu amplification [4], PTEN mutation/loss [5] and PI3K p110α mutations [6]. These p110α mutations cause increased PI3K activity [6, 7] are oncogenic [7, 8] and are prevalent at high frequency in breast cancers [9–11]. The presence of p110α mutations in breast tumors was found to be associated with estrogen receptor and progesterone receptor expression, and inversely correlated with PTEN expression, but did not affect prognosis or survival [12]. Phosphoinositide signals in breast cancer therefore point to therapeutic opportunities. Since the genetic diversity of commonly used breast cancer cell lines reflects many of the alterations found in breast cancer, these cell lines represent an ideal system to assess the potential for therapeutic agents which act on the PI3K pathway [13, 14].
The paradigms for clinical use of protein kinase inhibitors suggest that their effectiveness will be dictated by the specificity profile of a given compound and the genetic context in which it is being applied. This is well illustrated when considering the dramatic impact that imatinib (gleevec) has had on recent cancer treatment. While imatinib was first approved for chronic myelogenous leukemia patients expressing an activated form of abl, the knowledge that it also inhibits platelet derived growth factor receptor (PDGFR) and c-kit led to successful treatment of gastrointestinal stomach cancers and hyperoesinophilic syndrome. This latter indication is especially revealing as it was not previously known that these patients harbored alterations in PDGFR, kit or abl, but their dramatic responses to this agent led to sequencing efforts showing activating translocations of the PDGFR gene [15]. Therefore, detailed knowledge of the target (and off-target) specificities of new molecular therapies is critical to understand potential side effects, as well as predict which tumors will be most likely to respond to these agents.
Historically, the availability of p110 inhibitors has been largely limited to a single natural product, wortmannin and the synthetic chromone, LY294002 which broadly act on the majority of PI3Ks along with other related kinases such as mammalian target of rapamycin (mTOR), and are toxic when administered systemically in vivo [16]. Potential therapies have therefore been slow because selective compounds have been lacking. Recently however, a number of new classes of isoform selective PI3K inhibitors have been reported [17–20]. The most selective molecule, IC87114, exhibits >100 fold selectivity for 110δ vs. all other PI3K family members, allowing for precise analysis of this isoform in neutrophil migration [21, 22] and oxidase activation [23]. This compound has also revealed important roles for p110δ in breast cancer cell chemotaxis [24], and in myeloid leukemia [25]. The next most selective molecule is TGX-221, which inhibits PI3Kβ with high specificity, allowing for the analysis of the role of this isoform in thrombosis [26]. Compounds with true selectivity for p110α have not thus far been reported although the tool set of compounds available has pinpointed a critical role for p110α in insulin signaling [17] also confirmed by genetic approaches [27].
The availability of isotype selective PI3K inhibitors allows fundamental questions regarding the role of individual p110 isoforms in control of cell biology to be addressed. These include: 1) in cells coexpressing p110α and 110β, are distinct signaling functions regulated by each isoform? 2) in cells expressing all four isoforms can selective inhibitors reveal unique sensitivities under distinct growth conditions or in a genotype specific manner? 3) is loss of PTEN vs. p110α activating mutations equivalent or different in terms of creating inhibitor sensitivities? And 4) between the two most closely related p110 isoforms (α and β) why have only p110α activating mutations been identified in human cancers?
We have generated and characterized a panel of the most potent reported inhibitors with respect to biochemical activity against 18 PI3Ks and protein kinases, as previously described [17]. With this set of PI3K inhibitors we can target virtually any member of the PI3K class I family, as well as select members of other PI3K related kinases such as DNA-PK, mTOR. Our goal is to utilize this panel of inhibitors as a family wide approach to probe the role of PI3K family members in regulating breast tumor cell proliferation. These compounds comprise a wide variety of chemotypes with varying cross selectivities among the p110 isoforms. The advantage of this approach is that compound specific pharmacology, which often masks the real targets of lead compounds is somewhat ameliorated because of the presence of multiple chemotypes with similar biochemical targets. Any compounds that exhibit different biological responses but display apparent equivalent biochemical specificity can be quickly identified. Thus, each inhibitor in the panel becomes a drug candidate itself and a control for other molecules in the panel. The compounds used in this study include; p110 delta selective PIK-23, the p110 beta selective compounds TGX-286 and PIK-108, and multi-targeted PI3K inhibitors PIK-75, PI-103, PIK-85, PIK-90, and PIK-124. For full in-vitro activity characterisation along with chemical structures of these compounds, see previously reported data [17], and Table 2.
Table 2.
In-vitro IC50 data for the isoform selective small molecule inhibitors determined in the presence of 10μM ATP. Most of this data was previously published in [17].
| Compound | PIK-23 | TGX-286 | PIK-75 | PIK-85 | PIK-90 | PIK-108 | PI-103 | PIK124 |
|---|---|---|---|---|---|---|---|---|
| PI3Ks | ||||||||
| p110α/p85α | >200 | 4.5 | 0.0058 | 0.044 | 0.011 | 2.6 | 0.0082 | 0.023 |
| p110β/p85α | 42 | 0.12 | 1.3 | 0.8 | 0.35 | 0.057 | 0.088 | 1.1 |
| p110δ/p85α | 0.097 | 1 | 0.51 | 0.08 | 0.058 | 0.26 | 0.048 | 0.34 |
| p110γ | 50 | 10 | 0.076 | 0.05 | 0.018 | 4.1 | 0.15 | 0.054 |
| PI3KC2α | >100 | >100 | ~10 | ND | 0.047 | ~100 | ~1 | 0.14 |
| PI3KC2β | 100 | ~100 | ~1 | ND | 0.064 | ~20 | 0.026 | 0.37 |
| PI3KC2y | >100 | ND | ND | ND | ND | ND | ND | ND |
| hsVPS34 | ~50 | 3.1 | 2.6 | ND | 0.83 | ~5 | 2.3 | 10 |
| PI4Ks | ||||||||
| PI4Kllα | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| PWKIIIα | >100 | >100 | >100 | 2.5 | 0.83 | ~.50 | >100 | >100 |
| PI4KIIIβ | >100 | >100 | >100 | 3.6 | 3.1 | >100 | >100 | >100 |
| PIKKs | ||||||||
| ATR | >100 | >100 | 21 | ND | 15 | >100 | 0.85 | 2 |
| ATM | >100 | >100 | 2.3 | ~50 | 0.61 | 35 | 0.92 | ND |
| DNA-PK | >100 | ~50 | 0.0017 | 0.061 | 0.013 | 0.12 | 0.0019 | 1.5 |
| mTORC1 | >100 | >100 | 1 | >100 | 1.05 | ND | 0.02 | ND |
| mTORC2 | >101 | ND | 0.16 | ND | ND | ND | 0.125 | ND |
| hSMG-1 | ND | ND | ND | ND | ND | ND | ND | ND |
| PIPKs | ||||||||
| PI4P5Klα | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| PI4P5KIβ | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| PI5P4KIIβ | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
A suppressor enhancer chemical genetic screen was performed in order to look for differential sensitivity between genetically diverse breast cancers with a view to identify new therapeutic opportunities for breast cancer. Our results display a surprising heterogeneity of responses to different compounds in a cell line dependent manner. We conclude that the effectiveness of these drugs as targeted therapies will likely be determined by the cellular genetic alterations, as well as differences in drug metabolism and/or adaptation properties of individual tumors. The diversity of genetic changes leading to different types of breast cancers means that molecular markers of therapeutic sensitivity will be necessary to best match up therapies with genetic backgrounds. Importantly, we demonstrate that while p110β activity is not required for PI3K signaling in normal and WT PTEN expressing cancer cells, PTEN loss sensitises to p110β inhibitors. In addition, breast cancer cell lines expressing mutant Ras are resistant to all PI3K inhibitors tested, which can be reversed using a combination approach of PI3K inhibitors and MEK inhibitors.
