Abstract
Symbiosis between microbes and their mammalian host is vital to maintaining homeostasis. Symbiotic microbes within the gastrointestinal tract provide an array of benefits to the host, including promotion of host immunity. A coordinated effort of the host and symbiotic microbes deters the colonization and survival of many invading pathogens. However, pathogens have devised strategies to overcome these mechanisms. Furthermore, some pathogens can hijack host hormones and bacterial autoinducers to induce virulence traits. Intra- and inter-species (bacteria: bacteria) and inter-kingdom (bacteria: host) communication orchestrates the complex relationship among symbiotic microbes, invading pathogens, and their mammalian host. Insight into this communication will provide a foundation for the development of targeted antimicrobial therapies.
INTRODUCTION
Infectious diseases wreak havoc on mankind. The mammalian immune system has in place a line of defense specialized in recognizing and eradicating invading pathogens; however, sometimes the pathogen evades these mechanisms and establishes disease in its host. Therapies like antibiotics and vaccination abet the immune system in its fight against pathogenic microbes. Over time, resistance to antibiotics has developed due to the intense selective pressure the antibiotics place on bacteria. Furthermore, while a number of vaccines have been successful, far too many infectious diseases still do not have efficacious vaccines. A desperate call for new therapeutics exists.
Understanding the complex relationship among the host, symbiotic microbes and invading pathogens will provide important insight for the rational design of therapeutics. Bacteria can communicate with one another through hormone-like signals to modulate their gene expression 1 in a process termed quorum sensing (QS) 2. Additionally, these bacterial signals can modify mammalian cell-signal transduction 3, and likewise host hormones can cross signal to regulate bacterial gene expression 4 in a process termed inter-kingdom signaling. Interference with the cell-to-cell signaling pathways that control bacterial virulence offers a promising new strategy in the treatment of bacterial infections. This review will discuss both the mechanisms employed by the host and symbiotic bacteria to impair pathogen virulence, as well as the conserved cell-to-cell signaling pathways implemented by pathogens that allow for exploitation of their host environment.
ANTIMICROBIAL STRATEGIES ENLISTED BY THE HOST AND SYMBIONTS
The human gut hosts an estimated 500–1000 species of bacteria 5,6. A mutually beneficial relationship exists between the human intestine and many of its symbionts: the human intestine provides nutrients to the resident bacteria while bacteria aid in the digestion of food and absorption of nutrients, produce vitamins such as biotin and vitamin K, regulate immune system function and hinder the colonization of pathogenic microorganisms 7. Two major bacterial phyla, Firmicutes and Bacteroidetes, and five minor bacterial phyla, Proteobacteria, Actinobacteria, Fusobacteria, Cyanobacteria and Verrucomicrobia, compose the intestinal gut flora in adult humans 8. Variations in gut microbiota, however, can result from genetic and environmental factors such as diet, living conditions, and birthplace 6.
In a concerted effort, the host and symbiotic bacteria employ the use of physical, chemical and cell-mediated antimicrobial strategies to prevent or impair pathogen survival and virulence. Physical barriers provide the first line of resistance against pathogens. The intestinal mucosal barrier, composed of a thick mucus layer, a layer of epithelial cells, and an underlying layer of cells composed predominantly by immune cells, provides both a physical and chemical barrier through the secretion of mucins, immunoglobulins, antimicrobial peptides, and lectins 9. Especially important in barrier function, immunoglobulins secreted by B-lymphocytes aid in the opsonization of microbes and prevent microbial penetration of the mucosal layer 9. Resident bacteria provide another crucial line of defense against the colonization of pathogens by competing for nutrition and attachment sites to the colonic epithelium, a mechanism known as the “barrier effect”10.
Intestinal epithelial cells produce two major classes of antimicrobial peptides: defensins and cathelicidins11,12. Defensins function by embedding into the microbial membrane to form pores that allow for the efflux of essential ions and nutrients 13. In addition to their antimicrobial activity, defensins and cathelicidins modulate the host immune response 14, in large part by forming local chemotactic gradients that promote the mobilization of leukocytes 12. Angiogenins represent another important class of antimicrobial proteins. While mouse Ang1 and human ANG exhibit bactericidal and fungicidal activity against systemic pathogens, Ang4 acts selectively in the gut against enteric pathogens. Secreted by Paneth cells into the gut lumen, Ang4 expression is induced by the intestinal bacteria Bacteroides thetaiotamicron 15. Also influenced by the intestinal flora, the mouse C-type lectin RegIIIγ and its human counterpart HIP/PAP exert their bactericidal activity against Gram-positive bacteria via an interaction with peptidoglycan 16 and are vital to antimicrobial protection in the mammalian gut 17.
