Abstract
Animal embryos have diverse anatomy and vary greatly in size. It is therefore remarkable that a common signaling pathway – BMP signaling – controls development of the dorsoventral (DV) axis throughout the Bilateria [1-8]. In vertebrates, spatially opposed expression of the BMP-family signaling proteins Bmp4 and Admp (anti-dorsalizing morphogenetic protein) can promote restoration of DV pattern following tissue removal [9-11]. bmp4 orthologs have been identified in all three groups of the Bilateria (deuterostomes, ecdysozoans, and lophotrochozoans) [12]. By contrast, the absence of admp orthologs in ecdysozoans such as Drosophila and C. elegans has suggested that a DV regulatory circuit of oppositely expressed bmp4 and admp genes represents an innovation specific to deuterostomes. Here we describe the existence of spatially opposed bmp and admp expression in a protostome. An admp ortholog (Smed-admp) is expressed at the ventral pole and laterally in adult Schmidtea mediterranea planarians, spatially opposing the dorsal-pole domain of Smed-bmp4 expression. Smed-admp is required for planarian regeneration following parasagittal amputation. Furthermore, Smed-admp promotes Smed-bmp4 expression and Smed-bmp4 inhibits Smed-admp expression, generating a regulatory circuit that buffers against perturbations of Bmp signaling. These results suggest that a Bmp/Admp regulatory circuit is a central feature of the Bilateria, used broadly for the establishment, maintenance, and regeneration of the DV axis.
Results and Discussion
Spatially opposed expression of Bmp and Admp genes in adult planarians
Planarians are flatworms famous for their regenerative capacities. The ability of planarians to regenerate entire adult animals from small tissue fragments makes them well suited for study of body axis polarization and patterning [13]. Furthermore, their phylogenetic position as a member of the protostome superphylum the Lophotrochozoa makes them ideal for identifying features that are conserved across the Bilateria. Planarians utilize a dorsally expressed bmp4 ortholog, Smed-bmp4 (in short, bmp4), to maintain and regenerate the DV axis [2-4]. We isolated a putative admp ortholog in the planarian Schmidtea mediterranea that is to our knowledge the first characterized in a protostome [14-16] (Figure S1 and see functional data below). We cloned two highly similar admp sequences (Smed-admp-1a and Smed-admp-1b, see Figure S1 and experimental procedures for details); it is unknown whether these sequences reflect the existence of distinct admp alleles or of highly similar admp paralogs. We refer to a single gene in this text as Smed-admp (in short, admp).
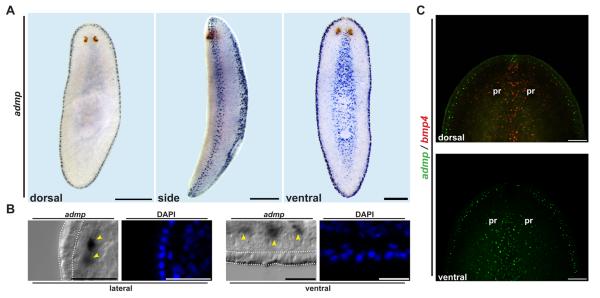
admp expression was detected in sub-epidermal cells on the ventral animal midline and around lateral animal edges at the dorsal/ventral boundary (Figures 1A and 1B). These ventral and lateral domains spatially oppose the bmp4 expression domain on the dorsal midline [2, 3]. Double-labeling with admp and bmp4 RNA probes revealed that expression of these genes does not detectably overlap (Figure 1C). admp expression opposing bmp4 expression in planarians is noteworthy, as it provides the first example of spatially opposed bmp and admp expression outside of the deuterostome lineage. Following head and tail amputation, lateral admp expression first appeared at wound sites by 48h whereas ventral admp expression decreased at 24h, 48h, and 60h before increasing again at 72h (Figure S2A). These data indicate that ventral admp expression is regulated following transverse amputation, possibly by wound-induced factors. admp expression was not detected dorsally at any point during regeneration (Figure S2A). Together, admp and bmp4 form complementary expression domains that identify the dorsal and ventral midlines as well as the lateral, dorsal-ventral boundary of animals.
Figure 1.
Smed-admp is expressed ventrally and laterally. (A) in situ hybridization with Smed-admp RNA probe displayed ventral and lateral expression. (B) Transverse sections (20 micron), differential interference contrast (DIC) images: admp-expressing cells were subepidermal (yellow arrowheads). White lines: epidermis. (C) Wild-type animals double-labeled with Smed-admp (green) and Smed-bmp4 (red) RNA probes. Pr: photoreceptors. Bars: 200 microns for (A), (C); 20 microns for (B). Anterior, up.
