Introduction
Primary tumours of the central nervous system (CNS) account for roughly two percent of human malignancies. The most common CNS tumours of adults are malignant gliomas. In general, malignant gliomas are not curable tumours, but their management has undergone change over the past two decades, with novel approaches to surgery, radiation and chemotherapy improving survival and quality of life for patients to variable degrees. The present review addresses how new molecular diagnostic advances have also played roles in changing the management of patients with malignant gliomas over the last twenty years.
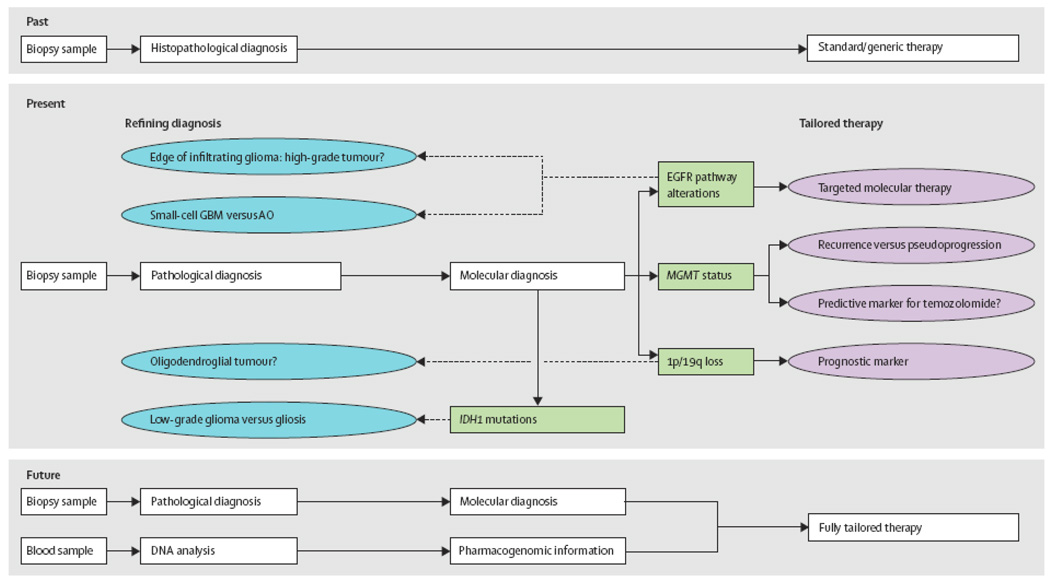
Until recently, treatment decisions regarding malignant gliomas began with establishing diagnosis by standard histopathology only. Histopathology has been a dynamic tool for nearly a hundred years, providing knowledge about the biology and behaviour of many neoplasms. The 2007 WHO classification, the most recent consensus approach to CNS tumor diagnosis, divides the malignant gliomas of adults into astrocytic tumors—the most malignant of which is termed glioblastoma—as well as oligodendrogliomas and oligoastrocytomas.1 The data gained from histopathologic examination of tumour tissue has been augmented, but not supplanted, by molecular approaches to tissue diagnosis.
Knowledge about the molecular biology of cancer—including CNS tumours—continues to increase markedly. Having a dynamic classification of tumours allows the integration of new markers that are discovered to help determine prognosis and likelihood of therapeutic response. This review focuses on those markers that have found utility in the evaluation of adult malignant gliomas—specifically on 1p/19q co-deletion, methylation of the O6 methylguanine methyltransferase (MGMT) gene promoter, evaluation of the epidermal growth factor receptor (EGFR) pathway and isocitrate dehydrogenase 1 (IDH1) gene mutations. The reader is also reminded that molecular approaches are also used in the assessment of other CNS tumours of neuroepithelial origin, notably in pediatric tumors such as medulloblastoma,2 atypical teratoid rhabdoid tumour3 and potentially in pilocytic astrocyoma.4
1p 19q loss in glioma
Background
Following empiric observations of favourable responses to chemotherapy in a high proportion of recurrent anaplastic oligodendrogliomas (AO III), Cairncross et al documented that AO III’s with loss of the short arm of chromosome 1 (1p) were preferentially chemosensitive when treated with a procarbazine-lomustine-vincristine (PCV) chemotherapy regimen. Moreover, they also found that patients whose tumours had co-deletions of 1p and the long arm of chromosome 19 (19q) had substantially improved survival times.5 These findings have been replicated many times over the past 10 years, and the correlations with therapeutic sensitivity have been extended to other agents such as temozolomide 6,7 and modalities like radiotherapy.8 Such data suggest the merit of this marker may not lie in specifically predicting chemosensitivity but rather in demonstrating tumour vulnerability to a broad range of therapeutic options. As a result, assessment of 1p and 19q status has been widely implemented in neuro-oncologic management of AO III patients.9
The prevalence of 1p/19q codeletion has been estimated at 80 to 90% of grade II oligodendrogliomas and 50 to 70% of grade III oligodendrogliomas, and around 70% overall.10 Notably, however, although the 1p and 19q regions have been extensively mapped and many genes evaluated as candidate tumour suppressor genes, no tumorigenic genes have been definitively implicated. In most cases, deletions appear to represent complete chromosomal arm loss, which may be the result of an unbalanced centromeric translocation of 1p and 19q.11,12 Although the responsible oncogenic genes on 1p and 19q remain unidentified, many correlations have been made regarding 1p/19q loss. For example, 1p/19q deleted tumours frequently show classic histology.13–15 and often have IDH1 and IDH2 mutations.16 1p/19q codeleted AO III preferentially display a proneural gene expression profile,17 This profile, which is partly characterized by expression of neuronal genes, is overrepresented among low grade gliomas and may predict for therapeutic response in glioblastoma.18 On the other hand, 1p and 19q loss correlates inversely with TP53 mutations, 10q deletions and amplification of EGFR.13 Tumour location is also associated with 1p/ 19q co-deletions: low grade oligodendroglioma and AO III in the frontal, parietal and occipital lobes are more likely to show loss than tumours involving the temporal lobe, insula and diencephalon.19,20,21
1p/19q loss is also found in mixed glial tumours (oligoastrocytomas), albeit in a lower proportion than “pure” oligodendroglioma (20 to 30%).22 Eoli et al demonstrated that in mixed gliomas, 1p loss is associated with prolonged progression free survival. Conversely, tumours without 1p loss show shortened times to recurrence, are more frequently located in the temporal lobe and have radiological features suggesting malignancy.23 Deletions involving 1p and 19q are uncommon in glioblastoma, but in the small number identified seem to predict for shortened survival, possibly reflecting true genomic instability.15
Thus, over the past 10 years, the frequent evaluation of oligodendroglial tumours for 1p/ 19q status has shown that this molecular profile denotes a clinically distinct tumour type with progression, prognosis and treatment responses that are different from other gliomas.10,24 Although the mechanism whereby 1p/19q codeletion generates these phenomena remains unclear, the clear differences between 1p/19q-codeleted tumors and other oligodendrogliomas has made testing useful and commonplace.