Symbiotic microbes provide another source of antimicrobial molecules within the mucosal barrier. Through the production of butyrate, a short-chain fatty acid (SCFA) that is the only known stimulus for cathelicidin expression 18, the enteric microflora aids in the stimulation of host innate immunity. SCFA have also been shown to decrease Shiga toxin expression in EHEC 19. Additionally, symbiotic microbes can interfere with pathogen survival and growth through the production of potent toxins called bacteriocins. For example, Lactobacillus salivarius UCC118 produces the bacteriocin ABP-118 that is active against food-borne pathogens, including Bacillus, Listeria, Enterococcus and Staphylococcus species 20,21.
Although both the host and symbiotic microbes have evolved mechanisms to prevent pathogen invasion and colonization, similarly pathogens have devised means to subvert and even exploit their environment.
QUORUM SENSING (QS)
Bacteria respond to hormone-like molecules called autoinducers to regulate specific target genes, a process known as QS 1,2. To date, four main categories of cell-to-cell signaling systems have been studied in detail. Gram-negative bacteria communicate in response to autoinducer-1 (AI-1) and autoinducer-3 (AI-3) while Gram-positive bacteria use an autoinducing polypeptide (AIP) system 22. Autoinducer-2 (AI-2) acts as a “universal” signal for interspecies communication and is found in both Gram-negative and Gram-positive cells 23. Due to space constraints, only the AI-1 and AI-3 cell signaling systems will be discussed in this review.
The LuxI/LuxR System
The foundation of QS, AI-1 and its cognate signaling system, the LuxI/LuxR system, were initially discovered in the bioluminescent marine bacterium Vibrio fisheri and its host, the Hawaiian Bobtail Squid Euprymna scolopes 1,24–26. LuxI synthesizes AI-1, an N-acyl homoserine lactone (AHL) that can diffuse freely across the bacterial membrane. The simultaneous production of AI-1 by a growing population of bacteria increases the local concentration of AI-1, and at high population density, AI-1 diffuses back into the cell. Inside the cell, AI-1 binds the transcriptional activator LuxR that, in its bound state to AI-1, can activate the transcription of the luxCDABEGH operon 27.
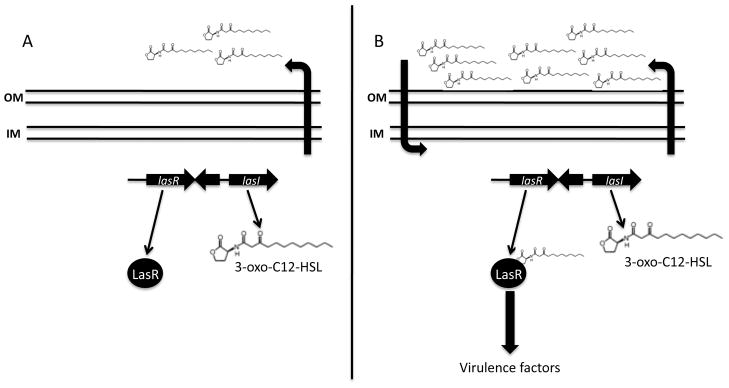
Homologues to the LuxI/LuxR system have been identified in many Gram-negative bacteria. For example, the opportunistic human pathogen, P. aeruginosa, has two homologous LuxI/LuxR systems: the LasI/LasR system (Figure 1) and the RhlI/RhlR system 28. P. aeruginosa produces two AHLs: 3-oxo-C12 HSL that acts on the las system and C4-HSL that acts on the rhl system. LasI produces 3-oxo-C12-HSL to activate LasR 29–31, which leads to the production of virulence factors like elastase 30 and pyoverdin 32. RhlI synthesizes C4-HSL to activate RhlR 33–35, allowing for the production of rhamnolipid biosurfactants 36 and a number of other virulence factors important in biofilm formation and pathogenesis 37.
Figure 1.
The LasI/LasR quorum sensing system in P. aeruginosa. (A) LasI synthesizes 3-oxo-C12 HSL, an AHL that freely diffuses across the bacterial membrane at low cell density; (B) At high cell density, 3-oxo-C12 HSL diffuses back into the cell, binds to the transcriptional activator LasR, and induces the expression of virulence genes.
