Abstract
DNA replication and repair in mammalian cells involves three distinct DNA ligases; ligase I (Lig1), ligase III (Lig3) and ligase IV (Lig4)1. Lig3 is considered a key ligase during base excision repair because its stability depends upon its nuclear binding partner Xrcc1, a critical factor for this DNA repair pathway2,3. Lig3 is also present in the mitochondria where its role in mitochondrial DNA (mtDNA) maintenance is independent of Xrcc14. However, the biological role of Lig3 is unclear as inactivation of murine Lig3 results in early embryonic lethality5. Here we report that Lig3 is essential for mtDNA integrity but dispensable for nuclear DNA repair. Inactivation of Lig3 in the mouse nervous system resulted in mtDNA loss leading to profound mitochondrial dysfunction, disruption of cellular homeostasis and incapacitating ataxia. Similarly, inactivation of Lig3 in cardiac muscle resulted in mitochondrial dysfunction and defective heart pump function leading to heart failure. However, Lig3 inactivation did not result in nuclear DNA repair deficiency, indicating essential DNA repair functions of Xrcc1 can occur in the absence of Lig3. Instead, we found that Lig1 was critical for DNA repair, but in a cooperative manner with Lig3. Additionally, Lig3 deficiency did not recapitulate the hallmark features of neural Xrcc1 inactivation such as DNA damage-induced cerebellar interneuron loss6, further underscoring functional separation of these DNA repair factors. Therefore, our data reveal that the critical biological role of Lig3 is to maintain mtDNA integrity and not Xrcc1-dependent DNA repair.
Lig3 has been ascribed repair functions during DNA replication, DNA strand break repair via the BER/single-strand break repair pathway, and has been implicated in double-strand break repair if non-homologous end-joining is disabled1,7–9. Given the close association between Xrcc1 and Lig3 and the substantial DNA repair deficiency present when Xrcc1 is inactivated, the link between Lig3 and DNA repair is compelling1–3,7,8.
In the mitochondria Lig3 functions during replication and repair of mtDNA, but in an Xrcc1-independent manner4,10–12. The highly oxidative environment of the mitochondria suggests that DNA repair will be important for mtDNA maintenance11–17, particularly as mtDNA point mutations or deletions can result in mitochondrial dysfunction13,18–20. Defects in mtDNA maintenance are the bases of multiple human syndromes. For example, distinct mutations of DNA polymerase gamma (PolG), a mitochondrial-specific polymerase that is essential for mtDNA replication, can result in diverse human syndromes involving ataxia, Parkinsonism or diabetes20–24. Consequently, Lig3 is potentially critical for both nuclear and mtDNA integrity.
In order to determine the biological role of Lig3, we generated Lig3Nes-cre mice, in which Lig3 was conditionally inactivated throughout the nervous system using Nestin-cre. While germline Lig3 deletion is lethal early during embryonic development5, Lig3Nes-cre mice were born alive. Although initially indistinguishable from wild type (WT) littermates, Lig3Nes-cre mice became growth retarded and profoundly ataxic by two weeks of age (Suppl. Movie 1), and didn’t survive beyond 20 days of age (Suppl. Fig. 1). Magnetic resonance imaging of postnatal day 14 (P14) Lig3Nes-cre mice revealed a smaller brain compared with controls (Fig. 1a). Lig3Nes-cre tissue had markedly reduced Lig3 levels in the brain, but not elsewhere (Fig. 1b), and despite Lig3 instability after Xrcc1 loss2,6, the converse did not occur (Fig. 1b). In contrast to WT and Xrcc1Nes-cre, the Lig3Nes-cre cerebellum was substantially smaller, showing both reduced proliferation and increased apoptosis of granule neuron progenitors, resulting in widespread loss of these cells (Fig. 1d and Suppl. Fig. 2). Other cell types such as oligodendrocytes were also affected in the Lig3Nes-cre brain (Suppl. Fig. 3).
Figure 1. Lig3 inactivation throughout the nervous system leads to a phenotype different to Xrcc1 loss.
(a) MRI analysis of the Lig3Nes-cre nervous system shows microcephaly and a small cerebellum (arrow) compared to WT. (b) Lig3 is deleted in various Lig3Nes-cre brain regions while Xrcc1 protein levels are unaffected. (c) Comparative Nissl staining of P14 cerebellum (Ce) from Lig3Nes-cre, Xrcc1Nes-cre and WT. (d) PCNA immunostaining shows disrupted neurogenesis in the P6 Lig3Nes-cre cerebellum (arrows). (e) Levels of the interneuron marker Pax2 are similar in control and Lig3Nes-cre tissue, but substantially reduced in the Xrcc1Nes-cre cerebellum. (f) Interneurons are present in the Lig3Nes-cre cerebellum but are absent from Xrcc1Nes-cre tissue (arrows) as determined by Pax2 immunostaining.
Given that Xrcc1 stabilizes Lig3, we determined if Lig3 loss also disrupts cerebellar interneurons, a hallmark phenotype after Xrcc1 inactivation6. However, the Lig3Nes-cre cerebellum contained a normal complement of interneurons as determined by Western blot of cerebellar extracts and tissue immunostaining using anti-Pax2, a marker for this neuronal type (Fig. 1e,f). Therefore, despite their close association, inactivation of Lig3 leads to a strikingly different neural phenotype to Xrcc1 loss.