Current laboratory testing
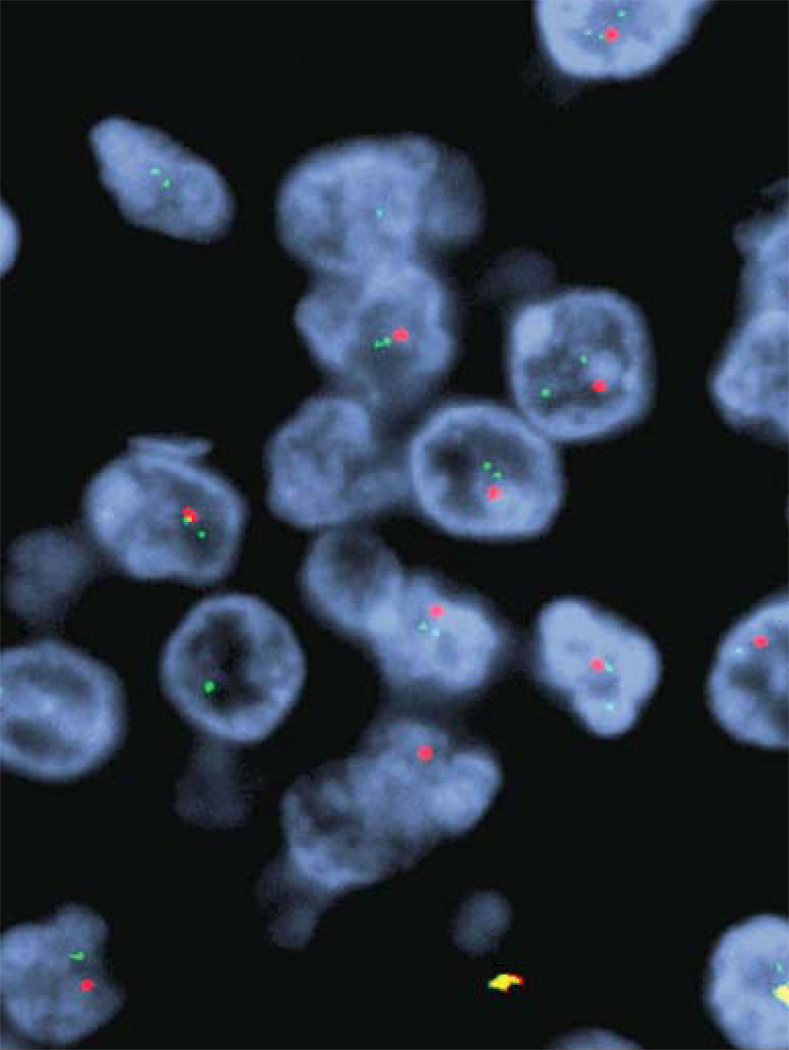
In general, most neuro-oncological institutions perform 1p/19q testing on tumours with oligodendroglial morphology, whether grade II or III and whether “pure” oligodendroglioma or “mixed” oligoastrocytoma. Two major methods exist for assessing loss of 1p and 19q: polymerase chain reaction (PCR)-based loss of heterozygosity (LOH) assays and fluorescence in situ hybridization (FISH). LOH assays are based on comparing multiple polymorphic alleles in tumor DNA versus normal blood DNA. Because not all alleles are informative, it is necessary to amplify multiple loci for each chromosomal arm; in addition, the assay requires available normal DNA, possibly delaying test results. Both of these drawbacks limit the applicability of LOH assays to studying 1p/19q status. FISH uses fluorescence-labeled probes to study chromosomes directly on tissue sections, which can be a time-consuming undertaking but which preserves tissue architecture and is therefore a common procedure available in many pathology laboratories. Until recently, studies had shown roughly comparable results between LOH and FISH assays.14 However, Snuderl et al have now demonstrated that additional information can be gleaned from the copy number information also revealed by FISH analysis, but not available in LOH studies: FISH assays allow assessment of polysomy, which may predict for reduced progression free survival.25
Other techniques may also impact testing in the near future. For example, array comparative genomic hybridization, a technique resulting in genome-wide differential labeling of reference and tumour DNA, provides an estimate of copy number at all chromosomal loci examined on a microarray, and can thus provide a ready means for detecting losses and assessing polysomy. Once this technique becomes less expensive and more widely available, it may replace FISH as the preferred technique.
Recommendations
Knowledge of 1p/19q status is now considered standard of care when managing oligodendroglial tumours26 and oncologists more often choose particular therapeutic options in anticipation of the longer survival and progression free survival times associated with this lesion.9 The following section explores the use of 1p/19q testing in four current settings: AO III, lower grade oligodendroglioma, "mixed" oligoastrocytic tumours, and histological diagnosis.
Large studies have now analyzed the potential role of 1p/19q loss in aiding management decisions in the setting of AO III. 1p/19q deletions were incorporated into two major therapeutic trials in patients with AO III. Both confirmed the prognostic and possible predictive role of these biomarkers at initial therapy.27,28 However, it remains unclear whether 1p/19q codeletion correlates with response to second line chemotherapy.7,25 Interpretation of the later stages of treatment in such trials is confounded by the effects of salvage therapy being given to all treatment groups.28,29 The major question, still being debated by the field is whether one can delay radiotherapy in patients whose AO III harbor 1p/19q loss; while the answer is not yet clear, a recent survey has shown that many North American neuro-oncologists are following this practice in selected patients.9 To refine the role of testing, future therapeutic trials of grade III gliomas may wind up recruiting and randomizing based on 1p/19q codeletion status rather than histology30
For lower-grade oligodendrogliomas, 1p/19q testing is also commonplace. 1p/19q codeletion predicts for increased progression free survival in temozolomide treated low grade gliomas,15,31 including radiotherapy naïve tumours32 and when utilized as monotherapy in tumours with 1p loss.31,33 One retrospective series of "expectantly managed" low grade oligodendroglial tumours found that 1p/19q codeletion status did not confer any prognostic advantage in progression free survival.34 Nonetheless, in an asymptomatic patient with a low grade oligodendroglioma harboring 1p/19q loss, a neuro-oncologist might argue that the likely slow growth and long survival allows for "watchful waiting" in the expectation of a superior response to therapy in the future. On the other hand, in a symptomatic patient with the same molecular signature, one would very likely opt for early therapy in the likelihood that the tumor will respond to therapy and patient symptoms will be ameliorated. At the present time, how 1p/19q status is used to manage patients with low grade oligodendrogliomas remains sub judice.
The diagnosis of "mixed" oligoastrocytic tumors is highly subjective, and varies from institution to institution, because of relatively lax diagnostic criteria. In the setting of "mixed" oligoastrocytic tumours, 1p/19q codeletions have been shown to have some influence on prognosis in some studies,23 but the data are probably not extensive or mature enough to affect therapeutic choices-- most notably because the entry criteria for oligoastrocytoma studies vary so much between institutions. Given this subjective diagnostic variability, the objective knowledge that an oligoastrocytoma has a genotype more like an astrocytoma versus more like an oligodendroglioma can provide some reassurance and guidance to clinicians.