Experimental Materials
Rabbit polyclonal Phospho PKB Serine 473 was generated as previously described [28]. Phospho-S6 (S235/236) and phospho-GSK3 S21/S9 were from cell signaling technology. β-actin antibody was from Sigma and PTEN antibody was from cascade biosciences. LY294002, UO126 and Rapamycin was obtained from Calbiochem. Hoescht dye, Alexa 488 rabbit secondary antibody was from Molecular probes. All other reagents were from Sigma unless otherwise stated.
Cell lines, Cell culture and transfection
Cell culture reagents were purchased from Invitrogen. BT474, BT549, BT20, HS578t, and T47D cells were maintained in RMPI with 10% fetal bovine serum (FBS). MDA-MB-468, MDA-MB-231, MCF7 and SKBR3 cells were maintained in DMEM with 10% FBS. MCF10A cells were cultured in media composed of 50:50 DMEM and F12 HAM supplemented with 20 ng/ml EGF, 100 ng/ml cholera toxin, 10.08 μg/ml insulin 500 ng/ml hydrocortisone and 5% FBS. All cell lines were obtained from the UCSF inter-SPORE collection as deposited with ATCC. Transfection of PTEN siRNA was performed using Oligofectamine™ reagent and either PTEN siRNA-1 (target sequence; AAGGCGTATACAGGAACAATA) or PTEN siRNA-1 (validated PTEN siRNA from Qiagen; Cat number SI00301504). Two rounds of transfection of siRNA at 100 nM were performed followed by treatment with either PIK-108 or PI-103 at 1 μM for 1 h prior to harvesting. To create BT-20 cells stably expressing KRasG12D, they were initially infected with retroviruses expressing the Ecotropic receptor and selected in medium containing 5 μg/ml blastocidin. Following selection for 2 weeks, these cells were subsequently infected with retroviruses containing KRasG12D, or empty vector, and maintained in 1 mg/ml G418 for 4 weeks to obtain pools of cells expressing activated Ras.
Western blotting
Cells were seeded on 6 well plates at 80% confluence prior to addition of p110 inhibitors and other compounds. Cells were harvested by scraping into 150 μl lysis buffer (1% NP40, 20 mM Tris, pH 7.5, 150 mM NaCl, 25 mM NaF, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM NaVO4, Complete protease inhibitor (1 pill in 20 ml, Roche, Indianapolis, IN)). Protein samples were quantitated using protein assay (Biorad laboratories). Typically 10 μg of protein per sample was separated by SDS PAGE, immunoblotted with indicated antibodies and visualized by enhanced chemiluminescence on film (Amersham). Western blot quantitations were performed using NIH Image™.
Immunofluorescence
Cells were seeded at 80% confluence on 96 well plates. Compounds were added as indicated in the figure legends and cells were then washed and fixed in 4% paraformaldehyde for 15min. Washes and incubations were performed in Phosphate Buffered Saline (PBS) unless otherwise stated. Cells were permeablized with 0.2% (v/v) Triton X-100 for 5 min, washed, and incubated in 1% (w/v) bovine serum albumin (BSA) for 20 min. Cells were incubated with primary antibody containing 1% (w/v) BSA for 1 h. Cells were then washed and incubated with fluorescent dye-conjugated secondary antibody for 1 h. Quantitation of Phospho S6 immunoflourescence was performed using the Cellomics arrayscan® HCS reader system and determined by the compartmental analysis bioapplication from Cellomics. Cell number was determined by counting nuclei stained with Hoescht.
FACS Analysis
Cells were seeded on 6 well plates at 80% confluence prior to addition of compounds as indicated in the figure legends. Adherent cells were collected by trypsinization and pooled with medium containing non-adherent cells, washed with PBS and subsequently fixed in 70% ethanol. A further wash with PBS containing 5% FBS was followed by resuspension in PBS containing propidium-iodide (10 μg/ml) and RnaseA (0.1 mg/ml). Analysis of 10,000 cells was performed on FACScan using Cell Quest software.
Results
A panel of breast cancer cell lines was selected to represent various genetic alterations seen in primary breast tumors such as PTEN mutation/loss, EGFR overexpression, ERBB2 overexpression, BRCA1 mutation, estrogen receptor expression and Ras mutation. These cell lines, BT474, SKBR3, BT20, T47D, MCF7, MDA-MB468, BT549, MDA-MB231 and Hs578t, are summarized in Table 1. MCF10A cells were used for comparative purposes as a non-tumorigenic, karyotypically normal cell line. Affymetrix gene expression profiling performed on these cell lines show that p110 α, β, and δ all show similar mRNA expression levels whereas p110γ, which is known to be more restricted in its tissue distribution [29] is expressed at low to undetectable levels (Luika Timmerman, Jennifer Yeh, Frank McCormick and Joe Gray, personal communication).
Table 1.
Breast cell lines studied; cell types and known genetic alterations.
| Cell Line | Cell Type | ER | Ras | PTEN | HER2 | PIK3CA |
|---|---|---|---|---|---|---|
| BT474 | Epithelial | +ve | Amplified | K111N | ||
| MCF-7 | +ve | E545K | ||||
| SKBR3 | +ve | Amplified | WT | |||
| T47D | +ve | H1047R | ||||
| BT20 | Trans/Basal | −ve | P539R H1047D |
|||
| MCF 10A | +ve | WT | ||||
| MDA-MB-468 | −ve | Δ44bp exon 3 | WT | |||
| Hs578t | Mesenchymal | +ve | HRas G12D |
WT | ||
| MDA-MB 231 | −ve | KRas G13D |
WT | |||
| BT549 | −ve | W274stop | WT |
Breast cancer cell lines display distinct biochemical sensitivities to p110 isoform-selective inhibitors
Of the eight compounds in our study, five were chosen for detailed analysis due to their target representation and promising activity in preliminary experiments. These include the potent p110α inhibitors PIK-75 and PI-103, the relatively selective p110β inhibitors TGX-286 and PIK-108, and PIK-85, which inhibits p110α γ and δ, and p110β less potently. The selectivity of these compounds as assessed by in vitro lipid kinase assays is shown in Table 2. Compounds were added to each of the cell lines at concentrations of 0.5 μM, 1 μM and 10 μM for 1 h to assess inhibition of PI3K signaling using phospho-specific antibodies against PKB/Akt (S473) and S6 (S235,236). S473 is generally associated with activated forms of PKB/Akt and is known to be phosphorylated in a PI3K dependent manner. However, as this site is directly phosphorylated by mTORC2 [30], PI3K inhibitors that also directly inhibit mTOR will have an effect on this site. S235,236 are generally phosphorylated by p70S6K, although under some circumstances can be phosphorylated by p90rsk [31, 32].