As Xrcc1Nes-cre and Lig3Nes-cre are phenotypically distinct, and because Lig3 inactivation can compromise mtDNA11, we determined if the Lig3Nes-cre neurodevelopmental defects were due to disruption of mitochondrial function. Using mitotracker to label mitochondria, we found a pronounced mitochondrial defect in primary Lig3Nes-cre cortical astrocytes (Fig. 2a). In both WT and Xrcc1Nes-cre astrocytes there was an elaborate distribution of actively streaming mitochondria. In stark contrast, mitochondria in Lig3Nes-cre astrocytes were static and showed an abnormal morphology and distribution (Fig. 2a and Suppl. Movie 2).
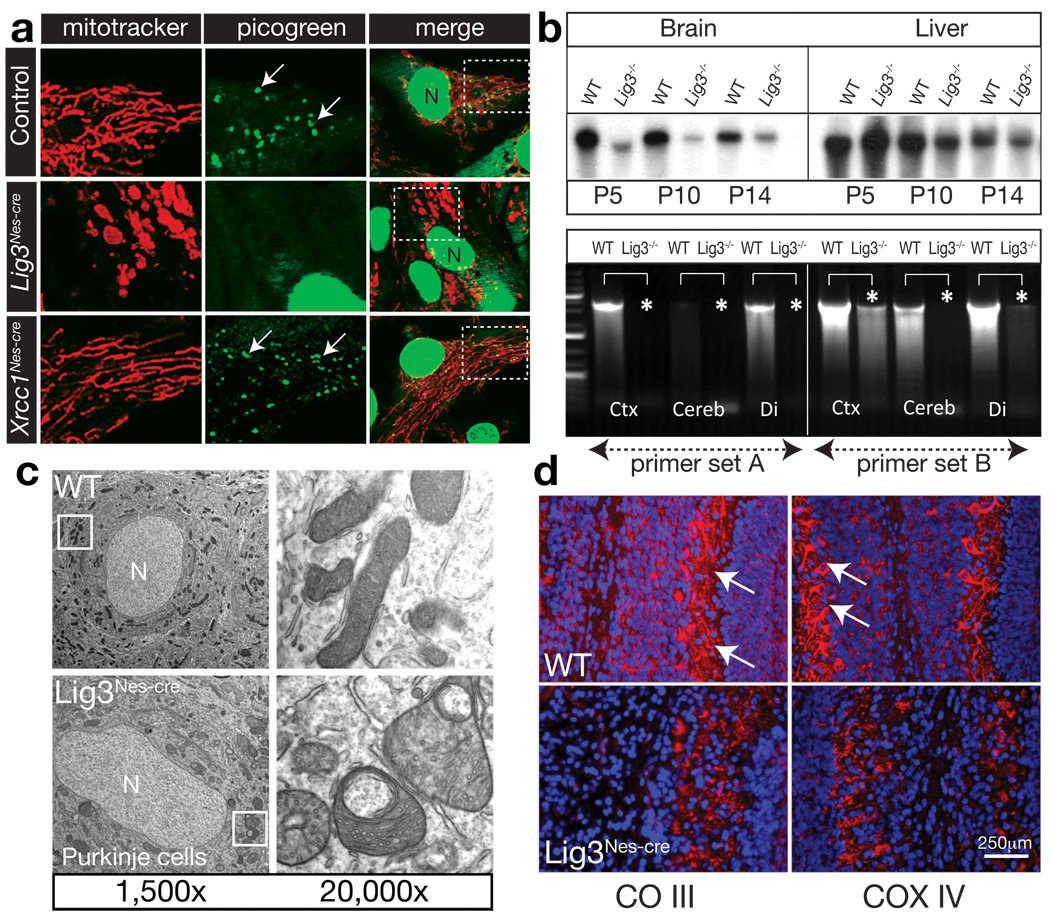
Figure 2. Mitochondrial function is disrupted in the Lig3Nes-cre brain.
(a) Mitotracker labeling reveals defects in mitochondria from Lig3Nes-cre but not WT or Xrcc1Nes-cre astrocytes. Picogreen staining shows reduced mtDNA in Lig3Nes-cre astrocytes; N is the nucleus. Boxed areas show the region expanded in adjacent panels. (b) Analysis of mtDNA via pulse field gel electrophoresis shows an age-dependent quantitative reduction in mtDNA from P5-P14 Lig3Nes-cre brain compared with controls. mtDNA levels in the Lig3Nes-cre liver were similar to control tissue. PCR of mtDNA revealed a reduction in mtDNA isolated from Lig3Nes-cre brain tissue (asterisks). Ctx is cortex, Cereb is cerebellum and Di is diencephalon. (c) Electron microscopy showed alterations in mitochondrial morphology in Lig3-deficient Purkinje cells compared to WT tissue. (d) Electron transport chain components complex III core 2 (CO III) and COX IV staining in vivo is reduced in P7 cerebellar Purkinje cell mitochondria; counterstain is DAPI.
Identification of mtDNA using pico-green showed a normal distribution in WT or Xrcc1Nes-cre cells (arrows), but an absence of mtDNA in Lig3Nes-cre astrocytes (Fig. 2a). We then assessed mtDNA integrity in vivo in the Lig3Nes-cre brain and found the amount of recovered mtDNA from this tissue was markedly decreased compared with that from equal amounts of WT tissue or Lig3Nes-cre liver (Fig. 2b). This was further confirmed using a PCR assay that amplified specific mtDNA regions, which showed a loss of mtDNA throughout the Lig3Nes-cre brain (Fig. 2b). Lig3Nes-cre astrocyte cultures also became acidic compared to WT cultures, suggesting lactic acid accumulation because of defective oxidative respiration, and accordingly, Lig3Nes-cre cells were defective in oxygen metabolism (Suppl. Fig. 4).