Lastly, one must ask whether the 1p/19q codeletion signature should define a histologic class of tumour (i.e., oligodendroglioma) because of its prognostic and predictive value. Although it is tempting to do so, the issue remains unclear. Evidence has emerged that subgroups exist within 1p/19q codeleted tumours. These include tumours showing polysomy of chromosomes 1 and 19 with shorter progression free survival more akin to tumours without codeletion, but no decrease in overall survival or response to salvage chemotherapy.25 Moreover, Walker et al recently showed that a sample of tumours with oligodendroglial morphology lacking 1p/19q codeletions contained a small subgroup with a positive response to therapy, implying that other mechanisms contribute to the improved prognosis associated with this histologic entity.35 Such evidence reinforces the definition of oligodendroglioma in the World Health Organization Classification of Tumours of the CNS, as a histologically, clinically and genetically defined lesion.1
MGMT in glioma
Background
Alkylating chemotherapeutic agents have long been used in the treatment of patients with malignant gliomas. At the present time, nearly all glioblastoma patients are treated with the orally administered alkylating agent temozolomide (TMZ).36 TMZ acts to methylate primarily the O6 position of the nucleotide guanine, resulting in cell death.37 Cells, however, have an inherent DNA repair mechanism that can counter the effects of TMZ: the constitutively expressed DNA repair enzyme MGMT will irreversibly transfer a methyl group from the O6 position of the modified guanine to a cysteine residue of the MGMT protein, mitigating against the cytotoxic effects of chemotherapy. However, this enzyme is present in finite quantities and approximately half of glioblastomas show decreased levels of MGMT protein, which could make these tumours more susceptible to the effects of TMZ. The primary mechanism whereby MGMT expression is downregulated in glioblastomas appears to be methylation of the MGMT gene promoter, which is a common way of silencing gene expression in tumours.37 Thus, the expression level of MGMT and the methylation status of the MGMT gene promoter should have predictive value in glioblastoma patients treated with alkylating agents such as TMZ.37,38 Indeed, a number of studies suggest such correlations in glioblastomas,39,40 paediatric glioblastoma41 and lower grade gliomas.42 MGMT promoter methylation status has also been shown to be a valid prognostic marker independent of TMZ use in elderly GBM patients.43 Of the many studies, the most notable was that of Hegi et al. who studied MGMT gene promoter status in tumours from patients entered into a large trial to evaluate the role of concomitant TMZ with radiation therapy versus radiation therapy alone.36 In 106 TMZ-treated patients (46 with methylated tumours; 60 with unmethylated tumours), 46% of patients with MGMT methylated tumours were alive at 2 years versus only 22·7% of tumours with unmethylated MGMT. Assessment of overall survival was confounded by many patients having salvage therapy that included alkylating agents, but this trend appeared to continue. These authors subsequently suggested that assays examining MGMT methylation status, in addition to providing prognostic information, may have utility as predictive tools to determine treatment with TMZ in glioblastoma.44 However, the group with unmethylated MGMT in this study also showed a survival benefit with TMZ therapy that, while inferior to that in MGMT methylated tumours, nearly reached significance (p=0·06).36 An explanation for this remains unclear, and could, among other reasons, reflect definitions of cut-off levels for separating methylated and unmethylated groups.45,46 However, this beneficial response to TMZ in MGMT unmethylated tumours, as well the current lack of a therapeutic alternative, suggests that treatment is unlikely to be withheld from patients on the basis of this test, especially given the oral bioavailability and well tolerated nature of the drug.
Notably, MGMT promoter methylation patterns may change in the time between primary diagnosis and recurrence, and change more frequently in tumours that initially show MGMT methylation.47 In this regard, the prognostic information conveyed by knowledge of the MGMT methylation status appears to apply only to primary tumours and not to recurrent tumours.45,47
Recently, the utility of MGMT methylation status in anaplastic (i.e. WHO grade III) gliomas was evaluated in the NAO-04 trial29 and the EORTC 26951 trial.48 Both trials found MGMT methylation status conferred prognostic benefit but did not specifically predict for response to therapy with alkylating agents. Interestingly MGMT promoter methylation has recently been re-examined in a cohort of patients with GBM treated with radiotherapy only.49 In this study, MGMT methylation appeared to predict for response to radiation therapy.49 Thus in the context of anaplastic glioma and possibly in GBM, the prognostic information conveyed by testing may not solely be attributable to MGMT promoter methylation enhancing the effect of therapy with alkylating agents, but may instead indicate a prognostically favorable molecular phenotype.45,49
The attention that surrounded the introduction of TMZ as the standard of care for glioblastomas patients in 2005, and with response to TMZ being associated with MGMT promoter methylation status, generated interest on the part of neuro-oncologists and patients to have MGMT testing performed. Although it was not going to alter therapy, many testing centers experienced pressure, generated largely by patients being informed by the internet, to set up the assay nonetheless.
More recently, it has been reported that patients whose glioblastomas have MGMT gene promoter methylation have a greater likelihood of demonstrating radiographic "pseudoprogression" when treated with TMZ than those whose tumours lack such methylation.45,50 "Pseudoprogression" is a phenomenon in which the neuroimaging appears to worsen following TMZ, but then improves with continued therapy. Thus, another role for MGMT promoter methylation testing could be evaluation of the likelihood of "pseudoprogression”.50
Current laboratory testing
Laboratory testing of MGMT status entails the use of methylation specific PCR, in which extracted DNA is treated with sodium bisulfite, resulting in substitution of unmethylated cytosine for uracil. Primers specific for methylated and unmethylated CpG rich areas of the MGMT gene promoter are then utilized to amplify the modified DNA sequences. The assay can be performed on paraffin-embedded tissue, but is dependent on tissue quantity and quality.51 Additional factors such as adequacy of bisulfite treatment, primer specificity and PCR conditions further input into determining the usefulness of the assay.51,52 Not infrequently, both methylated and unmethylated sequences are found in glioblastoma tissue, attributed to either inclusion of non-neoplastic tissue or tumour heterogeneity. Other bisulfite-based techniques including quantitative MSP, bisulfite sequencing, methylation-sensitive single strand conformation polymorphism analysis and mass spectrophotometer-based quantitative analysis are available,26 some of which have been utilized to assess MGMT methylation status.51,53 Non-bisulfite- based semi-quantitative techniques have also emerged, such as methylation specific-multiplex ligand-dependent probe amplification (MS-MLPA).54
Given that promoter methylation typically silences gene transcription and protein synthesis, assessment of protein expression directly in cells via immunohistochemistry, a widely available technique, would seem a logical way to assess MGMT status. However, immunohistochemical MGMT expression has been found to be heterogeneous both within tumours and within small regions of tumour.55 Reports have attempted to stratify staining intensity to identify methylated tumours,55 but the significance of these indices of staining intensities is currently not clear. Moreover, discordant results have been reported between genetic and protein assessments of MGMT status.56 At the present time, immunohistochemistry is not recommended for evaluating MGMT status in glioblastomas.
Recommendations
Given that TMZ and radiation therapy remain the standard of care for all glioblastoma patients, regardless of MGMT promoter methylation status, upfront diagnostic testing for MGMT is not necessary. Nonetheless, MGMT promoter methylation status provides prognostic information that many patients and oncologists will wish to have. Moreover, knowledge of MGMT promoter methylation status will be important for ascertaining the effects of novel therapy on patients who are enrolled in clinical trials, and must be included in all glioblastoma trials.