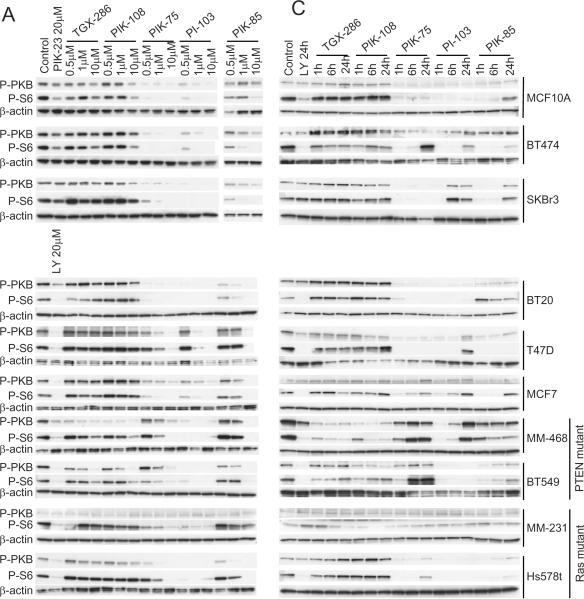
A surprising degree of heterogeneity in biochemical responses was observed across the different cell lines examined to p110 inhibitors. In general, p110α inhibitors were the most effective at inhibiting phosphorylation of these target proteins (Fig. 1A). PIK-75 and PI-103 are the most potent p110α inhibitors, followed by PIK-85. In seven of 10 cell lines examined, the inhibition of PKB/Akt and S6 phosphorylation was consistent with p110α driving the activity of this pathway, with PIK-75 and PI-103 being equipotent at inhibiting the phosphorylation of these proteins, PIK-85 being less potent, and TGX-286 and PIK-108 being ineffective. This includes the cell line chosen for its normal karyotype and non-transformed morphology (MCF10A), as well as cell lines BT474, SKBR3, BT20, T47D, MCF7 and HS578t. In contrast MDA-MB-231 cells, which showed very low basal PI3K pathway activity, were resistant to all p110 inhibitors tested when analyzing PKB/Akt phosphorylation. Surprisingly, S6 phosphorylation was still sensitive to p110α inhibitors PIK-75 and PI-103 in this cell line. MDA-MB-468 cells and BT549 cells showed a distinct profile of inhibition, with PI-103 (a p110α and p110β inhibitor) being more potent than PIK-75 (a potent p110α inhibitor, but not p110β inhibitor) at inhibiting PKB/Akt and S6 phosphorylation. Moreover, selective p110β inhibitors, such as TGX-286 and PIK-108, were able to inhibit the phosphorylation of PKB/Akt exclusively in MDA-MB-468 and BT549 cells. These cell lines harbor PTEN mutations [33], unlike the other eight cell lines in the panel which express wild type PTEN. While both PKB/Akt and GSK3 phosphorylation was inhibited by p110β inhibitors in PTEN mutant cell lines, S6 phosphorylation remained unchanged (Fig. 1B). As shown for MCF10A, SkBr3 and BT474, no inhibition of PI3K signaling as assessed by PKB/Akt phosphorylation was apparent in any cell line when using p110δ-selective compounds such as PIK-23. However, S6 phosphorylation was sometimes inhibited, suggesting a potential p110δ-dependent but PKB/Akt-independent pathway for regulating S6 phosphorylation in breast cancer cells. As expected, LY294002 was effective at inhibiting PI3K signaling in all cell lines examined (Fig. 1A). The PKB phosphorylation in response to the p110 inhibitors was quantitated, and the in cell IC50 values calculated. These are displayed in Fig. 1B, providing a summary of the effects of the p110 inhibitors in the panel of breast cancer cell lines. Therefore, PKB/Akt and S6 phosphorylation are dependent on either p110α or p110β activity, in a manner that correlates with PTEN status.
Figure 1. Dose-dependent and temporal biochemical responses of different breast cancer cell lines to increasing concentrations of isotype-selective PI3K inhibitors.
(A) MCF10A, BT474, SKBR3, BT20, T47D, MCF7, MDA-MB-468, BT549, MDA-MB-231 and Hs578t cells were cultured in six well dishes as described in Materials and Methods. The indicated concentrations of TGX-286, PIK-108, PIK-75, PI-103 and PIK-85 were added to the culture medium, and one hour later cells were harvested into lysis buffer. Proteins were separated by SDS-PAGE, and Western blotted using antibodies against phospho-PKB/Akt (Ser-473), phospho-S6 (Ser-235,236) and β-actin. Proteins were visualized by horse radish peroxidase coupled secondary antibodies and enhanced chemiluminescence (B) Graph displaying the in cell IC50 values for phospho-PKB/Akt (Ser-473), phospho-GSK3 (Ser-9/Ser-21) and phospho-S6 (Ser-235,236) in the different cell lines against the p110 isoform selective inhibitors. (C) MCF10A, BT474, SKBR3, BT20, T47D, MCF7, MDA-MB-468, BT549, MDA-MB-231 and Hs578t cells were cultured in six well dishes as described in Materials and Methods. 1 μM of TGX-286, PIK-108, PIK-75, PI-103 and PIK-85 were added, and the cells harvested into lysis buffer 1, 6 and 24 hours later. Proteins were separated by SDS-PAGE and analyzed by Western blotting with the indicated antibodies.
Distinct pharmacodynamic properties of p110 inhibitors in different cell lines
We next analyzed the temporal responses to p110 inhibitors. The inhibition of PKB/Akt and S6 phosphorylation was differentially sustained among cell lines studied (Fig. 1C). Some cell lines (for example BT549) showed transient inhibition of PKB/Akt and S6 phosphorylation by PIK-75, which returned by 6 h, whereas in other cell lines PIK-75 inhibition was sustained (for example T47D, MCF10A, BT20). Similar disparate results were also seen for PI-103 (transient inhibition in MDA-MB-468, SKBr3 and MCF7 versus sustained inhibition in BT549 and BT20). Whether this represents differences between cell lines in drug metabolism or in feedback compensation is currently unknown. However, such disparate pharmacodynamic responses are likely to influence the biological responses to these compounds. An increase in PKB phosphorylation in response to TGX-286 is apparent from Fig.1C. We believe this is an artifact of antibody labelling or `edge effect' since this finding was not replicated in Fig.1A and in other experiments.
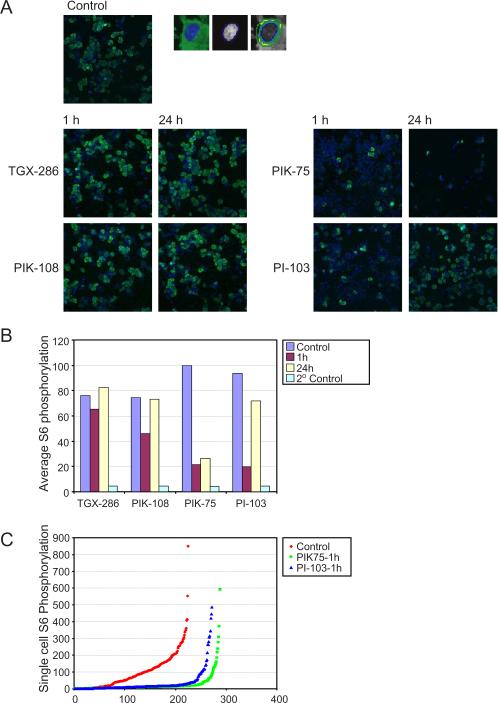
The phosphorylation status of S6 was also monitored by immunoflourescence. This was performed using the Cellomics arrayscan® HCS reader system and intensity of fluorescence determined by the compartmental analysis bioapplication. This analysis confirmed that PI-103 and PIK-75 inhibit S6 phosphorylation in SKBr3 cells after 1 h, which was sustained with PIK-75 but transient for PI-103 (Fig. 2 A,B). p110β inhibitors TGX-286 and PIK-108 showed only a minimal inhibition of S6 phosphorylation, similar to the results seen by Western blotting in this cell line. A cursory visual inspection of the immunofluorescence data suggested that PIK-75 and PI-103 appeared to reveal an “all or nothing” pattern of inhibition whereby the S6 phosphorylation is either fully inactivated or fully maintained, in a manner reminiscent of the switch-like behavior seen in the ERK pathway under some conditions [34]. However when intensity values for single cells were quantitated and graphed, there is in fact a graded range of fluorescence values that are inhibited in a uniform manner (Fig. 2C).
Figure 2. Recovery of breast cancer cells following treatment with isotype-selective PI3K inhibitors by immunofluorescence.
(A) SKBr3 cells were plated in 96 well dishes and treated 18 h later with 1 μM TGX-286, PIK-108, PIK-75 and PI-103 for 1 and 24 h. Cells were fixed and permeablized as described in Materials and Methods, and incubated with antibodies against phospho-S6. Fluorescently labeled secondary antibodies and Hoechst were then added to visualize phospho-S6 (green) and DNA (blue) respectively. Quantitation of cytoplasmic Phospho S6 immunoflourescence was performed using the Cellomics arrayscan® HCS reader system using the compartmental analysis bioapplication from Cellomics. Hoechst fluorescence was used to define nuclei, and a 2 μM ring was drawn around the region of Hoechst fluorescence to define cytoplasmic localization. A single image of four collected for each condition is shown. (B) Quantitation of average fluorescence from 200 cells in the presence of the indicated inhibitors at the indicated time points, as well as the fluorescence from addition of secondary antibody alone, is shown. (C) The phospho-S6 fluorescence of individual cells treated with PIK-75 and PI-103 for 1 h, or no drug is shown.