We then directly assessed mitochondria ultrastructure in the Lig3Nes-cre brain using electron microscopy. We found morphological abnormalities of the mitochondria at postnatal times coinciding with the emergence of ataxia. By P7, cerebellar Purkinje neurons showed mitochondria with distorted cristae structure and broad changes in mitochondrial morphology (Fig. 2c).
We also assessed components of the mtDNA-encoded electron transport chain, which are often affected in dysfunctional mitochondria22. By P5, there was a marked reduction of complex III (CO III), and COX IV immunostaining in Purkinje cells (Fig. 2d), and elsewhere throughout the brain including hippocampal structures and the cortices (Suppl. Fig. 5). The biochemical activity of complex I was also reduced in Lig3Nes-cre brain (Suppl. Fig. 5). Consistent with increased free radical production from dysfunctional mitochondria increased tyrosine nitrosylation was found in the Lig3Nes-cre brains (Suppl. Fig. 6).
We also generated Lig3Ckmm-cre mice, in which the muscle creatine kinase promoter drives cre-mediated deletion of Lig3, and confirmed diminished Lig3 levels in cardiac and skeletal muscle, while levels in the brain were normal (Fig. 3a). All Lig3Ckmm-cre mice died abruptly between 3.5 to 4.5 weeks of age (Fig. 3b). Although we did not find any histological difference between Lig3 mutant and wild type skeletal muscle, Lig3Ckmm-cre mice showed heart defects associated with cardiac Lig3 inactivation. Overall heart size of the mutants was larger than controls, and dilation of the ventricle and atrium and thinning of the ventricle walls was apparent (Suppl. Fig. 7). We also identified disruption in connexin-43, depletion of cardiac troponin-I and defective intercalated disc structure, suggesting that force generation and transmission during cardiac muscle contraction is defective (Fig. 3c and Suppl. Fig. 7). Electron microscopy of mutant heart muscle also revealed disruption of myofiber structure and pronounced defects in mitochondrial morphology (Fig. 3d and Suppl. Fig. 7).
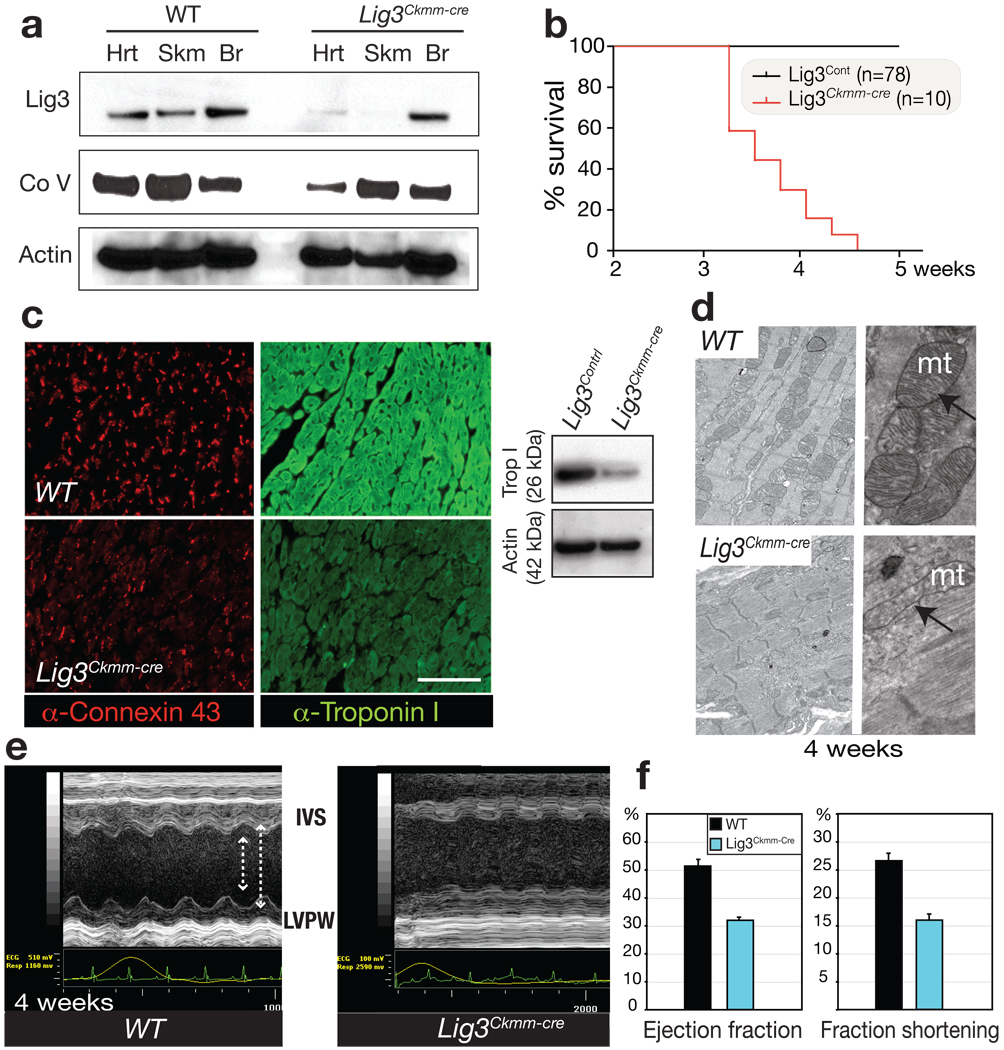
Figure 3. Lig3 inactivation causes cardiac failure associated with defective mitochondrial function.