As mentioned above MGMT promoter methylation status can be useful in the differential diagnosis of true tumour progression versus "pseudoprogression" seen on imaging. In this scenario, for a tumour with MGMT promoter methylation, "pseudoprogression" appears more likely and thus therapy with TMZ is often continued despite the "worse" appearance of the neuroimaging. Conversely, a worsening appearance on imaging, in a patient whose glioblastoma lacks MGMT promoter methylation, might prompt consideration to change therapy.50,57
Epidermal growth factor receptor (EGFR) pathway testing
Background
Traditional chemotherapeutic approaches non-specifically target dividing cells through a variety of mechanisms. Molecularly targeted therapies, on the other hand, aim for specific inhibition of aberrant or amplified proteins that directly drive tumour cell growth. Recent successful examples have included imatinab to inhibit the tyrosine kinase fusion protein BCR-ABL in chronic myeloid leukaemia, as well as the C-KIT oncogene in gastrointestinal stromal tumours,58,59 and trastuzamab in the treatment of HER2-positive breast carcinoma.60
A number of growth factors and their receptors are upregulated in malignant gliomas, particularly in glioblastomas. For example, upregulation of epidermal growth factor receptor (EGFR)-mediated signaling is present in around 30% of gliomas61,62 and 60% of glioblastoma,63 with overexpression in glioblastomas generally driven by EGFR gene amplification.64,65 In addition, about half of EGFRoverexpressing glioblastomas feature genetically mutant EGFR molecules, the most frequent of which is EGFRvIII, which constitutively activates the EGFR-PI3K pathway due to deletion of the ligand binding domain.66 Additional missense mutations have been identified in the exons encoding extracellular EGFR domains.67,68 Moreover, other described activating mutations in exons encoding extracellular EGFR have been shown in vitro to drive oncogenesis and can be inhibited by small molecule tyrosine kinase inhibitors.68
The prognostic utility of EGFR amplification is not clear, with a number of studies producing contradictory findings.69 EGFR amplification may be prognostically related to patient age70 and EGFRvIII expression may identify a subgroup of tumours with more aggressive behaviour.71
The presence of EGFR overexpression and EGFR mutation in glioblastomas raises the possibility that EGFR-targeted agents could be used to treat those patients with glioblastomas that feature these aberrations-- a similar situation to that of some non-small cell lung carcinomas that harbour activating EGFR mutations and that show remarkable responses to the EGFR inhibitors erlotinib and gefitinib. However, this optimistic scenario is clouded in the setting of glioblastoma for a number of biological reasons. Many glioblastomas feature dysregulation of signaling cascades downstream of EGFR, such as that mediated by the PTEN tumour suppressor gene and involving the PI3K pathway and AKT. This phenomenon may contribute to loss of reactivity to the down regulation of EGFR.72 In addition, other growth factors such as PDGF also exercise their effect by modulation of these pathways. Finally, many glioblastoma cells may feature activation of multiple growth factor pathways, suggesting the need to use cocktails of molecular targeting agents in glioblastoma patients.73
The results in the literature examining EGFR targeting agents looked promising initially. Two studies published in 2005 sought to clarify if assessment of EGFR status was useful in aiding decision making to use the small molecule kinase inhibitors gefitinib and erlotinib. Mellinghoff et al showed that co-expression of PTEN and EGFRvIII was associated with increased sensitivity to erlotinib, whereas tumours that lacked PTEN expression failed to respond to erlotinib.74 Haas-Kogan et al. suggested that glioblastomas with diffuse EGFR immunopositivity that lacked p-AKT stained were likely to show a clinical response to erlotinib.75 Both studies implied that those tumours that respond to small molecule kinase inhibitors overexpress EGFR or express EGFRvIII; and have an intact PTEN-AKT pathway. A curious feature of these two studies is that, although the immunohistochemical assays measured similar molecules, they did not necessarily represent the same biological endpoints. More problematically, subsequent trials did not show major responses or survival benefits in glioblastoma patients treated with these agents.76,77 Further studies have since evaluated erlotinib used concurrently with TMZ and radiotherapy.78,79 These trials have produced mixed results, one showing neither overall benefit nor identifying a vulnerable subgroup of tumours that might respond to therapy;78 the other suggesting a role for this regimen in patients with tumours showing MGMT promoter methylation and intact PTEN.79
At the present time, therefore, assessment of the EGFR signaling pathway in planning therapy is not clinically indicated, since current standard therapies are not directed specifically at this pathway. Nonetheless, the paradigm illustrated by the EGFR-non-small cell lung cancer story and hinted at in the EGFR-glioblastoma papers from 2005 is a critical one that is likely to surface repeatedly over the next decade. As new molecularly targeted agents reach clinical trials, alone and in combination, these will be tried in the setting of glioblastoma and it is highly likely that use of these novel agents will be guided by molecular testing of the respective signaling pathways, either at a DNA or a protein level. New methods should enable inquiry along multiple signaling pathways in single tumors on a routine basis. For example, at the Massachusetts General Hospital, a full range of mutational analyses are now carried out in standard paraffin-embedded samples using the Applied Biosystems SNaPshot system, incorporating multiplex PCR, single base extension and capillary electrophoresis. Such prospective and timely analysis already allows patient tailored therapy based on genotyping of known oncogenes and tumour suppressor genes.80,81
Current laboratory testing
In glioblastomas, amplification of EGFR occurs largely as extrachromosomal, double minute fragments. This makes confirmation of major copy number gain by FISH practical and accessible to many pathology laboratories. Moreover, FISH assays maintain in situ information, avoiding false negative assays due to intratumoural heterogeneity.82,83 Other techniques such as quantitative-PCR and RT-PCR also identify EGFR amplification;84 in addition, these and other molecular techniques can identify mutations in EGFR.84 Immunohistochemical assessment for EGFR expression is widely available, but of less clear value (see below). EGFR expression can be found in lower grade astrocytomas and other malignant gliomas. Moreover, scoring of EGFR immunopositivity can be variable85 and there may be discrepancies between EGFR amplification as determined by FISH and increased EGFR expression as determined using immunohistochemistry.86,87 Scoring of EGFR immunopositivity is also dependent on the antigenic specificity of the individual antibody clone used. Notably, however, antibodies specific for the novel antigenic epitope generated from the exonic deletion event in EGFRvIII permit the specific detection of cells expressing this variant protein.88
Recommendations
In the future, as new agents are developed to target EGFR in glioblastoma, EGFR amplification and mutation status could become an important variable to be assessed in patients involved in clinical trials of such agents. At the present time, EGFR testing has found utility in adult neuro-oncology in two situations. These involve the pathological characterization of glioblastomas in two settings: the diagnosis of small cell glioblastoma and the differential diagnosis of the edge of a glioblastoma versus an anaplastic astrocytoma.
Some glioblastomas may be composed of densely packed monotonous, small cells that may not stain strongly for the common glial immunohistochemical marker GFAP, resulting in diagnostic confusion with high grade oligodendroglial tumours. EGFR gene amplification is quite common in such "small cell" glioblastomas, being found in about 71% in one series.89 Thus, in such diagnostically challenging cases, the presence of EGFR gene amplification favors a diagnosis of small cell glioblastoma while 1p/19q loss would favor a diagnosis of anaplastic oligodendroglioma.
Another vexing scenario for the neuropathologist is the small biopsy that samples the edge of a glioblastoma. Although the biopsy does not demonstrate histological features sufficient for the diagnosis, the clinical and neuroimaging features may suggest glioblastoma. In such cases, the finding of EGFR gene amplification in the infiltrating tumor cells strongly suggests the presence of a tumor that is likely to act along the lines of a glioblastoma.90 In essence, in this latter scenario, EGFR gene copy number status is being used as a surrogate for grade in estimating prognosis. EGFR copy number status can be readily determined by FISH in both of these diagnostically challenging scenarios.85–87 Thus, in these two settings, we recommend diagnostic assessment of EGFR copy number using FISH.26 EGFR gene amplification implies, in both situations, that the tumor at hand will follow a course more similar to that of a glioblastoma.