Breast cancer cell lines show distinct responses following p110 isoform inhibition
In order to assess the biological effects of these compounds, cell numbers were examined at various times following addition of p110 inhibitors. Most of the cell lines were inhibited by many of the compounds to a greater or lesser extent, as illustrated in Figure 3A for MCF10A cells. In general, p110α selective inhibitors had the greatest ability to reduce cell number in a dose-dependent manner (Fig. 3A). However, other cell types such as Hs578t cells showed much greater resistance to decreases in cell number in response to these inhibitors (Fig. 3A), despite their sustained biochemical inhibition in response to PI-103 and PIK-85 (Figs 1B and 3B). Following 48 h of treatment, Hs578t cells were resistant to all compounds except PIK-75, which strongly inhibited cell number (Fig. 3A). Paradoxically, in this cell line S6 phosphorylation rapidly recovered to untreated levels following treatment with PIK-75, but remained inhibited by PI-103 up to 24 h (Figs 1B and 3B). This dissociation between PI3K inhibition and cell number suggests that PIK-75 decreased cell numbers due to effects on targets other than PI3K and is consistent with previously reported selectivity data for PIK-75 [17].
Figure 3. Effects of PI3K isoform selective inhibitors on breast cancer cell number.
(A) MCF10A and Hs578t cells were plated in 96 well dishes, and the indicated concentrations of TGX-286, PIK-108, PIK-75, PI-103, PIK-85, PIK-90 and PIK124 added to the medium the following day. Cells were incubated for a further 48 h (replacing media with fresh compound every 24 h), then fixed and permeablized. Hoechst was added to the cells, and the number of nuclei quantitated using cellomics arrayscan software. (B) Hs578t cells grown in 96 well dishes were treated with 1 μM PIK-75, PI-103 and PIK-108 for 24 h. Cells were fixed, permeablized, and incubated with anti phospho-S6 antibodies. After incubation with fluorescent secondary antibodies and Hoecht, S6 phosphorylation was quantitated using Cellomics arrayscan software. Average values per cell following treatment with ZK 75, PI-103 and PIK-108 are shown. (C) 1 μM of TGX-286, PIK-108, PIK-75, PI-103, PIK-85, PIK-90 and PIK124 were added to the indicated cell lines for 24, 48 and 72 h (replacing media with fresh compound every 24hrs). Cells were fixed and permeablized, and cell numbers quantitated using Cellomics arrayscan software by Hoechst fluorescence. Numbers are expressed as a percentage of untreated cells, and colour-coded for degree of inhibition
The effects of PI3K inhibitors on cell number were performed on eight of the 10 cell lines at 1 μM (MCF7 and BT474 were difficult to obtain consistent cell number data due to cell losses incurred by the washing and fixation protocol). Whereas PIK-75 consistently showed the greatest ability to induce loss of cell number in all cells examined, the responses to other compounds were more heterogeneous. MDA-MB-468, MCF10A, BT20 and T47D cells displayed time dependent inhibition of cell number with several compounds, whereas MDA-MB-231, Hs578t, SKBR3 and BT549 were more resistant (Fig. 3C). In some cases the pattern of cell number inhibition displayed properties reflective of biochemical sensitivity and pharmacodynamic properties. For example MDA-MB-468 (p110β inhibitors), MCF10A, BT20 and T47D cell lines displayed potent and sustained biochemical inhibition to their respective p110 inhibitors (Fig. 1B), whereas SKBR3 cells showed transient inhibition and MDA-MB-231 cells showed less potent biochemical inhibition (Fig. 1B), and are among the most biologically resistant cell lines (Fig. 3C). Exceptions to these observations include BT549 and Hs578t, which both showed strong and sustained inhibition by PI-103 yet were resistant to cell number decreases in response to this compound. Hs578t cells harbor an activating mutation in H-Ras, and will be discussed later.
p110 isoform selective inhibitors differentially induce cell death and/or cell cycle changes in breast cancer cell lines
Whilst changes in cell number analyzed by automated cell counting were a useful indication to the differential effects on different cell lines, we found the results to be somewhat variable, and in addition do not give any suggestion as to the underlying cause of cell number decreases. We therefore next analyzed the responses to p110 inhibitors in more detail using flow cytometry. As previously mentioned PIK-75 and PI-103 are potent inhibitors of p110α and reduce PKB/Akt and S6 phosphorylation, although PIK-75 is much more efficient at reducing cell number compared with PI-103 (Fig. 3). Using flow cytometry in SKBR3 cells, PIK-75 induced a significant sub-G1 population (Fig. 4A), consistent with the induction of apoptosis. Equivalent and even higher concentrations of PI-103 did not induce such an effect. These data support the concept that the potent effects of PIK-75 in reducing cell number are due to apoptosis, which occurs in a PI3K-independent manner.
Figure 4. Effects of PI3K isoform selective inhibitors on cell cycle progression in breast cancer cell lines; loss of PTEN induces sensitivity to p110β inhibition.
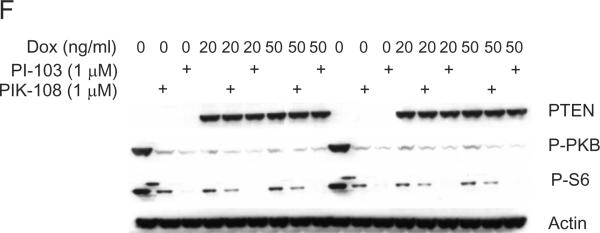
(A) SKBr3 cells were grown in six well dishes, and treated with the indicated concentrations of PIK-75 or PI-103. 24 h later, cells were fixed, permeablized and treated with 10 μg/ml propidium iodide and 0.1 mg/ml RnaseA.10,000 cells were analyzed for DNA content by flow cytometry, and the proportion of cells showing less than 2n DNA content are indicated. (B) MCF10A, BT474, MDA-MB-468 and MDA-MB-231 cells were grown in six well dishes and treated with 1 μM PIK-75, PI-103, TGX-286 and PIK-108 for the indicated time periods (replacing media with fresh compound every 24 h). Cells were fixed, permeablized and treated with 10 μg/ml propidium iodide and 0.1 mg/ml RnaseA, and 10,000 cells analyzed for DNA content by flow cytometry. The proportion of cells showing less that 2n DNA content are indicated in black, 2n DNA content indicated in dark grey, 4n DNA content indicated in light grey, and 2n<DNA content<4n indicated in white. (C) MCF10A, BT474, MDA-MB-468 and MDA-MB-231 cells were grown in six well dishes and treated with 1 μM of the indicated compounds for 24 h. Then the medium was replaced with 1 μM fresh compound in the presence of 70 ng/ml nocodazole for a further 24 h. Cells were fixed, permeablized and treated with 10 μg/ml propidium iodide and 0.1 mg/ml RnaseA, and 10,000 cells analyzed for DNA content by flow cytometry. Colour coding for DNA content is the same as for (B). (D) The indicated cell lines were grown in 96 well dishes and treated with 0.5 μM and 1 μM PIK-108. 48 h later (replacing media with fresh compound every 24 h) cells were fixed, permeablized, and treated with Hoechst for analysis of cell number. (E) MDA-MB-231 cells were cultured in six well dishes as described in Materials and Methods. Two rounds of transfection of PTEN siRNA was performed as indicated on consecutive days and as described in Materials and Methods. 1 μM of PIK-108 or PI-103 was added to the culture medium as indicated and one hour later cells were harvested into lysis buffer. Proteins were separated by SDS-PAGE, and Western blotted using antibodies against phospho-PKB/Akt (Ser-473), PTEN and β-actin. Proteins were visualized by horse radish peroxidase coupled secondary antibodies and enhanced chemiluminescence. (F) MDA-MB-468 cells expressing doxycyclin-inducible PTEN were cultured in DMEM+10% FBS in 6 well dishes, and treated with either doxycycline or solvent (methanol) for 24 hours. PIK-108 and PI-103 were added to 1 μM final concentration for 1 hour, and then cell lysates were harvested. Lysates were separated by SDS-PAGE, and Western blotted using the indicated antibodies. Lanes 1–9 and 10–18 represent duplicate samples.