(a) Lig3 protein levels are reduced in Lig3Ckmm-cre heart and skeletal muscle. A reduction of complex V subunit α also occurs in Lig3Ckmm-cre heart. (b) Lig3Ckmm-cre mice die between 3.5 and 4.5 weeks of age; n is the number of animals. (c) Altered connexin-43 distribution and depletion of cardiac troponin I occurs in 4-week-old Lig3Ckmm-cre heart. A Western blot shows decreased cardiac troponin I levels. (d) Electron microscopy of cardiac muscle shows disruption in myofiber structure and defects in mitochondrial morphology. (e) Echocardiographic analysis of Lig3Ckmm-cre mice shows movement of the interventricular septum (IVS) and the posterior walls of the left ventricle (LVPW) were decreased in the Lig3Ckmm-Cre heart (arrows). Traces below the echocardiogram show ECG readings indicate abnormal heart rhythm. (f) Cardiac contractility was also significantly depressed with decreased ejection fraction and fraction shortening indicating that the pump function of the Lig3Ckmm-Cre heart is defective. Three individual animals of each genotype were used; error bars represent standard deviation.
We monitored heart function using transthoracic ultrasound echocardiography (ECG) in Lig3Ckmm-cre and WT animals beginning from 3 weeks of age. M-mode echocardiography showed that the diastolic and systolic movement of the left ventricle wall and interventricular septum was decreased in Lig3Ckmm-cre mice, which was associated with an ECG reading reflecting an abnormal heart rhythm (Fig. 3e). Fractional shortening and ejection fraction (Fig. 3f), two ways of measuring cardiac contractility, were also decreased significantly in Lig3Ckmm-cre mice, indicating heart pump function was severely affected, a phenomenon often leading to heart failure.
Although Lig3 is linked to DNA repair via interaction with and stabilization by Xrcc12,6, the role for this ligase during nuclear DNA repair is unclear. Therefore, we initially compared nuclear DNA repair in WT, Lig3Nes-cre and Xrcc1Nes-cre astrocytes after treatment with various genotoxins. We found that DNA repair after H2O2 and ionizing radiation (IR) in quiescent Lig3Nes-cre astrocytes was similar to WT cells while Xrcc1Nes-cre cells showed a marked repair defect (Fig. 4a,b). One exception was UV damage, where DNA repair in Lig3Nes-cre cells was reduced compared to controls; although, even in this situation Xrcc1Nes-cre cells showed a substantially greater DNA repair defect (Suppl. Fig. 8). This may indicate that Lig3 is selectively required to repair specific types of DNA lesions25.
Figure 4. Lig3 is not essential for nuclear DNA repair.
(a) Alkaline comet analysis shows DNA strand break repair after hydrogen peroxide treatment is similar between WT and Lig3Nes-cre quiescent primary astrocytes, but is defective in Xrcc1Nes-cre cells, as is repair after ionizing radiation (b); R30/R60 is recovery from the genotoxin after 30 or 60 minutes. Assays were run in triplicate from at least three separate experiments; error bars in all panels represent standard error of the mean. (c) Western blot analysis indicates specific shRNA-mediated knockdown of mammalian DNA ligases. Non-target scrambled (Scr) and Nbs1 shRNAs were included as controls, and Ponceau staining indicates relative protein transfer. (d) Lig1 is required for DNA repair after H2O2 damage; dual inactivation of Lig1 and Lig3 reveals cooperation between these ligases during repair. Tdp1−/− MEFS are a repair-deficient control. Photograph indicates cells are quiescent based on p27+ and PCNA− immunoreactivity. (e) Representative individual comet tail moments of shRNA-mediated knockdowns after H2O2 treatment. (f) Lig1 and Lig4 are required for DNA repair after IR. Lig1 and Lig3 cooperate in the repair of IR-induced DNA damage. (g) DNA repair competency after H2O2 treatment is restored using a shLig1-resistant version of Lig1 cDNA (Lig1shR).
As Lig3 loss did not substantially impact DNA repair after H2O2 or IR, we considered if a sub-pathway of BER, termed long-patch-repair (LPR)7 might functionally compensate for defects in short patch BER. This is the situation when another repair factor, Aprataxin (Aptx) is defective26. We used aphidicolin to acutely inhibit the LPR polymerases Polδ and Polε, but found no DNA repair defect in Lig3Nes-cre cells after methylmethane sulphonate (MMS) (Suppl. Fig. 8). In contrast to Lig3Nes-cre cells, quiescent Aptx−/− astrocytes had a reduced rate of DNA repair after MMS in the presence of aphidicolin, indicating LPR inhibition (Fig. 4d).