Smed-admp is required for lateral planarian regeneration
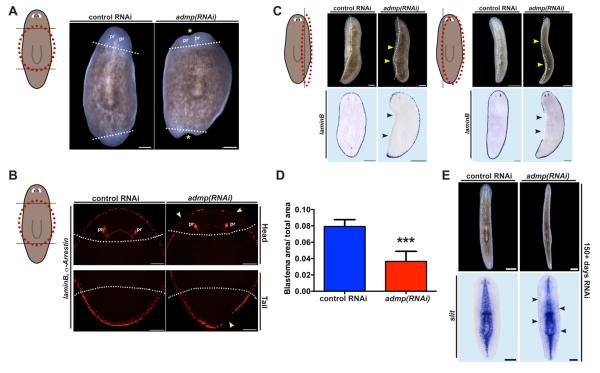
To investigate the role of admp in regeneration, we inhibited admp expression with RNA interference (RNAi) and amputated animal heads and tails. Planarian regeneration involves new tissue outgrowth at wound sites called a blastema [13]. admp(RNAi) fragments displayed regeneration defects including indented head and tail blastemas, a hallmark phenotype of planarian Bmp-pathway dysfunction [2, 3], as well as uncoordinated movement (Figure 2A, and Movies S1 and S2). in situ hybridization with a marker for lateral-edge cells identified defects in regeneration of lateral DV boundary tissue (Figure 2B). We conclude that admp is required for the regeneration of tissues at lateral animal edges, at the midpoint between dorsal and ventral poles, following transverse amputation.
Figure 2.
Smed-admp is required for planarian regeneration. (A) Transversely amputated admp(RNAi) animals displayed aberrant midline regeneration (yellow asterisks, n=14/20). Pr: photoreceptors. Bars: 200 microns. (B) Transversely amputated Smed-admp(RNAi) animals failed to regenerate DV boundary, laminB+, cells (yellow arrowheads, n=7/7). Scale bars, 200 microns. (C) admp(RNAi) animals failed to regenerate a missing side (yellow arrowheads, n=22/26 thin and 19/19 thick fragments) and lateral laminB+ tissue (black arrowhead, n=13/16 thin and 19/19 thick fragments) following parasagittal amputation. Bars: top, 500 microns; bottom, 200 microns. (D) Blastema size: unpigmented tissue at amputated sides (thick fragments), from anterior pharynx tip to tail, divided by worm area (difference with control was significant, p=0.0006, unpaired t-test). (E) Non-amputated admp(RNAi) animals became thinner than control animals following 155 days of RNAi and aberrantly expressed the ventral midline marker slit [31] at lateral positions following 163 days of RNAi (n=7/7, black arrowheads). RNAi of admp was shown to be effective and specific (see Figures S2B and S2C). White lines: approximate blastema boundary. Dorsal view, anterior up.
Bmp signaling is crucial for lateral planarian regeneration following sagittal amputations [2, 3]. Parasagittal amputation produces two left-right asymmetric fragments: a thin fragment that must regenerate an appropriately sized bmp4 expression domain de novo, and a thick fragment that must reposition and rescale its bmp4 expression domain to accommodate new animal dimensions. Parasagittal amputations therefore present a stringent test of establishment and scaling of DV as well as medial-lateral (ML) pattern. admp(RNAi) thin fragments were able to regenerate some structures along the anteroposterior (AP) axis within pre-existing tissue (photoreceptors and pharynx); however, they failed to regenerate a new side and corresponding lateral marker expression (Figure 2C). admp(RNAi) thick fragments also failed to regenerate a side (Figures 2C, 2D, and S2D). Furthermore, non-amputated admp(RNAi) animals displayed aberrant body dimensions and ML marker expression following several months of admp inhibition (Figures 2E, S3, and Movie S3), suggesting that admp is crucial for maintaining body form and proper ML pattern during animal homeostasis. We conclude that admp is required for lateral planarian regeneration and ML pattern maintenance.
Smed-admp promotes Smed-bmp4 expression
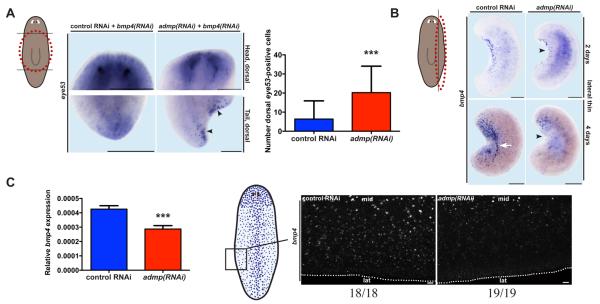
The indented head and tail blastemas and the failed lateral regeneration in admp(RNAi) animals are consistent with a defect in Bmp signaling [2, 3]. bmp4 promotes dorsal tissue maintenance and regeneration; we therefore investigated the role of admp in DV patterning. Whereas animals inhibited for admp expression alone did not show dorsal expansion of ventral markers, admp(RNAi) animals exposed to a low dose of bmp4 dsRNA became more ventralized near wound sites than did control animals exposed to the same bmp4 dsRNA dose (Figure 3A). These results indicate that animals depleted of admp activity become hypersensitive to small decreases in Bmp signaling level during DV axis regeneration.