We proceeded to perform further flow cytometry analysis on MCF10A, BT474, MDA-MB-231 and MDA-MB-468 cells over 24, 48 and 72 h with PIK-75, PI-103, TGX-286 or PIK-108 (Fig. 4B). Similar to the effects seen in SKBr3 cells, PIK-75 induced a large sub-G1 population that occurred rapidly in BT474 cells, and more delayed in MCF10A and MDA-MB-468 cells. In contrast, PIK-75 caused a strong accumulation of cells with a 4n DNA content in MDA-MB-231 cells. Neither PI-103, TGX-286 or PIK-108 showed strong effects on DNA content in these cells under these conditions.
In order to determine any potential additional effects on the cell cycle of TGX-286, PIK-108, PIK-75 and PI-103, we used nocodazole (a microtubule depolymerization agent which synchronizes cells in G2) following addition of compound. This protocol accentuates any effects on G1 arrest by arresting the cells at the G2/M transition, due to destabilized microtubules. In contrast, cells arrested in G1 by p110 inhibitors will not transition through to G2. These studies revealed that PI-103 induced a G1 arrest in BT474 and MCF10A cells although not in MDA-MB-231 cells or in MDA-MB-468 cells (Fig. 4C).
The PTEN mutant cells MDA-MB-468 and BT549 were the only cell lines in this study in which PKB/Akt phosphorylation was inhibited by the addition of the p110β selective compounds TGX-286 and PIK-108 (Fig. 1). Strikingly a G1 arrest was uniquely observed in MDA-MB-468 cells after TGX-286 and PIK-108 treatment, further supporting the finding that these cells are sensitive to p110β inhibition (Fig. 4C). Interestingly MDA-MB-468 cells also experienced a dose-dependent reduction in cell number upon addition of these compounds whereas five other cell lines remained broadly unaffected by TGX-286 and PIK-108 (Fig. 4D). BT549 cells however, which also exhibit biochemical sensitivity to TGX-286 and PIK-108, did not exhibit these responses to cell number, although this could be the result of lower sensitivity when compared with MDA-MB-468 cells (Fig. 1, and data not shown). We tested the hypothesis that loss of PTEN may induce sensitivity to p110β inhibition by using siRNA to PTEN in the biochemically resistant line, MDA-MB-231. siRNA to PTEN in these cells induced an increase in PKB phosphorylation, as expected. Moreover, this increase was abrogated following addition of PIK-108 or PI-103 (Fig. 4E). To determine whether the converse was also true, namely that expression of PTEN in a PTEN-null cell line would make these cells resistant to p110β inhibitors, we used MDA-MB-468 cells expressing a doxycycline-inducible PTEN. Unfortunately, under these conditions, expression of PTEN almost fully inhibited PKB/Akt S473 phosphorylation (while having relatively minor effects on P-S6 phosphorylation), making conclusions difficult to draw (Fig. 4F).
Ras mutations are an important determinant of response to PI3K inhibitors
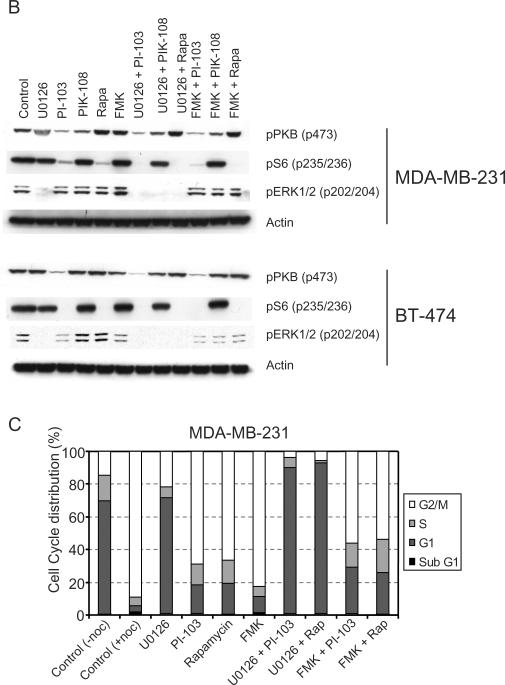
We next explored the observations that MDA-MB-231 cells were insensitive to cell cycle changes in response to PI-103 addition (Fig. 4C) and that Hs578t cells were insensitive to cell number inhibition across a range of PI3K inhibitors (Fig. 3A), despite responding biochemically to these compounds (Fig. 1). Both of these cell lines possess Ras mutations (K-Ras and H-Ras respectively) [35, 36]. We therefore tested whether activation of pathways downstream of Ras were responsible for resistance to PI3K inhibitors in these cells, and whether inhibition of these pathways could cooperate with inhibition of PI3K. Analysis of DNA content using the nocodazole protocol in both of these cell lines showed a marked insensitivity to the p110α/p110β inhibitor PI-103, in contrast to the WT Ras-expressing cell lines BT474, BT20 and MCF-10A (Fig. 5A). PIK-108 was also sufficient to cause a partial G1 arrest in the PTEN mutant cell line MDA-MB-468, as seen previously. Inhibition of MEK (using U0126) caused a G1 arrest in all cell lines examined, although the extent was variable (Fig. 5A). Strikingly, in the mutant Ras cell lines, U0126 and PI-103 caused a synergistic effect on G1 arrest (Fig. 5A). Furthermore, since PI-103 has been shown to have potent activity against mTOR [17], we tested the extent to which PI-103 was acting through inhibition of PI3K or mTOR. Addition of rapamycin to MDA-MB-231 or Hs578t cells was ineffective at causing a G1 arrest, similar to PI-103. In contrast, rapamycin caused a G1 arrest (to varying degrees) in all cells expressing WT Ras. Rapamycin also synergized with U0126 in the mutant Ras expressing cell lines, and caused a G1 arrest identical to that seen with the combination of U0126 and PI-103. Such synergy was not apparent in BT20 or MDA-MB-468 cells. This synergy was also not apparent in BT474 or MCF10A cells, although this is due to the fact that either compound alone was very effective in these cells. Therefore, the status of Ras mutations is likely to play an important role in determining resistance to PI3K inhibition, but this resistance can be reversed using MEK inhibitors.
Figure 5. Effects of combined PI3K isoform selective inhibitors and MEK inhibition on cell cycle progression in breast cancer cell lines.
(A) MDA-MB-231, Hs578t, BT20, BT474, MCF10A and MDA-MB-468 cells were grown in six well dishes and treated with the indicated compounds for 24 h (1 μM PI3K inhibitor, 10 μM U0126 and 100 nM rapamycin as indicated). Then the medium was replaced with fresh compound at the same concentration in the presence of 70 ng/ml nocodazole for a further 24 h. Cells were fixed, permeabilized and treated with 10 μg/ml propidium iodide and 0.1 mg/ml RnaseA, and 10,000 cells analyzed for DNA content by flow cytometry. Colour coding for DNA content is the same as for Figure 4B. (B) MDA-MB-231 cells and BT-474 cells were treated with the indicated compounds for 1 h and then harvested for cell lysates. Proteins were separated by SDS-PAGE, and analyzed for phosphorylation by Western blotting with the indicated antibodies.