To further determine repair capability and potential functional compensation by other DNA ligases in the absence of Lig3, we used lentiviral-mediated shRNA in primary MEFs to inactivate Lig1, Lig3 and Lig4 (Fig. 4c). Importantly, under the conditions we established for shRNA-mediated ligase knockdown, minimal cell lethality was observed (not shown). Using this approach, we found that similar to Lig3Nes-cre cells, Lig3-depleted cells were also competent to repair H2O2-induced DNA damage in quiescent cells (Fig. 4d). In contrast, inactivation of Lig1, but not Lig4, resulted in a marked defect in the ability to repair H2O2-induced DNA damage (Fig. 4d). Moreover, dual inactivation of Lig1 and Lig3 resulted in an exacerbated repair deficiency that was similar to Tdp1−/− cells, indicating functional cooperation between Lig1 and Lig3 after H2O2 treatment (Fig. 4d,e). We then compared other genotoxins including IR and MMS and found that similar to damage induced by H2O2, individual loss of Lig1 but not Lig3, strongly affected DNA repair (Fig. 4f and Suppl. Fig. 8). In the case of IR, where both DNA single- and double-strand breaks occur, Lig4 inactivation also markedly reduced repair. Again, a cooperative defect was found after dual inactivation of Lig1 and Lig3 during repair of IR-induced lesions. The DNA repair defect associated with Lig1 inactivation was specific, as a modified Lig1 cDNA that was shRNA-resistant (Lig1shR) restored repair capacity of the shLig1-targeted cells after H2O2 and MMS (Fig. 4g and Suppl. Fig. 8).
While Xrcc1-mediated stabilization of Lig3 in the nucleus suggests Lig3 is important during DNA repair2,7,8, our data show that Lig3 is generally dispensable for Xrcc1-mediated DNA repair. On the other hand, our data show that Lig1 is clearly the operative ligase that repairs DNA lesions in quiescent cells usually ascribed to Lig3. However, it is likely that the repair of specific subsets of DNA lesions do involve Lig3 given the increased repair deficiency after coordinate inactivation of Lig1 and Lig3. While some controversy exists over the role of Lig1 during DNA repair27,28, our data clearly identify Lig1 as a key player in this process. Increased sensitivity of repair assays may aid in further delineating Lig3 function during nuclear DNA repair.
The mitochondrial defects associated with Lig3 inactivation could occur at the level of mtDNA replication and/or repair. Generation of a mouse with a defect in the proof-reading function of PolG lead to accumulation of mtDNA mutations20. However, while these animals were viable they showed a variety of age-related defects associated with mtDNA mutations, and many survived greater than a year without developing the phenotypes we found after Lig3 inactivation in the brain or heart. In contrast, mitochondrial transcription Factor A is essential for mtDNA replication, and its inactivation in the heart using Ckmm-cre29 produced a similar phenotype to the Lig3Ckmm-cre mice, suggesting defective mtDNA replication underpins the Lig3 mutant phenotype.
Although the Lig3-deficient nervous system (and heart) develops in a relatively normal manner, we assume mtDNA is nonetheless compromised during development of these tissues. The pronounced decline in the newborn Lig3Nes-cre brain (and Lig3Ckmm-cre heart) may be a combination of mtDNA defects acquired during organogenesis and stresses associated with the physiological and respiratory changes at birth30, and the high demand for oxidative metabolism.
Thus, our data point to disparate biological roles for Lig3 and Xrcc1, with each primarily important in separate cellular compartments to ensure the integrity of mitochondrial and nuclear DNA, respectively. Additional analyses of Lig3 function during nuclear DNA repair and mtDNA maintenance will provide important insights into genome stability, mitochondrial biology and tissue homeostasis. Lig3 mutations may also underpin some of the many mitochondrial diseases for which the causative gene defect is unknown.
Methods Summary
Generation of Lig3 mutant mice
Embryonic stem cells were electroporated with a targeting vector incorporating LoxP sites flanking exons 6–14 of Lig3 to obtain mice with a floxed Lig3 allele. Intercrossing these with Nes-cre or Ckmm-cre mice generated Lig3Nes-cre or Lig3Ckmm-cre animals.
Mitochondrial analysis
MitoTracker Red and Picogreen were used to label astrocyte mitochondria. For EM, tissues were paraformaldehyde-fixed with osmium tetroxide post-fixing, and stained with uranyl acetate. mtDNA was assessed using pulsed field gel electrophoresis and PCR.
Cardiac analysis
A VEVO-770 instrument (VisualSonics, Toronto, Canada) was used for echocardiography analysis. ECG was determined and M-mode analysis was done to measure cardiac contractility.
DNA repair analysis
Lig3Nes-cre, Xrcc1Nes-cre, Aptx−/− and control astrocytes were prepared as described previously6. MEFs were established from E13.5 embryos. RNA interference was used to inactivate DNA ligases in primary MEFs. ShRNA-encoding lentiviral particles were produced via co-expression of pLKO.1-puro plasmids (TRC1, Sigma) with Mission Lentiviral packaging DNA (Sigma, SHP001) in 293T cells. The Comet Assay IV system (Perceptive Instruments) was used to assay DNA damage as described6,26.