IDH testing
Background
In a recent genome-wide survey, mutations in isocitrate dehydrogenase 1 (IDH1) were identified at high frequency in younger patients with secondary glioblastoma91 and subsequently in lower-grade diffuse gliomas.16,92 The somatic mutations identified at codon 132 were present in 12% of all glioblastomas and appeared to confer a prognostic advantage, even after adjustment for age.91 A follow-up study showed improved outcome for IDH1 and IDH2 mutated tumours, with median overall survival 31 months versus 15 months for glioblastoma lacking mutations and 65 versus 20 months for anaplastic astrocytoma.16 How these alterations cause oncogenesis is uncertain, although recently the role that metabolic pathways may have in contributing to oncogenic activation has been examined.93,94
Potential applications
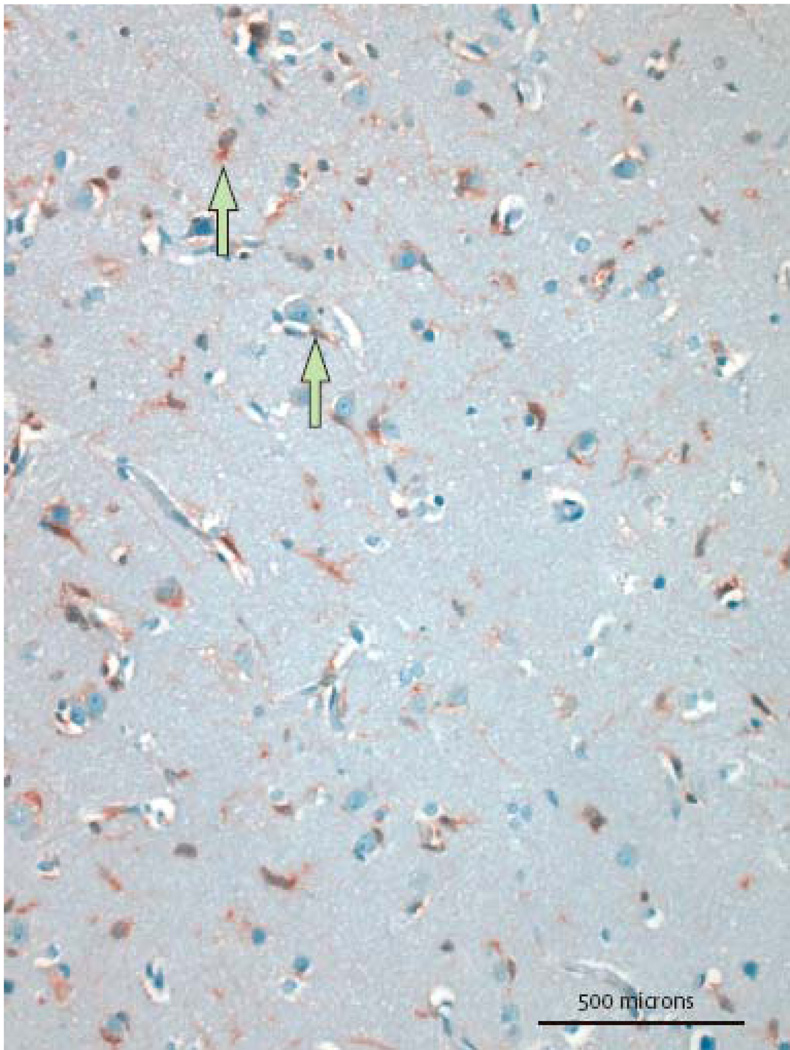
The role of IDH mutations as prognostic indicators is still being defined. From a diagnostic point of view, however, it is interesting that IDH1 and IDH2 mutations occur rarely in primary glioblastomas (0·05%), but at higher frequencies in secondary glioblastoma (84·6%) and lower grade gliomas (including diffuse astrocytomas (83·3%), anaplastic astrocytoma (69·2%), oligodendroglioma (80·4%), anaplastic oligodendrogliomas (86·1%) and oligoastrocytomas(100%)).16 This is of diagnostic interest since lower grade, infiltrating gliomas may prove diagnostically challenging, especially on small biopsies in which the differential diagnosis includes reactive gliosis. The presence of mutant IDH1/2 clearly has potential to identify infiltrating glial tumours and aid in this clinically essential differential diagnosis.95 One study has already demonstrated this using a PCR-based assay, showing mutations in the tumors but not in a series of reactive conditions.96 Following the development of an antibody to mutant R132H IDH1 protein95 which could discriminate single infiltrating cells expressing wild type and mutant IDH1,97 it was demonstrated that immunohistochemistry utilizing an antibody specific for the mutant R132H IDH1 protein could successfully identify infiltrating cells of low grade infiltrating glioma in 9 of 21 cases, while not staining at all in 20 cases of gliosis. Moreover, when combined with p53 immunhistochemistry, 14 of 21 tumours could be positively identified and distinguished from gliosis.98 Thus, the detection of IDH mutation, whether by DNA sequencing or by simple immunohistochemistry will prove a powerful addition to the neuropathological armamentarium in evaluating small, diagnostically challenging biopsies.
Concluding remarks
In this review, we have endeavored to cover those markers that have achieved a place in current diagnostic adult neuro-oncology. In doing so we have avoided discussion of those markers that are sometimes requested by clinicians and patients based on more limited data; while such markers may assume a place in the diagnostic armamentarium in the future, the current literature does not support their routine use at the present time. Moreover, we have covered preferred technical approaches to evaluating certain markers, but it is important to realize that the field of molecular diagnostics is moving rapidly, and the advent of technologies such as next-generation sequencing may “disrupt” our current thinking about preferred approaches.
The IDH story discussed above also demonstrates the rapidity with which molecular assays are being brought into common use. For instance, over the past 15 years, the diagnosis of the rare pediatric atypical teratoid/rhabdoid tumor transitioned from using FISH for chromosome 22q loss, to INI1 gene sequencing, to INI1 immunohistochemistry. The IDH1 story in gliomas, however, has progressed over one year, from initial description of the mutations to implementation of a practical immunohistochemical assay.
This progress is likely to quicken even more. The Cancer Genome Atlas was initiated by the National Cancer Institute of the National Institutes of Health in 2006 to accelerate our molecular understanding of cancer. This is to be achieved by large scale integrative genomic and epigenomic analyses at multiple levels, studying gene copy number, mutation and expression. The first tumor to be studied was glioblastoma.99 Other large-scale projects have pursued similar approaches.91 These systematic examinations of the glioblastoma genome have already provided a wealth of information, reiterating and elaborating upon prior findings, and creating avenues for new insights. Also, second generation sequencing technology is becoming increasingly accessible for researchers and clinicians in the high throughput analysis of cancers.100 Coupled with improved bioinformatic tools and filtering algorithms, new sequencing technology could prove extremely useful in elucidating genetic alterations such as gene fusion events and aberrant RNA-editing.
TCGA and similar projects will quickly further understanding of the molecular basis of neoplasia. This progress in turn portends rapid expansion in the development of molecular assays, with the potential to improve our ability to predict tumour behaviour in individual patients. These exciting developments are the first steps toward personalized medicine in the setting of malignant glioma.
Search Strategy and Research Criteria
References for this review were identified by a series of Pubmed searches from January 1998 to January 2010. Search terms included “glioma”, "astrocytoma," "oligodendroglioma”, “oligoastrocytoma”, “mixed glioma”, “glioblastoma”, “1p" ,"19q", “MGMT”, “IDH1” and “EGFR”. Articles from the authors’ own files were also identified and utilized. Only papers published in English were reviewed.
Figure 1.
Figure 2.
Figure 3.
Table 1.
Recommendations for Molecular Markers in Malignant Gliomas
| Known Biologic Mechanism |
Laboratory Testing Ready |
Research Testing for Clinical Trials |
Clinical Testing for Patients |
|
|---|---|---|---|---|
| 1p19q LOH | − | + | + | + |
| MGMT | + | − | + | +/− |
| EGFR pathway assessment | + | +/− | + | − |
| IDH mutation testing | +/− | + | +? | + |
EGFR, epidermal growth factor receptor; LOH, loss of heterozygosity; MGMT, O6-Methylguanine DNA methyltransferase; IDH, isocitrate dehydrogenase
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to declare.
Contributor Information
Michael Jansen, Pathology Service, Massachusetts General Hospital; and Histopathology Department, Cork University Hospital.
Stephen Yip, Pathology Service, Massachusetts General Hospital; Department of Pathology & Laboratory Medicine and Centre for Translational and Applied Genomics, British Columbia Cancer Agency.