Interestingly, both E. coli and S. Typhimurium encode for a LuxR homologue named SdiA but do not have a LuxI homologue 38,39. The absence of a LuxI homologue indicates that neither E. coli nor S. Typhimurium can produce its own AI-1. Instead, presence of SdiA may allow for these bacteria to respond to AI-1 made by other bacteria, including AI-1 produced by the resident flora in the gastrointestinal (GI) tract where both of these pathogens colonize.
Pathogenesis in the Gut
The GI tract is a diverse and dynamic environment, home to large communities of bacterial flora and constantly threatened by opportunistic and pathogenic microbes. Within the GI tract, bacteria communicate with one another and with their host to coordinate expression of key genes. Hormonal communication between the host and microorganisms is termed inter-kingdom signaling 40. One example of an inter-kingdom signaling system is the AI-3/epinephrine/norepinephrine signaling system 4.
Commensal E. coli, as well as the intestinal bacterial species EHEC, enteropathogenic E. coli (EPEC), K. pneumoniae, Shigella sp., Salmonella sp., and Enterobacter cloacae, produce AI-3 41. The mammalian hormones epinephrine and norepinephrine are also found in the intestine 42. While epinephrine’s and norepinephrine’s primary role are to modulate smooth muscle contraction, submucosal blood flow, and chloride and potassium secretion 43, enteric pathogens like EHEC O157:H7 can hijack these host hormones to induce virulence genes and promote colonization in the intestine 4.
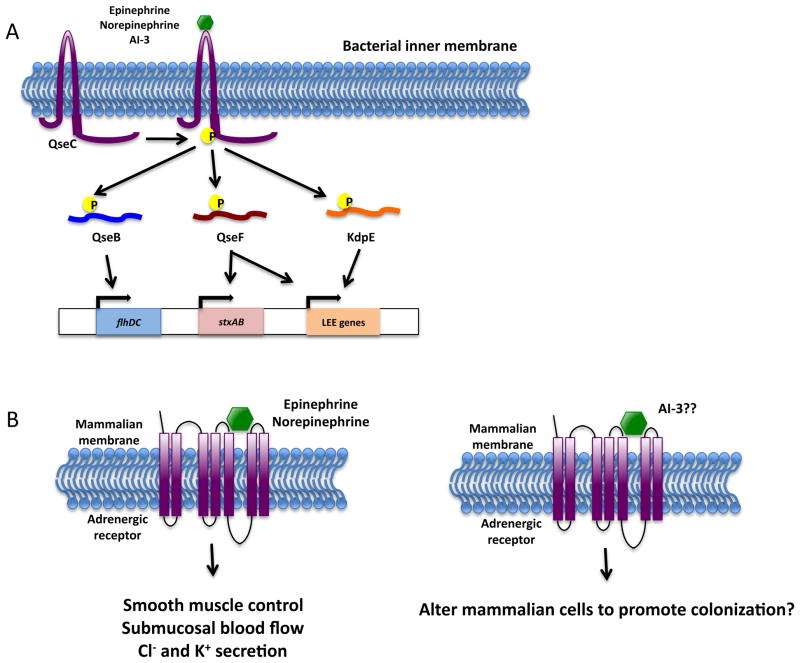
The QseBC two-component QS system can detect AI-3, epinephrine and norepinephrine44. Upon sensing its signal, the histidine sensor kinase QseC stimulates an intrinsic autophosphorylation activity that results in phosphorylation of a conserved histidine residue present in the cytoplasm (Figure 2A). The phosphate group is then transferred to a conserved aspartate residue on three response regulators: QseB, KdpE, and QseF 45. Once phosphorylated, the response regulators differentially regulate gene expression. QseB activates the master flagellar regulator genes flhDC to regulate flagella biosynthesis and motility 46. KdpE controls potassium uptake, osmolarity, and the formation of attaching and effacing (AE) lesions 45. QseF regulates the bacterial SOS stress response, as well as the formation of AE lesions 45. The genes responsible for AE lesion formation are contained within the locus of enterocyte effacement (LEE), a chromosomal pathogenicity island responsible for much of EHEC’s virulence; in addition, the LEE encodes for a type III secretion system and secreted effectors 47–49.
Figure 2.