Full Methods
Ligase 3 mice
A targeting vector was designed to conditionally delete exons 6–14 using recombineering. A single LoxP site was inserted between exons 14 and 15 and a neomycin-thymidine kinase selection cassette flanked by Frt sites with a single LoxP site was inserted between exons 5 and 6. The targeting construct was transfected into embryonic stem (ES) cells via electroporation and selection with G418, homologous recombinants were screened by Southern blotting using both 3’ and 5’ probes outside the targeted region. Positive clones were transfected a second time with a Flt recombinase expressing vector. Cells were selected using FIAU to remove cells retaining the thymidine kinase and further screened by PCR using primers flanking the LoxP site or the LoxP/Frt remaining after recombination. Positive clones were blastocyst-injected to generate chimeric mice. The male chimeras were bred with C57BL/6 females to establish mice carrying the conditional Lig3 allele. Further breeding with cre mice under the control of the Nestin promoter (JAX#003771: B6.Cg-Tg(Nes-cre)1Kln/J) resulted in deletion throughout the nervous system, while breeding with the Ckmm2-cre mice (JAX#006475: B6.FVB(129S4)-Tg(Ckmm-cre)5Khn/J) deleted Ligase 3 in skeletal and cardiac muscle.
Histology
Mice were perfused with 4% (w/v) buffered paraformaldehyde and cryoprotected in buffered 25% sucrose (w/v) solution. Brains were dissected, cut sagittally and sectioned at 10µm using an HM500M cryostat (Microm). Nissl staining was carried out with 1% (w/v) thionin. Hematoxylin and eosin staining was done according to standard procedures. Immunohistochemical and immunocytochemical staining of tissues and cells were carried out with the antibodies listed below. For colorimetric visualization of positive signals, sections were incubated with antibodies overnight at room temperature after quenching endogenous peroxidase using 0.6% (v/v) H2O2 in methanol. Slides were washed with phosphate-buffered saline (PBS) three times, followed by incubation with biotinylated secondary antibody and avidin-biotin complex (Vectastain Elite kit, Vector Labs). Antibodies were used after citrate buffer–based antigen retrieval. Immunoreactivity was visualized with the VIP substrate kit (Vector Labs) using the manufacturer's protocol. After staining, sections were counterstained with 0.1% (w/v) methyl green, dehydrated, and mounted in DPX (Fluka). For fluorescent signals of immunoreactivity, FITC- or Cy3-conjugated secondary antibodies (Jackson Immunologicals) were used and counterstained with 4'6-diamidino-2-phenylindole (DAPI) or propidium iodide (Vector Laboratories). For immunohistochemistry and immunocytochemistry, we used the following antibodies: calbindin (mouse, 1:2,000, Sigma), PCNA (mouse, 1:500, Santa Cruz Biotechnology), Pax2 (rabbit, 1:500, Zymed #71–6000), CO III-core 2 (mouse, 1:2000, MitoSciences), Ki67 (rabbit, 1:250, Vector Laboratories), COX IV (mouse, 1:500, Abcam), cardiac Troponin-I (mouse, 1:50 fluorescence and 1:500 Western blots, Abcam) and connexin-43 (rabbit, 1:50, Cell Signaling). Apoptosis was detected using the TUNEL assay with the Apoptag kit (Chemicon) according to the manufacturer's protocol.
Western blot analysis
Western blot analysis was performed with tissues (cortex, cerebella, liver and spleen) from both P5 control mice (Lig3LoxP/+;Nes-cre) and conditional knockout mice (Lig3LoxP/LoxP;Nes-cre). Protein extracts were prepared by using lysis buffer (50mM Tris-HCl, 200mM NaCl, 0.2% NP-40, 1% Tween-20 (v/v), 1mM NaF, 1mM sodium vanadate, 50mM β-glycerophosphate, 2mM PMSF, and Complete protease inhibitor (Roche) and quantified by Bradford assay (Bio-Rad). Proteins (50µg per lane) were separated through a 4–12% (w/v) Bis-Tris SDS polyacrylamide gel (Invitrogen) and transferred onto nitrocellulose membrane (Bio-Rad). Blots were sequentially immunostained with mouse anti-Lig3 antibody (1:500, BD Biosciences) followed by horseradish peroxidase–conjugated donkey anti-mouse (1:2,000, GE Healthcare) and detected using ECL Plus chemiluminescence reagent (GE Healthcare). To detect XRCC1 protein levels, the immunoblots were stained with rabbit anti-Xrcc1 antibody6 (1:1000) and processed as described above. Immunoblots were also stained with mouse complex V subunit-α antibody (mouse, 1:1000, Mitoscience) and Pax2 (1/5000, Abcam #79389) and processed as described above. Actin antibody (1:1,000, Santa Cruz Biotech) was used as a protein-loading control.
Mitochondrial analysis
For mitochondria staining, primary astrocytes were incubated with prewarmed (37°C) culture medium described below containing MitoTracker Red (1:200, Invitrogen) for one hour under normal growth conditions followed by media replacement for an additional 20mins. For mtDNA staining, the astrocytes were incubated with prewarmed culture medium containing Picogreen (3µl/ml, Invitrogen) for one hour under normal growth conditions. Cells were imaged and processed using a Nikon C1Si confocal microscope and EZCi imaging software (Nikon USA, Melville, N.Y.) using a 561 DPSS laser (Melles Griot, Albuquerque, NM). Complex I activity assays were done with brain samples using a MitoSciences microplate assay kit (www.mitosciences.com) according to the manufacturers directions. Cellular oxygen consumption was measured using an Oxygen Biosensor system 96-well plate (BD) according to the manufacturer’s directions.