David N. Louis, Pathology Service and Cancer Center, Massachusetts General Hospital; and Department of Pathology, Harvard Medical School.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO Classification of tumours of the central nervous system. Lyon, France: IARC Press; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamont JM, McManamy CS, Pearson AD, Clifford SC, Ellison DW. Combined histopathological and molecular cytogenetic stratification of medulloblastoma patients. Clin Cancer Res. 2004 Aug 15;10(16):5482–5493. doi: 10.1158/1078-0432.CCR-03-0721. [DOI] [PubMed] [Google Scholar]
- 3.Sigauke E, Rakheja D, Maddox DL, Hladik CL, White CL, Timmons CF, et al. Absence of expression of SMARCB1/INI1 in malignant rhabdoid tumors of the central nervous system, kidneys and soft tissue: an immunohistochemical study with implications for diagnosis. Mod Pathol. 2006 May;19(5):717–725. doi: 10.1038/modpathol.3800581. [DOI] [PubMed] [Google Scholar]
- 4.Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008 Nov 1;68(21):8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998 Oct 7;90(19):1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 6.Brandes AA, Tosoni A, Cavallo G, Reni M, Franceschi E, Bonaldi L, et al. Correlations between O6-methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: a prospective GICNO study. J Clin Oncol. 2006 Oct 10;24(29):4746–4753. doi: 10.1200/JCO.2006.06.3891. [DOI] [PubMed] [Google Scholar]
- 7.Kouwenhoven MC, Kros JM, French PJ, Biemond-ter Stege EM, Graveland WJ, Taphoorn MJ, et al. 1p/19q loss within oligodendroglioma is predictive for response to first line temozolomide but not to salvage treatment. Eur J Cancer. 2006 Oct;42(15):2499–2503. doi: 10.1016/j.ejca.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 8.Bauman GS, Ino Y, Ueki K, Zlatescu MC, Fisher BJ, Macdonald DR, et al. Allelic loss of chromosome 1p and radiotherapy plus chemotherapy in patients with oligodendrogliomas. Int J Radiat Oncol Biol Phys. 2000 Oct 1;48(3):825–830. doi: 10.1016/s0360-3016(00)00703-3. [DOI] [PubMed] [Google Scholar]
- 9.Abrey LE, Louis DN, Paleologos N, Lassman AB, Raizer JJ, Mason W, et al. Survey of treatment recommendations for anaplastic oligodendroglioma. Neuro Oncol. 2007 Jul;9(3):314–318. doi: 10.1215/15228517-2007-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairncross G, Jenkins R. Gliomas with 1p/19q codeletion: a.k.a. oligodendroglioma. Cancer J. 2008 Nov–Dec;14(6):352–357. doi: 10.1097/PPO.0b013e31818d8178. [DOI] [PubMed] [Google Scholar]
- 11.Griffin CA, Burger P, Morsberger L, Yonescu R, Swierczynski S, Weingart JD, et al. Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol. 2006 Oct;65(10):988–994. doi: 10.1097/01.jnen.0000235122.98052.8f. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006 Oct 15;66(20):9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 13.Nutt CL. Molecular genetics of oligodendrogliomas: a model for improved clinical management in the field of neurooncology. Neurosurg Focus. 2005 Nov;19(5):E2. doi: 10.3171/foc.2005.19.5.3. [DOI] [PubMed] [Google Scholar]
- 14.Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G, et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999 Jul 15;18(28):4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 15.Smith JS, Perry A, Borell TJ, Lee HK, O'Fallon J, Hosek SM, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000 Feb;18(3):636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 16.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009 Feb 19;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducray F, Idbaih A, de Reynies A, Bieche I, Thillet J, Mokhtari K, et al. Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Mol Cancer. 2008;7:41. doi: 10.1186/1476-4598-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sulman EP, Guerrero M, Aldape K. Beyond grade: molecular pathology of malignant gliomas. Semin Radiat Oncol. 2009 Jul;19(3):142–149. doi: 10.1016/j.semradonc.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Zlatescu MC, TehraniYazdi A, Sasaki H, Megyesi JF, Betensky RA, Louis DN, et al. Tumor location and growth pattern correlate with genetic signature in oligodendroglial neoplasms. Cancer Res. 2001 Sep 15;61(18):6713–6715. [PubMed] [Google Scholar]
- 20.Laigle-Donadey F, Martin-Duverneuil N, Lejeune J, Criniere E, Capelle L, Duffau H, et al. Correlations between molecular profile and radiologic pattern in oligodendroglial tumors. Neurology. 2004 Dec 28;63(12):2360–2362. doi: 10.1212/01.wnl.0000148642.26985.68. [DOI] [PubMed] [Google Scholar]
- 21.Kouwenhoven MC, Gorlia T, Kros JM, Ibdaih A, Brandes AA, Bromberg JE, et al. Molecular analysis of anaplastic oligodendroglial tumors in a prospective randomized study: A report from EORTC study 26951. Neuro Oncol. 2009 Feb 17; doi: 10.1215/15228517-2009-011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aldape K, Burger PC, Perry A. Clinicopathologic aspects of 1p/19q loss and the diagnosis of oligodendroglioma. Arch Pathol Lab Med. 2007 Feb;131(2):242–251. doi: 10.5858/2007-131-242-CAOQLA. [DOI] [PubMed] [Google Scholar]
- 23.Eoli M, Bissola L, Bruzzone MG, Pollo B, Maccagnano C, De Simone T, et al. Reclassification of oligoastrocytomas by loss of heterozygosity studies. Int J Cancer. 2006 Jul 1;119(1):84–90. doi: 10.1002/ijc.21759. [DOI] [PubMed] [Google Scholar]
- 24.Reifenberger G, Louis DN. Oligodendroglioma: toward molecular definitions in diagnostic neuro-oncology. J Neuropathol Exp Neurol. 2003 Feb;62(2):111–126. doi: 10.1093/jnen/62.2.111. [DOI] [PubMed] [Google Scholar]
- 25.Snuderl M, Eichler AF, Ligon KL, Vu QU, Silver M, Betensky RA, et al. Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clin Cancer Res. 2009 Oct 15;15(20):6430–6437. doi: 10.1158/1078-0432.CCR-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yip S, Iafrate AJ, Louis DN. Molecular diagnostic testing in malignant gliomas: a practical update on predictive markers. J Neuropathol Exp Neurol. 2008 Jan;67(1):1–15. doi: 10.1097/nen.0b013e31815f65fb. [DOI] [PubMed] [Google Scholar]
- 27.Cairncross G, Berkey B, Shaw E, Jenkins R, Scheithauer B, Brachman D, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol. 2006 Jun 20;24(18):2707–2714. doi: 10.1200/JCO.2005.04.3414. [DOI] [PubMed] [Google Scholar]
- 28.van den Bent MJ, Carpentier AF, Brandes AA, Sanson M, Taphoorn MJ, Bernsen HJ, et al. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol. 2006 Jun 20;24(18):2715–2722. doi: 10.1200/JCO.2005.04.6078. [DOI] [PubMed] [Google Scholar]
- 29.Wick W, Hartmann C, Engel C, Stoffels M, Felsberg J, Stockhammer F, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009 Dec 10;27(35):5874–5880. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 30.Idbaih A, Omuro A, Ducray F, Hoang-Xuan K. Molecular genetic markers as predictors of response to chemotherapy in gliomas. Curr Opin Oncol. 2007 Nov;19(6):606–611. doi: 10.1097/CCO.0b013e3282f075f3. [DOI] [PubMed] [Google Scholar]
- 31.Kaloshi G, Benouaich-Amiel A, Diakite F, Taillibert S, Lejeune J, Laigle-Donadey F, et al. Temozolomide for low-grade gliomas: predictive impact of 1p/19q loss on response and outcome. Neurology. 2007 May 22;68(21):1831–1836. doi: 10.1212/01.wnl.0000262034.26310.a2. [DOI] [PubMed] [Google Scholar]