There was also a discrepancy in the inhibition of S6 phosphorylation in Ras mutant vs Ras WT cell lines. While rapamycin was sufficient to fully inhibit S6 phosphorylation in the WT Ras expressing cell lines BT-474 (Fig. 5B) and BT20 (data not shown), there was residual phosphorylation of S6 in the mutant Ras expressing MDA-MD-231 cell line. We hypothesized that this could be due to a Ras driven ERK/p90rsk contribution to S6 phosphorylation in MDA-MB-231 cells, similar to that seen in cells lacking p70S6K [31], and in 293E cells expressing NRas Q61L or stimulated with phorbol esters [32]. Consistent with this, the combination of U0126 with PI-103 or rapamycin completely abolished S6 phosphorylation in these cells. Similarly, the selective p90rsk inhibitor fmk [37] also cooperated with U0126 or rapamycin in reducing S6 phosphorylation (Fig. 5B). However, fmk was not sufficient to cause any effect on cell cycle arrest, nor did it cooperate with PI-103 or rapamycin (Fig. 5C).
To determine whether expression of mutant Ras was sufficient for causing partial resistance to the ability of PI-103 and rapamycin to inhibit S6 phosphorylation, we stably expressed KRasG12D in BT20 cells. Although the expression levels of exogenous mutant KRas were quite low relative to endogenous Ras isoforms, they were sufficient to elevate ERK phosphorylation under starved conditions (Fig. 6A). Expression of KRasG12D was also sufficient to cause a minor resistance to inhibition of S6 phosphorylation following treatment with either PI-103 or rapamycin (Fig. 6B). Moreover, expression of KRasG12D reduced the G1 arrest induced by treatment with PI-103, although an additive or synergistic interaction with U0126 was not apparent from these experiments (Fig. 6C).
Figure 6. Expression of mutant Ras results in signaling to S6 phosphorylation and resistance to G1 arrest by PI3K inhibitors.
(A) BT-20 cells were infected with retroviruses expressing empty vector or KRasG12D, and selected in neomycin for 4 weeks. Parental, vector or KRasG12D expressing cells were serum starved overnight, and then stimulated with 0.5 or 10% FBS for 1 hour and then harvested for cell lysates. (B) Cells from (A) were treated with the indicated compounds for 1 h and then harvested for cell lysates. Proteins were separated by SDS-PAGE, and analyzed for phosphorylation by Western blotting with the indicated antibodies. (C) The cell lines shown were treated with the indicated compounds for 24 h, and treated as described for Fig. 5A.
Discussion
The data presented herein describe the biochemical and biological effects of PI3K isotype selective small molecule inhibitors on a panel of breast cancer cell lines. We demonstrate that these are highly effective at inhibiting the PI3K pathway, at inhibiting cell number and inducing cell death and/or cell cycle arrest, in a cell type-dependent manner depending on distinct genetic characteristics.
The importance of increased PI3K signaling in breast cancer is highlighted by the finding that p110α is mutated at high frequency in this disease [9–11]. In particular these have been shown to be activating mutations, making this isoform of great interest with regard to the usefulness of p110α selective small molecules [6]. The p110α mutation status in the breast cancer cell lines studied here has recently been determined, both by the Sanger Center, (data on their website) and in [12]. BT20, BT474, T47D and MCF7 contain p110α mutations, and BT549, Hs578t, MDA-MB-468, MDA-MB-231, SKBr3 express wild type p110. We show that while p110α inhibitors are generally effective at the inhibition of PI3K signaling both biochemically and biologically, there was no clear increase in sensitivity seen in cell lines expressing mutant p110α. Moreover, it was also not obvious that cells expressing mutated p110α showed increased basal phosphorylation of either PKB/Akt or S6. A possible explanation for this is that while four of the 10 cell lines showed p110α mutations, all of the cancer cell lines analyzed had mutations in either p110α, PTEN, Ras or amplification of ErbB2, all of which could cause elevated PI3K pathway activity. Another explanation attests to whether the presence of a particular genetic alteration automatically implies that the altered gene is a good target for therapy. Whilst this has proven true for oncogenes such as bcr-abl and kit [38], current evidence suggests that mutations in EGFR may not predict response to EGFR inhibitors in lung cancer [39, 40], and B-Raf mutations in melanoma do not currently appear to predict response to sorafenib [41], a compound which inhibits B-Raf activity.
It should be noted that MCF10A cells, which are karyotypically normal and are not transformed, were equally sensitive to p110α inhibitors, and were in fact the most sensitive to MEK inhibitors, as measured by flow cytometry. p110α inhibitors were also recently shown to inhibit glucose uptake in non-transformed adipocytes and myotubes [17]. It remains unclear whether strong effects on `normal' cells in culture will translate into systemic toxicity in the clinic.
Loss of PTEN is well documented to be associated with a poor clinical outcome in breast cancer [42, 43]. The unique sensitivity of MDA-MB-468 and BT549 cells to the p110β selective inhibitors, TGX-286 and PIK-108 is therefore of particular interest. These are the only cell lines studied which respond to these compounds when examining PKB/Akt and S6 phosphorylation. In addition, these compounds also cause a G1 arrest in MDA-MB-468 cells. Therefore, while “normal” and WT PTEN expressing tumor cells are equally sensitive to p110α inhibitors (with the exception of cell lines expressing mutant Ras, discussed below), PTEN mutant tumor cells may become more dependent on additional isoforms such as p110β. This correlation was extended to an isogenic system in which knockdown of PTEN in MDA-MB-231 cells using siRNA was sufficient to confer responsiveness to p110β inhibitors. It is unclear why PTEN mutated/deleted cells show a particular isotype dependence, although cellular localization of PTEN and particular p110 isoforms could play a role. In this regard, it has been reported that both PTEN [44] and p110β [45] can be localized to the nucleus. Although somewhat preliminary, these results may suggest a window of therapeutic opportunity in tumors expressing mutant PTEN to p110β inhibition. Interestingly, a particular reliance of p110β in prostate tumor cells has been previously noted by other investigators [46]. PC3 cells, which express a mutant form of PTEN, show a high basal level of PKB/Akt phosphorylation, which is reduced upon inhibition of p110β expression, but not p110α. p110β inhibition also resulted in inhibition of invasive growth through matrigel in these cells, in contrast to p110α inhibitors [46]. While PTEN mutations are relatively rare in breast tumors, decreases in PTEN levels are more frequent and have been shown to have prognostic significance in this disease. Tumors that have low levels of PTEN without mutations may therefore also be candidates for p110β inhibition. In addition, preliminary experiments in prostate cancer (data not shown) and glioblastoma [47] cell lines show a similar correlation between PTEN mutations and sensitivity to p110β inhibitors. These tumors display a greater incidence of PTEN mutations and may represent a wider arena for this class of drugs.
Distinct pharmacodymamic responses were observed among the inhibitors tested and the cell lines studied. Although we have not analyzed the stability of these compounds, their structures do not suggest any aqueous instability, and most are effective over a 72 hour time period in at least one or more cell lines. In addition, other cellular screens have shown phenotypes consistent with inhibition of their respective targets [17]. One explanation for such heterogeneous pharmacodynamic responses could be that different cell lines metabolize drugs at differing rates. An alternative explanation could be that different cell types have the ability to switch their dependence for a particular PI3K isoform after one is inhibited. These hypotheses could be tested by re-adding either the same compounds, and/or different ones at later time points to see whether this is sufficient to reduce the restored PKB/Akt phosphorylation. Given that PI-103 also inhibits p110β, one might have expected this compound to also induce a G1 arrest in MDA-MB-468 cells. However, pharmacodynamic data from Figure 2 shows that in MDA-MB-468 cells, the effects of PI-103 on PI3K inhibition are considerably more transient when compared to TGX-286 and PIK-108 which could explain this apparent discrepancy. These data further illustrate the importance of pharmacodynamics in drug responses and highlight the need for reliable pharmacodynamic markers in treated patients.