Mitochondria were isolated from brain tissue using the Qproteome™ Mitochondria Isolation Kit, (QIAGEN) according to the manufacturer’s directions, and mtDNA was isolated using the Mitochondrial DNA Isolation Kit (BioVision) according to the manufacturer’s directions. PCR of mtDNA was done using the Expand Long Range, dNTPack (Roche) with the following conditions: Initial denaturation at 92°C for 30sec followed by annealing at 56°C for 30sec and elongation at 68°C (at 60s/kb) for 39 cycles, with a final 68oC elongation for 10mins. Primers used: ‘Set A’, forward, 5’-CCT TCA TCC TTC TCT CCC TAT GAG GA, reverse 5’-GGT TGT TTG ATC CTG TTT CGT GGA and ‘Set B’, forward, 5’-CCC AGC TAC TAC CAT CAT TCA AGT, reverse 5’-CAG TAT GCT TAC CTT GTT ACG ACT.
Mitochondrial DNA isolated from equal amounts of Lig3 mutant or control tissue were lysed in 1mg/ml proteinase K, 1% (w/v) N-lauroylsarcosine, (pH9.5) in LM-agarose plugs overnight and subjected to pulsed field gel electrophoresis. Mitochondrial DNA was separated for 24 hours with gradual changes in pulses from 0.05 to 20secs at 3.0V/cm at an angle of 120° in 1% megabase agarose gel (Bio-Rad) with the CHEF Mapper Pulsed Field Electrophoresis system (Bio-Rad). DNA was visualized with ethidium bromide or Southern blot analysis using radioactive mtDNA as a probe.
Isolation of primary cells
Primary astrocytes were prepared from P2 mouse brains as described previously6. Cortices were dissociated by passage through a 5ml pipette and cells were resuspended in Dulbecco's modified Eagle's medium and Ham's nutrient mixture F-12 (1:1 DMEM/F12, Gibco-BRL) supplemented with 10% fetal bovine serum (v/v), 1× glutamax, 100U/ml penicillin, 100µg/ml streptomycin and 20ng/ml epidermal growth factor (Millipore). Primary astrocytes were established in Primeria T-25 tissue culture flasks (Falcon) at 37°C in a humidified CO2-regulated (5%) incubator.
Primary mouse embryonic fibroblasts (MEFs) were prepared from E13.5 embryos. Embryos were minced using dissection scissors, passed through a 23 gauge syringe followed by a 70 µm cell strainer. Cells were resuspended in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (v/v), 1× glutamax, 100 U/ml penicillin, 100 µg/ml streptomycin and β-mercaptoethanol and established in T-25 tissue culture flasks (Falcon) at 37°C in a humidified CO2-regulated (5%) incubator.
Magnetic resonance imaging
Two week-old mutant and control mice (age and sex matched) were subjected to magnetic resonance imaging analysis. All animals were scanned on a 7 Tesla Bruker Clinscan animal magnetic resonance imaging scanner (Bruker BioSpin MRI GmbH) as described previously6.
Electron microscopy
Mice were perfused with 4% paraformaldehyde, 100mM cacodylate, 2.5% sucrose, (pH 7.4), and fixed overnight. Samples were post fixed in osmium tetroxide, stained in 2% uranyl acetate, dehydrated, and embedded in Epon Beem Capsules. 80 nm en face sections were prepared on a Leica UCT ultramicrotome and collected onto 200 mesh copper grids. Images were collected on JoeL JEM-1200 EXII TEM equipped with AMTV600 digital camera.
DNA repair analysis
Quiescent primary astrocytes or MEFs (2×105 cells/ml/sample) were treated with either 150µM H2O2 for 10 min on ice, methylmethane sulphonate (MMS) for 10min at various concentrations at 37°C or γ–irradiation (either 10 or 20Gy, Cs137). Following H2O2 and IR treatments, cells were incubated for various times in drug-free medium at 37°C. Cells were trypsinized and suspended in pre-chilled 1× PBS, mixed with an equal volume of 1.2% low-melting point agarose (Invitrogen) maintained at 42°C and immediately layered onto frosted glass slides (Fisher) pre-coated with 0.6% agarose and maintained in the dark at 4°C for all further steps. Slides were immersed in pre-chilled lysis buffer (2.5 M NaCl, 10mM Tris-HCl, 100 mM EDTA (pH 8.0), 1% (v/v) Triton X-100, 3% (v/v) DMSO, pH10) for 1 h, washed with pre-chilled distilled water (twice for 10 min each) and placed into pre-chilled alkaline electrophoresis buffer (50 mM NaOH, 1mM EDTA, 1% DMSO) for 45 min. Electrophoresis was carried out at 95mA (astrocytes) or 190 mA (MEFs) for 25 min, followed by neutralization in 0.4 M Tris-HCl (pH 7.0). Comets were stained with SYBR Green (1:10,000 in 1× PBS, Sigma) for 10 min. A minimum of 100 comet tail moments were measured using the Comet Assay IV system (Perceptive Instruments) coupled to an Axioskop2 plus microscope (Zeiss) at 200× magnification.
Lentivirus particles were used to deliver and express shRNAs to knock down murine ligases 1, 3 and 4, with Nbs1 and a non-target scrambled shRNA used as controls. All MISSION® shRNA plasmid DNA constructs were obtained from SIGMA-ALDRICH (St Louis, MO). The pLKO.1-puro parental vector expressed the TRC1 shRNAs and included: shLig1–5’-CCG GGC CCG GAC ATT TGA GAA GAT TCT CGA GAA TCT TCT CAA ATG TCC GGG CTT TTT G, shLig3–5’-CCG GCC ACC GAA CAG AAG CTC AAT ACT CGA GTA TTG AGC TTC TGT TCG GTG GTT TTT G and shLig4–5’-CCG GCC TCT CAG TAT GTT TCG ACA TCT CGA GAT GTC GAA ACA TAC TGA GAG GTT TTT G.