- 32.Levin N, Lavon I, Zelikovitsh B, Fuchs D, Bokstein F, Fellig Y, et al. Progressive low-grade oligodendrogliomas: response to temozolomide and correlation between genetic profile and O6-methylguanine DNA methyltransferase protein expression. Cancer. 2006 Apr 15;106(8):1759–1765. doi: 10.1002/cncr.21809. [DOI] [PubMed] [Google Scholar]
- 33.Hoang-Xuan K, Capelle L, Kujas M, Taillibert S, Duffau H, Lejeune J, et al. Temozolomide as initial treatment for adults with low-grade oligodendrogliomas or oligoastrocytomas and correlation with chromosome 1p deletions. J Clin Oncol. 2004 Aug 1;22(15):3133–3138. doi: 10.1200/JCO.2004.10.169. [DOI] [PubMed] [Google Scholar]
- 34.Weller M, Berger H, Hartmann C, Schramm J, Westphal M, Simon M, et al. Combined 1p/19q loss in oligodendroglial tumors: predictive or prognostic biomarker? Clin Cancer Res. 2007 Dec 1;13(23):6933–6937. doi: 10.1158/1078-0432.CCR-07-0573. [DOI] [PubMed] [Google Scholar]
- 35.Walker C, Haylock B, Husband D, Joyce KA, Fildes D, Jenkinson MD, et al. Clinical use of genotype to predict chemosensitivity in oligodendroglial tumors. Neurology. 2006 Jun 13;66(11):1661–1667. doi: 10.1212/01.wnl.0000218270.12495.9a. [DOI] [PubMed] [Google Scholar]
- 36.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005 Mar 10;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 37.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000 Nov 9;343(19):1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 38.Silber JR, Bobola MS, Ghatan S, Blank A, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase activity in adult gliomas: relation to patient and tumor characteristics. Cancer Res. 1998 Mar 1;58(5):1068–1073. [PubMed] [Google Scholar]
- 39.Jaeckle KA, Eyre HJ, Townsend JJ, Schulman S, Knudson HM, Belanich M, et al. Correlation of tumor O6 methylguanine-DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis-chloroethylnitrosourea: a Southwest Oncology Group study. J Clin Oncol. 1998 Oct;16(10):3310–3315. doi: 10.1200/JCO.1998.16.10.3310. [DOI] [PubMed] [Google Scholar]
- 40.Martinez R, Schackert G, Yaya-Tur R, Rojas-Marcos I, Herman JG, Esteller M. Frequent hypermethylation of the DNA repair gene MGMT in long-term survivors of glioblastoma multiforme. J Neurooncol. 2007 May;83(1):91–93. doi: 10.1007/s11060-006-9292-0. [DOI] [PubMed] [Google Scholar]
- 41.Pollack IF, Hamilton RL, Sobol RW, Burnham J, Yates AJ, Holmes EJ, et al. O6-methylguanine-DNA methyltransferase expression strongly correlates with outcome in childhood malignant gliomas: results from the CCG-945 Cohort. J Clin Oncol. 2006 Jul 20;24(21):3431–3437. doi: 10.1200/JCO.2006.05.7265. [DOI] [PubMed] [Google Scholar]
- 42.Everhard S, Kaloshi G, Criniere E, Benouaich-Amiel A, Lejeune J, Marie Y, et al. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006 Dec;60(6):740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- 43.Gerstner ER, Yip S, Wang DL, Louis DN, Iafrate AJ, Batchelor TT. Mgmt methylation is a prognostic biomarker in elderly patients with newly diagnosed glioblastoma. Neurology. 2009 Nov 3;73(18):1509–1510. doi: 10.1212/WNL.0b013e3181bf9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hau P, Stupp R, Hegi ME. MGMT methylation status: the advent of stratified therapy in glioblastoma? Dis Markers. 2007;23(1–2):97–104. doi: 10.1155/2007/159242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weller M, Stupp R, Reifenberger G, Brandes AA, van den Bent MJ, Wick W, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010 Jan;6(1):39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 46.Iafrate AJ, Louis DN. "MGMT for pt mgmt": is methylguanine-DNA methyltransferase testing ready for patient management? J Mol Diagn. 2008 Jul;10(4):308–310. doi: 10.2353/jmoldx.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandes AA, Franceschi E, Tosoni A, Bartolini S, Bacci A, Agati R, et al. O6-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro Oncol. 2010 Mar;12(3):283–288. doi: 10.1093/neuonc/nop050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Bent MJ, Dubbink HJ, Sanson M, van der Lee-Haarloo CR, Hegi M, Jeuken JW, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2009 Dec 10;27(35):5881–5886. doi: 10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010 Feb;12(2):116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brandes AA, Franceschi E, Tosoni A, Blatt V, Pession A, Tallini G, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008 May 1;26(13):2192–2197. doi: 10.1200/JCO.2007.14.8163. [DOI] [PubMed] [Google Scholar]
- 51.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. Biotechniques. 2002 Sep;33(3):632. doi: 10.2144/02333rv01. 4, 6–49. [DOI] [PubMed] [Google Scholar]
- 52.Preusser M. MGMT analysis at DNA, RNA and protein levels in glioblastoma tissue. Histol Histopathol. 2009 Apr;24(4):511–518. doi: 10.14670/HH-24.511. [DOI] [PubMed] [Google Scholar]
- 53.Maxwell JA, Johnson SP, Quinn JA, McLendon RE, Ali-Osman F, Friedman AH, et al. Quantitative analysis of O6-alkylguanine-DNA alkyltransferase in malignant glioma. Mol Cancer Ther. 2006 Oct;5(10):2531–2539. doi: 10.1158/1535-7163.MCT-06-0106. [DOI] [PubMed] [Google Scholar]
- 54.Jeuken JW, Cornelissen SJ, Vriezen M, Dekkers MM, Errami A, Sijben A, et al. MS-MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007 Oct;87(10):1055–1065. doi: 10.1038/labinvest.3700664. [DOI] [PubMed] [Google Scholar]
- 55.Nakasu S, Fukami T, Baba K, Matsuda M. Immunohistochemical study for O6-methylguanine-DNA methyltransferase in the non-neoplastic and neoplastic components of gliomas. J Neurooncol. 2004 Dec;70(3):333–340. doi: 10.1007/s11060-004-9170-6. [DOI] [PubMed] [Google Scholar]
- 56.Preusser M, Charles Janzer R, Felsberg J, Reifenberger G, Hamou MF, Diserens AC, et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. 2008 Oct;18(4):520–532. doi: 10.1111/j.1750-3639.2008.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fabi A, Russillo M, Metro G, Vidiri A, Di Giovanni S, Cognetti F. Pseudoprogression and MGMT status in glioblastoma patients: implications in clinical practice. Anticancer Res. 2009 Jul;29(7):2607–2610. [PubMed] [Google Scholar]
- 58.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006 Dec 7;355(23):2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 59.Schnadig ID, Blanke CD. Gastrointestinal stromal tumors: imatinib and beyond. Curr Treat Options Oncol. 2006 Nov;7(6):427–437. doi: 10.1007/s11864-006-0018-5. [DOI] [PubMed] [Google Scholar]
- 60.Lohrisch C, Piccart M. An overview of HER2. Semin Oncol. 2001 Dec;28(6 Suppl 18):3–11. [PubMed] [Google Scholar]
- 61.Humphrey PA, Wong AJ, Vogelstein B, Friedman HS, Werner MH, Bigner DD, et al. Amplification and expression of the epidermal growth factor receptor gene in human glioma xenografts. Cancer Res. 1988 Apr 15;48(8):2231–2238. [PubMed] [Google Scholar]
- 62.Agosti RM, Leuthold M, Gullick WJ, Yasargil MG, Wiestler OD. Expression of the epidermal growth factor receptor in astrocytic tumours is specifically associated with glioblastoma multiforme. Virchows Arch A Pathol Anat Histopathol. 1992;420(4):321–325. doi: 10.1007/BF01600211. [DOI] [PubMed] [Google Scholar]