The epinephrine/norepinephrine/AI-3 inter-kingdom signaling system. (A) The histidine sensor kinase QseC senses the mammalian hormones epinephrine and norepinephrine and the bacterial autoinducer AI-3. Activation of QseC stimulates an intrinsic autophosphorylation activity, allowing QseC to then transfer its phosphate group to one of three response regulators: QseB, QseF, and KdpE. The response regulators differentially regulate gene expression. (B) Epinephrine and norepinephrine bind to mammalian adrenergic receptors and play a role in smooth muscle control, submucosal blood flow, and chloride and potassium secretion. It remains unknown if AI-3 can bind to mammalian adrenergic receptors and alter mammalian function.
QseC is a functional analogue of an adrenergic receptor, and its activity can be blocked by the α-adrenergic antagonist phentolamine 44. Interestingly, the α2-adrenergic receptor is expressed at high levels in the proximal and transverse colon of the human intestine, the site at which EHEC and commensal flora colonize 50. It remains unknown if AI-3 can bind to mammalian adrenergic receptors (Figure 2B). If so, this could be another mechanism by which EHEC exploits the intestinal environment to promote its colonization.
The importance of QseC is highlighted by its conservation in a number of bacterial species, including Salmonella sp., Shigella flexneri, Vibrio parahaemolyticus, Yersinia pestis, and Francisella tularensis 51. In a mutant strain of S. typhimurium deficient in QseC, S. typhimurium has impaired flagellar motility and reduced invasion and survival in macrophages52. Furthermore, mice deficient in dopamine β-hydroxylase that are unable to produce epinephrine or norepinephrine have different susceptibility to Salmonella infection, indicating a role for AI-3/epinephrine/norepinephrine in inter-kingdom signaling 52.
Pathogenesis in the Lungs
P. aeruginosa is an opportunistic human pathogen that colonizes the lungs of cystic fibrosis patients. P. aeruginosa produces two AHLs as QS signaling molecules, 3-oxo-C12-HSL and C4-HSL, as discussed previously, that act on the las and rhl systems, respectively 29–31,33–35. In addition to the AHL-based QS systems, P. aeruginosa uses an autoinducer regulatory system based on 2-alkyl-4(1H)-quinolones (AQs). The AQ biosynthetic enzymes of P. aeruginosa enable the generation of a diverse range of related AQ molecules: 2-heptyl-4-hydroxyquinoline N-oxide (HQNO), 4-hydroxy-2-heptylquinoline (HHQ), and Pseudomonas quinolone signal (3,4-dihydroxy-2-heptylquinoline [PQS]) 53,54. PQS induces the expression of pqsABCDE 55 and is required for the expression of phzA1-G1, the gene responsible for pyocyanin (PCN) production. Release of PCN induces neutrophil apoptosis and epithelial cell damage 56, allowing P. aeruginosa to subvert immune surveillance and gain residency in the lungs. PQS also controls the expression of the lasB (elastase) gene 57, and a synergistic effect is achieved in the presence of both PQS and C4-HSL 58. Furthermore, PQS increases expression of the lecA gene that encodes for PA-I lectin (PA-IL) 59.
In addition to bacterial autoinducers, P. aeruginosa activates virulence genes in response to the opioid dynorphin 60. Endogenous opioids are among the first signals released during times of stress 61. Similar to EHEC’s ability to respond to the stress hormones epinephrine and norepinephrine, P. aeruginosa exploits the host during a weakened state for its own benefit. Dynorphin synergizes with PQS to increase expression of pqsABCDE and induce expression of the AQs HQNO and HHQ. Additionally, dynorphin enhances virulence against the probiotic Lactobacillus species and the nematode Caenorhabditis elegans 60.
In addition to P. aeruginosa, AQs have been identified in a number of species of Burkholderia, including B. ambifaria, B. thailandensis, and B. pseudomallei. These organisms produce 3-methyl derivatives of PHQ, HHQ and NHQ termed 4-hydroxy-3-methyl-2-alkylquinolones 62. Because AQs also act as antibiotics, as evidenced by the ability of AQs produced by P. aeruginosa to inhibit the growth of Staphylococcus aureus and Candida albicans63, it remains to be determined if Burkholderia sp. use AQs solely as antibiotics or if they also use AQs as signaling molecules.
CONCLUSIONS
Bacterial communities reside on the skin and on every mucosal surface in the human body. While many bacteria benefit the host’s well being, opportunistic and pathogenic bacteria await times of stress to exploit their host. Defining pathways unique to opportunistic and pathogenic bacteria will allow for the development of therapies that target specifically the virulence associated with these bacteria.