Lentivirus was made via co-transfection of pLKO.1-puro plasmids with Mission Lentiviral packaging DNA (Sigma, SHP001) into subconfluent 293T cells using Fugene 6 (Roche). Lentivirus was collected 48 hours later and filtered (0.45µm pore size) and primary MEFs (1.25 × 105 cells in 6cm2 plates grown for 20 hrs) were transduced with individual or a combination of lentiviral particles for 36 hours at 37°C with polybrene (10 µg/ml). All data reflects experiments performed in triplicate. Western blots were used to confirm shRNA knockdowns; antibodies used were Nbs1 (1/500, Cell Signaling), Lig1 (1/1000, Santa Cruz Biotech), Lig3 (1/500, BD Biosciences) and Lig4 (1/400, Abcam).
Generation of an shLig1-resistant Lig1 cDNA (Lig1shR) was done using the following primers: L1shRforward-5’-CTT ACC TGG CTG TGG CAA GCA CGT TCG AGA AAA TCG AGG AGG TGT CTG C and L1shRreverse-5’-GCA GAC ACC TCC TCG ATT TTC TCG AAC GTG CTT GCC CAC GCC AGG TAA G. These silent mutations were introduced into human Lig1 cDNA using the Quikchange II Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s instructions. The modified Lig1 construct was co-transfected into MEFs with pmaxGFP plasmid (4:1 ratio) using Nucleofector methodology (Lonza) and then FACS-sorted to select for GFP-positive cells (~30%). Lig1shRtransfected cells were then treated with shScr, shLig1 or a combination of shLig1/shLig3 lentivirus and assayed for DNA repair after genotoxic stress using the comet assay, as described above.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Hartwell Center for biotech support, the Transgenic core facility for blastocyst injections and the ARC for animal husbandry. We also acknowledge the Light Microscopy Core, the Electron Microscopy core and Chris Calabrese and the Animal Imaging core for MRI and echocardiography analysis. PM is supported by the NIH (NS-37956, CA-21765), the CCSG (P30 CA21765) and the American Lebanese and Syrian Associated Charities of St. Jude Children’s Research Hospital. S.K. is a Neoma Boadway AP Endowed Fellow.
Footnotes
Author Contributions
Y.G., S.K. and Y.L performed all experiments characterizing the Lig3-deficient mouse and contributed to writing the manuscript. Y.G. and H.R. generated the targeted ES cells for blastocyst injection and were responsible for colony production and maintenance with assistance from S.K and Y.L. S.K. and J.Z. established the functional analyses of ligases. J.R. provided pathology analysis. P.M. was project leader and produced the final version of the manuscript.
Supplemental Data
Eight supplemental figures and two movies are associated with this manuscript.
References
- 1.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ljungquist S, Kenne K, Olsson L, Sandstrom M. Altered DNA ligase III activity in the CHO EM9 mutant. Mutat Res. 1994;314:177–186. doi: 10.1016/0921-8777(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 4.Lakshmipathy U, Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 2000;28:3880–3886. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puebla-Osorio N, Lacey DB, Alt FW, Zhu C. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol Cell Biol. 2006;26:3935–3941. doi: 10.1128/MCB.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci. 2009;12:973–980. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 10.Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakshmipathy U, Campbell C. Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res. 2001;29:668–676. doi: 10.1093/nar/29.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinz KG, Bogenhagen DF. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol Cell Biol. 1998;18:1257–1265. doi: 10.1128/mcb.18.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnan KJ, et al. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 14.LeDoux SP, et al. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 15.Liu P, et al. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomkinson AE, Bonk RT, Linn S. Mitochondrial endonuclease activities specific for apurinic/apyrimidinic sites in DNA from mouse cells. J Biol Chem. 1988;263:12532–12537. [PubMed] [Google Scholar]
- 17.Van Houten B, Woshner V, Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 2006;5:145–152. doi: 10.1016/j.dnarep.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Tyynismaa H, Suomalainen A. Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep. 2009;10:137–143. doi: 10.1038/embor.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 21.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 23.Chan DC. Mitochondrial dynamics in disease. N Engl J Med. 2007;356:1707–1709. doi: 10.1056/NEJMp078040. [DOI] [PubMed] [Google Scholar]
- 24.Chan SS, Copeland WC. DNA polymerase gamma and mitochondrial disease: understanding the consequence of POLG mutations. Biochim Biophys Acta. 2009;1787:312–319. doi: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moser J, et al. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Reynolds JJ, et al. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia 1. Mol Cell Biol. 2009;29:1354–1362. doi: 10.1128/MCB.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentley DJ, et al. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J Cell Sci. 2002;115:1551–1561. doi: 10.1242/jcs.115.7.1551. [DOI] [PubMed] [Google Scholar]
- 28.Sleeth KM, Robson RL, Dianov GL. Exchangeability of mammalian DNA ligases between base excision repair pathways. Biochemistry. 2004;43:12924–12930. doi: 10.1021/bi0492612. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, et al. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 30.Nakai A, et al. Developmental changes in mitochondrial activity and energy metabolism in fetal and neonatal rat brain. Brain Res Dev Brain Res. 2000;121:67–72. doi: 10.1016/s0165-3806(00)00025-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.