- 63.Omuro AM, Faivre S, Raymond E. Lessons learned in the development of targeted therapy for malignant gliomas. Mol Cancer Ther. 2007 Jul;6(7):1909–1919. doi: 10.1158/1535-7163.MCT-07-0047. [DOI] [PubMed] [Google Scholar]
- 64.Bigner SH, Humphrey PA, Wong AJ, Vogelstein B, Mark J, Friedman HS, et al. Characterization of the epidermal growth factor receptor in human glioma cell lines and xenografts. Cancer Res. 1990 Dec 15;50(24):8017–8022. [PubMed] [Google Scholar]
- 65.McLendon RE, Turner K, Perkinson K, Rich J. Second messenger systems in human gliomas. Arch Pathol Lab Med. 2007 Oct;131(10):1585–1590. doi: 10.5858/2007-131-1585-SMSIHG. [DOI] [PubMed] [Google Scholar]
- 66.Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rao RD, James CD. Altered molecular pathways in gliomas: an overview of clinically relevant issues. Semin Oncol. 2004 Oct;31(5):595–604. doi: 10.1053/j.seminoncol.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 68.Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006 Dec;3(12):e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heimberger AB, Suki D, Yang D, Shi W, Aldape K. The natural history of EGFR and EGFRvIII in glioblastoma patients. J Transl Med. 2005 Oct 19;3:38. doi: 10.1186/1479-5876-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001 Aug 15;93(16):1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- 71.Jeuken J, Sijben A, Alenda C, Rijntjes J, Dekkers M, Boots-Sprenger S, et al. Robust detection of EGFR copy number changes and EGFR variant III: technical aspects and relevance for glioma diagnostics. Brain Pathol. 2009 Oct;19(4):661–671. doi: 10.1111/j.1750-3639.2009.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sulis ML, Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003 Sep;13(9):478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 73.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007 Oct 12;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 74.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005 Nov 10;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 75.Haas-Kogan DA, Prados MD, Tihan T, Eberhard DA, Jelluma N, Arvold ND, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005 Jun 15;97(12):880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 76.Rich JN, Reardon DA, Peery T, Dowell JM, Quinn JA, Penne KL, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004 Jan 1;22(1):133–142. doi: 10.1200/JCO.2004.08.110. [DOI] [PubMed] [Google Scholar]
- 77.Kesari S, Ramakrishna N, Sauvageot C, Stiles CD, Wen PY. Targeted molecular therapy of malignant gliomas. Curr Neurol Neurosci Rep. 2005 May;5(3):186–197. doi: 10.1007/s11910-005-0046-8. [DOI] [PubMed] [Google Scholar]
- 78.Brown PD, Krishnan S, Sarkaria JN, Wu W, Jaeckle KA, Uhm JH, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol. 2008 Dec 1;26(34):5603–5609. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009 Feb 1;27(4):579–584. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pao W, Kris MG, Iafrate AJ, Ladanyi M, Janne PA, Wistuba II, et al. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res. 2009 Sep 1;15(17):5317–5322. doi: 10.1158/1078-0432.CCR-09-0913. [DOI] [PubMed] [Google Scholar]
- 81.Hayden EC. Personalized cancer therapy gets closer. Nature. 2009 Mar 12;458(7235):131–132. doi: 10.1038/458131a. [DOI] [PubMed] [Google Scholar]
- 82.Okada Y, Hurwitz EE, Esposito JM, Brower MA, Nutt CL, Louis DN. Selection pressures of TP53 mutation and microenvironmental location influence epidermal growth factor receptor gene amplification in human glioblastomas. Cancer Res. 2003 Jan 15;63(2):413–416. [PubMed] [Google Scholar]
- 83.Aldape KD, Ballman K, Furth A, Buckner JC, Giannini C, Burger PC, et al. Immunohistochemical detection of EGFRvIII in high malignancy grade astrocytomas and evaluation of prognostic significance. J Neuropathol Exp Neurol. 2004 Jul;63(7):700–707. doi: 10.1093/jnen/63.7.700. [DOI] [PubMed] [Google Scholar]
- 84.Arjona D, Bello MJ, Alonso ME, Aminoso C, Isla A, De Campos JM, et al. Molecular analysis of the EGFR gene in astrocytic gliomas: mRNA expression, quantitative-PCR analysis of non-homogeneous gene amplification and DNA sequence alterations. Neuropathol Appl Neurobiol. 2005 Aug;31(4):384–394. doi: 10.1111/j.1365-2990.2005.00653.x. [DOI] [PubMed] [Google Scholar]
- 85.Kersting C, Packeisen J, Leidinger B, Brandt B, von Wasielewski R, Winkelmann W, et al. Pitfalls in immunohistochemical assessment of EGFR expression in soft tissue sarcomas. J Clin Pathol. 2006 Jun;59(6):585–590. doi: 10.1136/jcp.2005.028373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kersting C, Tidow N, Schmidt H, Liedtke C, Neumann J, Boecker W, et al. Gene dosage PCR and fluorescence in situ hybridization reveal low frequency of egfr amplifications despite protein overexpression in invasive breast carcinoma. Lab Invest. 2004 May;84(5):582–587. doi: 10.1038/labinvest.3700077. [DOI] [PubMed] [Google Scholar]
- 87.Dei Tos AP, Ellis I. Assessing epidermal growth factor receptor expression in tumours: what is the value of current test methods? Eur J Cancer. 2005 Jul;41(10):1383–1392. doi: 10.1016/j.ejca.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 88.Wikstrand CJ, Hale LP, Batra SK, Hill ML, Humphrey PA, Kurpad SN, et al. Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res. 1995 Jul 15;55(14):3140–3148. [PubMed] [Google Scholar]
- 89.Burger PC, Pearl DK, Aldape K, Yates AJ, Scheithauer BW, Passe SM, et al. Small cell architecture--a histological equivalent of EGFR amplification in glioblastoma multiforme? J Neuropathol Exp Neurol. 2001 Nov;60(11):1099–1104. doi: 10.1093/jnen/60.11.1099. [DOI] [PubMed] [Google Scholar]
- 90.Mott RT, Turner KC, Bigner DD, McLendon RE. Utility of EGFR and PTEN numerical aberrations in the evaluation of diffusely infiltrating astrocytomas. Laboratory investigation. J Neurosurg. 2008 Feb;108(2):330–335. doi: 10.3171/JNS/2008/108/2/0330. [DOI] [PubMed] [Google Scholar]
- 91.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008 Sep 26;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009 Oct;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 93.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009 Dec 10;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009 Apr 10;324(5924):261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Capper D, Weissert S, Balss J, Habel A, Meyer J, Jager D, et al. Characterization of R132H Mutation-specific IDH1 Antibody Binding in Brain Tumors. Brain Pathol. 2009 Oct 27; doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horbinski CKJ, Kelly LM, et al. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin-fixed, paraffin-embedded glioma tissues. J Neuropathol Exp Neurol. 2009;68:1319–1325. doi: 10.1097/NEN.0b013e3181c391be. [DOI] [PubMed] [Google Scholar]
- 97.Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009 Nov;118(5):599–601. doi: 10.1007/s00401-009-0595-z. [DOI] [PubMed] [Google Scholar]
- 98.Camelo-Piragua S, Jansen M, Ganguly A, Kim JC, Louis DN, Nutt CL. Mutant IDH1-specific immunohistochemistry distinguishes diffuse astrocytoma from astrocytosis. Acta Neuropathol. 2009 doi: 10.1007/s00401-009-0632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008 Nov;92(5):255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]