The finding that Hs578t cells are relatively resistant in terms of cell proliferation to the drugs studied whereas PI3K signaling is inhibited indicates that these cells can dispense with PI3K signals for cell proliferation. One explanation for this could be that these cells have activation of parallel pathways, which are able to compensate for the loss of PI3K signals. For example, Hs578t cells are known to express activated forms of the H-Ras oncogene [36]. Another resistant cell line (both biochemically and biologically) is MDA-MB-231 which is known to harbor K-Ras mutations [35]. The clinical importance of these observations is apparent by the finding that Ras mutations confer resistance to additional signal transduction inhibitors such as anti-EGFR therapies [39, 48]. Importantly, in these two breast cancer cell lines examined, the resistance to PI3K inhibitors could be overcome by the simultaneous addition of a MEK inhibitor. Part of the reason for this could be the additional input to S6 S235,236 phosphorylation through the MEK-ERK-p90rsk pathway seen in the context of activated KRas. This observation has previously been seen in HEK 293E cells expressing activated NRas [32]. However, this residual phosphorylation could also be abolished using the p90rsk inhibitor fmk, without having any large effect on cell cycle profile, suggesting that additional MEK-dependent pathways downstream of Ras are important for this. Therefore the rational combination of targeted therapies in the appropriate genetic contexts is likely to extend the patient populations that will respond to these agents. We have previously shown that high PKB/Akt phosphorylation predicts poor response to EGFR inhibitors in patients with glioblastoma [49]. These patients would therefore be candidates for combination therapy with a PI3K or PKB/Akt inhibitor. In support of this a strong synergistic response between LY294002 and gefitinib was previously demonstrated in glioblastoma xenograft tumors [50].
Although PIK-75 was very effective at killing all cell types examined, several lines of evidence suggest that this compound is unlikely to mediate its cell death responses through inhibition of PI3K. First, PI-103 is as potent at inhibiting p110α, but does not cause such rapid and potent inhibition of cell number. Second, PIK-75 results in inhibition of cell number in MDA-MB-231 cells, which are not biochemically affected by this compound. Third, PIK-75 causes apoptosis, in contrast to PI-103 (as shown for SKBr3 cells Fig. 3b). Fourth, in some cell lines PIK-75 appears to be causing arrest of the cell cycle at G2/M. This is in contrast with the G1 arrest as seen here with PI-103, and also more typically seen with previous studies showing inhibition of PI3K [51]. Given the potent and somewhat selective ability of PIK-75 to induce cell death, the task of finding its additional cellular target(s) remains an interesting question.
The cell lines described here may well reflect the behavior of tumor sub-types with respect to p110 isoform inhibition. Our findings that some small molecules described are able to inhibit PI3K and induce cell cycle arrest differentially among the breast cancer cell lines studied, indicates that these compounds may be able to operate within a therapeutic window. The behavior of sensitive and resistant cell lines described here in xenograft models will further support the validity of these compounds as therapeutics. Taken together, our data reinforces the importance of PI3K in maintaining normal cell growth and suggests that p110 selective drugs may be of value as targeted therapies.
Acknowledgements
We are grateful to Benoit Bilanges, Sam Tolman and Clodagh O'Shea, for critical reading of the manuscript and also to Pablo Rodriguez-Viciana for useful comments and reagents. We would also like to thank Jack Taunton for providing us with fmk p90rsk inhibitor. This work was supported by the UCSF Breast Cancer SPORE, NCI CA58207, and a grant from the Sandler program in asthma research (KS).
Abbreviations
- (PI3K)
Phosphoinositide 3-kinase
- (PDK1)
3-phosphoinositide dependent kinase 1
- (PKB/Akt)
protein kinase B
- (p70S6K)
p70 ribosomal protein S6 kinase
- (PDGFR)
platelet derived growth factor receptor
- (mTOR)
mammalian target of rapamycin
- (FBS)
fetal bovine serum
Footnotes
Financial Disclosure NT is currently a paid employee of PIramed Inc. DS is currently a paid employee of Genentech Inc.
References
- 1.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. Biochem. J. 2000;346(Pt 3):561–576. [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 5.Mills GB, Lu Y, Fang X, Wang H, Eder A, Mao M, Swaby R, Cheng KW, Stokoe D, Siminovitch K, Jaffe R, Gray J. The role of genetic abnormalities of PTEN and the phosphatidylinositol 3-kinase pathway in breast and ovarian tumorigenesis, prognosis, and therapy. Semin. Oncol. 2001;28:125–141. doi: 10.1016/s0093-7754(01)90290-8. [DOI] [PubMed] [Google Scholar]
- 6.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 7.Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, Ohta M, Jazag A, Guleng B, Tateishi K, Asaoka Y, Matsumura M, Kawabe T, Omata M. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 8.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. U S A. 2005 doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol. Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 10.Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 11.Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- 12.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 13.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res. Treat. 2004;83:249–289. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- 15.Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, Cross NC, Tefferi A, Malone J, Alam R, Schrier SL, Schmid J, Rose M, Vandenberghe P, Verhoef G, Boogaerts M, Wlodarska I, Kantarjian H, Marynen P, Coutre SE, Stone R, Gilliland DG. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N. Engl. J. Med. 2003;348:1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 16.Norman BH, Shih C, Toth JE, Ray JE, Dodge JA, Johnson DW, Rutherford PG, Schultz RM, Worzalla JF, Vlahos CJ. Studies on the mechanism of phosphatidylinositol 3-kinase inhibition by wortmannin and related analogs. J. Med. Chem. 1996;39:1106–1111. doi: 10.1021/jm950619p. [DOI] [PubMed] [Google Scholar]
- 17.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, Balla T, Weiss WA, Williams RL, Shokat KM. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finan PM, Thomas MJ. PI 3-kinase inhibition: a therapeutic target for respiratory disease. Biochem. Soc. Trans. 2004;32:378–382. doi: 10.1042/bst0320378. [DOI] [PubMed] [Google Scholar]
- 19.Knight ZA, Chiang GG, Alaimo PJ, Kenski DM, Ho CB, Coan K, Abraham RT, Shokat KM. Isoform-specific phosphoinositide 3-kinase inhibitors from an arylmorpholine scaffold. Bioorg. Med. Chem. 2004;12:4749–4759. doi: 10.1016/j.bmc.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 20.Raynaud FI, Eccles S, Clarke PA, Hayes A, Nutley B, Alix S, Henley A, Di-Stefano F, Ahmad Z, Guillard S, Bjerke LM, Kelland L, Valenti M, Patterson L, Gowan S, de Haven Brandon A, Hayakawa M, Kaizawa H, Koizumi T, Ohishi T, Patel S, Saghir N, Parker P, Waterfield M, Workman P. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 2007;67:5840–5850. doi: 10.1158/0008-5472.CAN-06-4615. [DOI] [PubMed] [Google Scholar]
- 21.Sadhu C, Dick K, Tino WT, Staunton DE. Selective role of PI3K delta in neutrophil inflammatory responses. Biochem Biophys Res Commun. 2003;308:764–769. doi: 10.1016/s0006-291x(03)01480-3. [DOI] [PubMed] [Google Scholar]