Since Alexander Fleming’s discovery of the antibiotic penicillin in 1928, a number of additional antibiotics have been isolated from living organisms or synthesized. Unfortunately, due to the intense selective pressure placed on bacteria by antibiotics, a number of multidrug resistant bacteria have emerged. Pathogens utilize an array of virulence strategies to promote colonization and cause disease in their hosts, including expression of adhesins, production of toxins and secretion of effectors through specialized secretion systems. An alternative strategy to antibiotics is the development of antimicrobial drugs that target microbial virulence instead of growth 64. Several such drugs are being tested in both laboratory and clinical settings against pathogens like Bacillus anthracis 65,66, S. typhimurium, EHEC, F. tularensis 51,64, S. aureus 67, and P. aeruginosa 68,69.
Because many of the QS pathways are conserved across bacterial species, a single therapy could be designed to target multiple pathogens. However, much research remains to be done to evaluate the risk of such therapies on resident flora that may also signal through these pathways. It is crucial that we seek new therapies against microbial pathogens as the incidence of resistance to current antibiotics rapidly rises. As we gain further understanding of the complex relationship among the host, symbiotic microbes, and invading pathogens, we will be better able to rationally design therapies that specifically target virulence traits incurred by pathogens.
References
- 1.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuqua C, Winans SC, Greenberg EP. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 3.Telford G, et al. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-L-homoserine lactone has immunomodulatory activity. Infect Immun. 1998;66:36–42. doi: 10.1128/iai.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci U S A. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. 1537100100 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 6.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. 1110591 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. 307/5717/1915 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. 07-PLBI-RA-0129 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGuckin MA, Eri R, Simms LA, Florin TH, Radford-Smith G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm Bowel Dis. 2009;15:100–113. doi: 10.1002/ibd.20539. [DOI] [PubMed] [Google Scholar]
- 10.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. S0140-6736(03)12489-0 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Lehrer RI, Ganz T, Selsted ME. Defensins: endogenous antibiotic peptides of animal cells. Cell. 1991;64:229–230. doi: 10.1016/0092-8674(91)90632-9. 0092-8674(91)90632-9 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Yang D, Chertov O, Oppenheim JJ. Participation of mammalian defensins and cathelicidins in anti-microbial immunity: receptors and activities of human defensins and cathelicidin (LL-37) JLeukoc Biol. 2001;69:691–697. [PubMed] [Google Scholar]
- 13.Hazlett L, Wu M. Defensins in innate immunity. Cell Tissue Res. 2010 doi: 10.1007/s00441-010-1022-4. [DOI] [PubMed] [Google Scholar]
- 14.Bowdish DM, Davidson DJ, Hancock RE. Immunomodulatory properties of defensins and cathelicidins. Curr Top Microbiol Immunol. 2006;306:27–66. doi: 10.1007/3-540-29916-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. ni888 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Lehotzky RE, et al. Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc Natl Acad Sci U S A. 2010;107:7722–7727. doi: 10.1073/pnas.0909449107. 0909449107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. 313/5790/1126 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schauber J, et al. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52:735–741. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carey CM, Kostrzynska M, Ojha S, Thompson S. The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J Microbiol Methods. 2008;73:125–132. doi: 10.1016/j.mimet.2008.01.014. S0167-7012(08)00034-1 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Claesson MJ, et al. Multireplicon genome architecture of Lactobacillus salivarius. Proc Natl Acad Sci U S A. 2006;103:6718–6723. doi: 10.1073/pnas.0511060103. 0511060103 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunne C, et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 2001;73:386S–392S. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- 22.Parker CT, Sperandio V. Cell-to-cell signalling during pathogenesis. Cell Microbiol. 2009;11:363–369. doi: 10.1111/j.1462-5822.2008.01272.x. CMI1272 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. mmi2532 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. 0092-8674(83)90063-6 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruby EG. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 27.Devine JH, Shadel GS, Baldwin TO. Identification of the operator of the lux regulon from the Vibrio fischeri strainATCC7744. Proc Natl Acad Sci U S A. 1989;86:5688–5692. doi: 10.1073/pnas.86.15.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relationships. Infect Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 31.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci U S A. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stintzi A, Evans K, Meyer JM, Poole K. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol Lett. 1998;166:341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x. S0378109798003528 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Latifi A, et al. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 34.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winson MK, et al. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J Bacteriol. 2001;183:5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes DT, et al. Chemical sensing in mammalian host-bacterial commensal associations. Proc Natl Acad Sci U S A. 2010;107:9831–9836. doi: 10.1073/pnas.1002551107. 1002551107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. nrmicro1836 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walters M, Sircili MP, Sperandio V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J Bacteriol. 2006;188:5668–5681. doi: 10.1128/JB.00648-06. 188/16/5668 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eldrup E, Richter EA. DOPA, dopamine, and DOPAC concentrations in the rat gastrointestinal tract decrease during fasting. Am J Physiol Endocrinol Metab. 2000;279:E815–822. doi: 10.1152/ajpendo.2000.279.4.E815. [DOI] [PubMed] [Google Scholar]
- 43.Horger S, Schultheiss G, Diener M. Segment-specific effects of epinephrine on ion transport in the colon of the rat. Am J Physiol. 1998;275:G1367–1376. doi: 10.1152/ajpgi.1998.275.6.G1367. [DOI] [PubMed] [Google Scholar]
- 44.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. 0604343103 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC) PLoS Pathog. 2009;5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clarke MB, Sperandio V. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2005;57:1734–1749. doi: 10.1111/j.1365-2958.2005.04792.x. MMI4792 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Elliott SJ, Yu J, Kaper JB. The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect Immun. 1999;67:4260–4263. doi: 10.1128/iai.67.8.4260-4263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarvis KG, et al. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci U S A. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. 1473 [pii] [DOI] [PubMed] [Google Scholar]
- 50.Valet P, et al. Characterization and distribution of alpha 2-adrenergic receptors in the human intestinal mucosa. J Clin Invest. 1993;91:2049–2057. doi: 10.1172/JCI116427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasko DA, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. 321/5892/1078 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreira CG, Weinshenker D, Sperandio V. QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. InfectImmun. 2010;78:914–926. doi: 10.1128/IAI.01038-09. IAI.01038-09 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lepine F, Deziel E, Milot S, Rahme LG. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim Biophys Acta. 2003;1622:36–41. doi: 10.1016/s0304-4165(03)00103-x. S030441650300103X [pii] [DOI] [PubMed] [Google Scholar]
- 54.Lepine F, Milot S, Deziel E, He J, Rahme LG. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom. 2004;15:862–869. doi: 10.1016/j.jasms.2004.02.012. S1044030504001680 [pii] [DOI] [PubMed] [Google Scholar]
- 55.Wade DS, et al. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005;187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. 187/13/4372 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10:599–606. doi: 10.1016/j.molmed.2004.10.002. S1471-4914(04)00260-6 [pii] [DOI] [PubMed] [Google Scholar]
- 57.Pesci EC, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diggle SP, et al. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. 3672 [pii] [DOI] [PubMed] [Google Scholar]
- 60.Zaborina O, et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 2007;3:e35. doi: 10.1371/journal.ppat.0030035. 06-PLPA-RA-0373R3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peterson PK, Molitor TW, Chao CC. The opioid-cytokine connection. J Neuroimmunol. 1998;83:63–69. doi: 10.1016/s0165-5728(97)00222-1. [DOI] [PubMed] [Google Scholar]
- 62.Vial L, et al. Burkholderia pseudomallei, B. thailandensis, and B. ambifaria produce 4-hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J Bacteriol. 2008;190:5339–5352. doi: 10.1128/JB.00400-08. JB.00400-08 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother. 1992;30:615–623. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- 64.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. nrd3013 [pii] [DOI] [PubMed] [Google Scholar]
- 65.Karginov VA, Nestorovich EM, Moayeri M, Leppla SH, Bezrukov SM. Blocking anthrax lethal toxin at the protective antigen channel by using structure-inspired drug design. Proc Natl Acad Sci U S A. 2005;102:15075–15080. doi: 10.1073/pnas.0507488102. 0507488102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shoop WL, et al. Anthrax lethal factor inhibition. Proc Natl Acad Sci U S A. 2005;102:7958–7963. doi: 10.1073/pnas.0502159102. 0502159102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kiran MD, et al. Discovery of a quorum-sensing inhibitor of drug-resistant staphylococcal infections by structure-based virtual screening. Mol Pharmacol. 2008;73:1578–1586. doi: 10.1124/mol.107.044164. mol.107.044164 [pii] [DOI] [PubMed] [Google Scholar]
- 68.Lesic B, et al. Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 2007;3:1229–1239. doi: 10.1371/journal.ppat.0030126. 07-PLPA-RA-0204 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muh U, et al. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob Agents Chemother. 2006;50:3674–3679. doi: 10.1128/AAC.00665-06. AAC.00665-06 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]