- 22.Puri KD, Doggett TA, Douangpanya J, Hou Y, Tino WT, Wilson T, Graf T, Clayton E, Turner M, Hayflick JS, Diacovo TG. Mechanisms and implications of phosphoinositide 3-kinase delta in promoting neutrophil trafficking into inflamed tissue. Blood. 2004;103:3448–3456. doi: 10.1182/blood-2003-05-1667. [DOI] [PubMed] [Google Scholar]
- 23.Condliffe AM, Davidson K, Anderson KE, Ellson CD, Crabbe T, Okkenhaug K, Vanhaesebroeck B, Turner M, Webb L, Wymann MP, Hirsch E, Ruckle T, Camps M, Rommel C, Jackson SP, Chilvers ER, Stephens LR, Hawkins PT. Sequential activation of class IB and class IA PI3K is important for the primed respiratory burst of human but not murine neutrophils. Blood. 2005;106:1432–1440. doi: 10.1182/blood-2005-03-0944. [DOI] [PubMed] [Google Scholar]
- 24.Sawyer C, Sturge J, Bennett DC, O'Hare MJ, Allen WE, Bain J, Jones GE, Vanhaesebroeck B. Regulation of breast cancer cell chemotaxis by the phosphoinositide 3-kinase p110delta. Cancer Res. 2003;63:1667–1675. [PubMed] [Google Scholar]
- 25.Sujobert P, Bardet V, Cornillet-Lefebvre P, Hayflick JS, Prie N, Verdier F, Vanhaesebroeck B, Muller O, Pesce F, Ifrah N, Hunault-Berger M, Berthou C, Villemagne B, Jourdan E, Audhuy B, Solary E, Witz B, Harousseau JL, Himberlin C, Lamy T, Lioure B, Cahn JY, Dreyfus F, Mayeux P, Lacombe C, Bouscary D. Essential role for the p110delta isoform in phosphoinositide 3-kinase activation and cell proliferation in acute myeloid leukemia. Blood. 2005;106:1063–1066. doi: 10.1182/blood-2004-08-3225. [DOI] [PubMed] [Google Scholar]
- 26.Jackson SP, Schoenwaelder SM, Goncalves I, Nesbitt WS, Yap CL, Wright CE, Kenche V, Anderson KE, Dopheide SM, Yuan Y, Sturgeon SA, Prabaharan H, Thompson PE, Smith GD, Shepherd PR, Daniele N, Kulkarni S, Abbott B, Saylik D, Jones C, Lu L, Giuliano S, Hughan SC, Angus JA, Robertson AD, Salem HH. PI 3-kinase p110beta: a new target for antithrombotic therapy. Nat. Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- 27.Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006 doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 28.Taylor V, Wong M, Brandts C, Reilly L, Dean NM, Cowsert LM, Moodie S, Stokoe D. 5' Phospholipid Phosphatase SHIP-2 Causes Protein Kinase B Inactivation and Cell Cycle Arrest in Glioblastoma Cells. Mol. Cell. Biol. 2000;20:6860–6871. doi: 10.1128/mcb.20.18.6860-6871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science. 2000;287:1046–1049. doi: 10.1126/science.287.5455.1046. see comments. [DOI] [PubMed] [Google Scholar]
- 30.Sarbassov dos D, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 31.Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, Sonenberg N, Blenis J. RAS/ERK Signaling Promotes Site-specific Ribosomal Protein S6 Phosphorylation via RSK and Stimulates Cap-dependent Translation. J. Biol. Chem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Lin YZ, LaPushin R, Cuevas B, Fang X, Yu SX, Davies MA, Khan H, Furui T, Mao M, Zinner R, Hung MC, Steck P, Siminovitch K, Mills GB. The PTEN/MMAC1/TEP tumor suppressor gene decreases cell growth and induces apoptosis and anoikis in breast cancer cells. Oncogene. 1999;18:7034–7045. doi: 10.1038/sj.onc.1203183. [DOI] [PubMed] [Google Scholar]
- 34.Ferrell JE, Jr., Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 35.Kozma SC, Bogaard ME, Buser K, Saurer SM, Bos JL, Groner B, Hynes NE. The human c-Kirsten ras gene is activated by a novel mutation in codon 13 in the breast carcinoma cell line MDA-MB231. Nucleic Acids Res. 1987;15:5963–5971. doi: 10.1093/nar/15.15.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraus MH, Yuasa Y, Aaronson SA. A position 12-activated H-ras oncogene in all HS578T mammary carcinosarcoma cells but not normal mammary cells of the same patient. Proc. Natl. Acad. Sci. U S A. 1984;81:5384–5388. doi: 10.1073/pnas.81.17.5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawyers CL. Opportunities and challenges in the development of kinase inhibitor therapy for cancer. Genes Dev. 2003;17:2998–3010. doi: 10.1101/gad.1152403. [DOI] [PubMed] [Google Scholar]
- 39.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Janne PA, Januario T, Johnson DH, Klein P, Miller VA, Ostland MA, Ramies DA, Sebisanovic D, Stinson JA, Zhang YR, Seshagiri S, Hillan KJ. Mutations in the Epidermal Growth Factor receptor and in KRAS are Predictive and Prognostic Indicators in Patients with Non-Small Cell Lung Cancer Treated With Chemotherapy Alone and in Combination with Erlotinib. J. Clin. Oncology. 2005;23 doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 40.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, Marrano P, da Cunha Santos G, Lagarde A, Richardson F, Seymour L, Whitehead M, Ding K, Pater J, Shepherd FA. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N. Engl. J. Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 41.Flaherty K. Phase II Trials of BAY 43-9006 Alone and in Combination With Chemotherpay Alone in Metastatic Melanoma. 6th World Congress on Melanoma; 2005. Abstract 049. [Google Scholar]
- 42.Tsutsui S, Inoue H, Yasuda K, Suzuki K, Higashi H, Era S, Mori M. Reduced expression of PTEN protein and its prognostic implications in invasive ductal carcinoma of the breast. Oncology. 2005;68:398–404. doi: 10.1159/000086981. [DOI] [PubMed] [Google Scholar]
- 43.Garcia JM, Silva JM, Dominguez G, Gonzalez R, Navarro A, Carretero L, Provencio M, Espana P, Bonilla F. Allelic loss of the PTEN region (10q23) in breast carcinomas of poor pathophenotype. Breast Cancer Res. Treat. 1999;57:237–243. doi: 10.1023/a:1006273516976. [DOI] [PubMed] [Google Scholar]
- 44.Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, Tempst P, Chi SG, Kim HJ, Misteli T, Jiang X, Pandolfi PP. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martelli AM, Borgatti P, Bortul R, Manfredini M, Massari L, Capitani S, Neri LM. Phosphatidylinositol 3-kinase translocates to the nucleus of osteoblast-like MC3T3-E1 cells in response to insulin-like growth factor I and platelet-derived growth factor but not to the proapoptotic cytokine tumor necrosis factor alpha. J. Bone Miner. Res. 2000;15:1716–1730. doi: 10.1359/jbmr.2000.15.9.1716. [DOI] [PubMed] [Google Scholar]
- 46.Schwarzer R, Tondera D, Arnold W, Giese K, Klippel A, Kaufmann J. REDD1 integrates hypoxia-mediated survival signaling downstream of phosphatidylinositol 3-kinase. Oncogene. 2005;24:1138–1149. doi: 10.1038/sj.onc.1208236. [DOI] [PubMed] [Google Scholar]
- 47.Chen JS, Zhou LJ, Entin-Meer M, Yang X, Donker M, Knight ZA, Weiss W, Shokat KM, Haas-Kogan D, Stokoe D. Characterization of structurally distinct, isoform-selective phosphoinositide 3'-kinase inhibitors in combination with radiation in the treatment of glioblastoma. Mol. Cancer Ther. 2008;7:841–850. doi: 10.1158/1535-7163.MCT-07-0393. [DOI] [PubMed] [Google Scholar]
- 48.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas-Kogan DA, Prados MD, Tihan T, Eberhard DA, Jelluma N, Arvold ND, Baumber R, Lamborn KR, Kapadia A, Malec M, Berger MS, Stokoe D. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J. Natl. Cancer Inst. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 50.Fan QW, Specht KM, Zhang C, Goldenberg DD, Shokat KM, Weiss WA. Combinatorial Efficacy Achieved Through Two-Point Blockade within a Signaling Pathway-A Chemical Genetic Approach. Cancer Res. 2003;63:8930–8938. [PubMed] [Google Scholar]
- 51.Gottschalk AR, Doan A, Nakamura JL, Haas-Kogan DA, Stokoe D. Inhibition of phosphatidylinositol-3-kinase causes cell death through a protein kinase B (PKB)-dependent mechanism and growth arrest through a PKB-independent mechanism. Int. J. Radiat. Oncol. Biol. Phys. 2005;61:1183–1188. doi: 10.1016/j.ijrobp.2004.12.024. [DOI] [PubMed] [Google Scholar]