Figure 3.
Smed-admp inhibits ventral fates and is required for normal Smed-bmp4 expression. (A) Smed-bmp4 dsRNA addition in Smed-admp(RNAi) animals caused ectopic dorsal eye53 expression (black arrowheads). Out of focus signal in control animals is from ventral cells. Difference in dorsal eye53-expressing cell numbers was significant, p<0.0001, unpaired t-test. (B) admp(RNAi) thin fragments had reduced Smed-bmp4 expression (black arrowheads, n=5/6 and n=9/15 at 2 and 4 days, respectively) and failed to reposition Smed-bmp4 expression as in control animals (white arrow). (C) Left, bmp4 expression depiction. Eight micron optical sections (identical exposures) from left, post-pharyngeal regions of intact admp(RNAi) and control RNAi animals. admp(RNAi) animals had reduced bmp4 expression (37/37 animals blindly scored correctly, three independent experiments). mid: medial dorsal, lat: lateral dorsal. Right, bmp4 expression was reduced in intact admp(RNAi) animals (by quantitative RT-PCR). Difference was significant, p<0.0001, paired t-test. RNAi of bmp4 was shown to be effective (see Figure S4B). White lines: approximate lateral animal edge. Bars: 200 microns for (A), (B); 20 microns for (C). Dorsal view, anterior up.
We next assessed whether admp influences bmp4 expression. Thin fragments produced by parasagittal amputation must re-express bmp4 during regeneration. admp(RNAi) thin fragments displayed reduced bmp4 expression and this expression domain did not reposition to reflect a new dorsal midline (Figure 3B). These results indicate that admp promotes bmp4 expression and controls the positioning of bmp4 expression during regeneration of left-right asymmetric fragments. Whether Admp signaling regulates bmp4 expression by direct action or through some other mechanism is unknown.
In addition to regenerating, planarians undergo extensive tissue turnover and growth as adults - processes that also require patterning genes for instructing new cell identities [17-19]. Consequently, if admp promotes bmp4 expression, non-amputated admp(RNAi) animals should display reduced bmp4 expression. Quantitative RT-PCR confirmed that bmp4 expression was reduced in intact admp(RNAi) animals (Figure 3C). This decrease in bmp4 expression was particularly apparent in cells more distal from the dorsal midline (Figure 3C). Together these data indicate that Admp signaling is required to maintain the appropriate level and broad spatial distribution of bmp4 expression during adult tissue maintenance and growth.
Smed-bmp4 inhibits Smed-admp expression
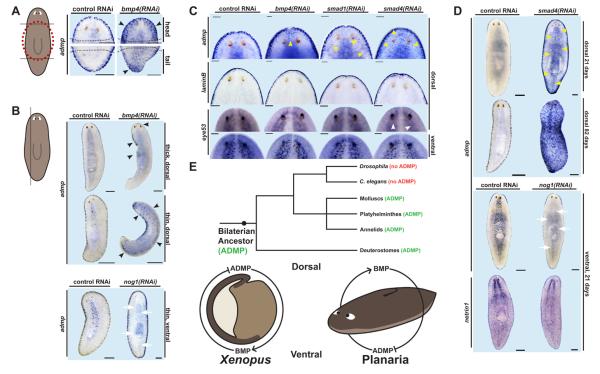
To determine whether admp is regulated by Bmp4 signaling, we examined the effect of Bmp pathway inhibition on admp expression. In both transversely and parasagittally amputated bmp4(RNAi) animals, admp expression was increased and expanded dorsally (Figures 4A and B). To conversely test whether an increase in Bmp signaling leads to a reduction in admp expression, we inhibited a ventrally expressed planarian noggin homolog (Smed-nog1 or nog1 in short) [3]. Noggins are well-characterized inhibitors of Bmp signaling [20]. Parasagittally amputated nog1(RNAi) animals indeed displayed a marked decrease in ventral admp expression (Figure 4B). Together these results indicate that admp expression is negatively regulated by Bmp signaling.
Figure 4.
admp expression is negatively regulated by Bmp signaling. (A) Transversely amputated bmp4(RNAi) animals had ectopic, dorsal admp expression (black arrowheads, n=14/14) in pre-existing tissue. 19 days of regeneration, dorsal view. (B) Parasagittally amputated bmp4(RNAi) animals had ectopic dorsal admp expression (Black arrowheads, n=5/5) in pre-existing tissue; parasagittally amputated Smed-nog1(RNAi) animals had reduced ventral admp expression (White arrows, n=20/20). 14 and 19 days of regeneration for bmp4(RNAi) and Smed-nog1(RNAi) animals, respectively. (C) Non-amputated animals inhibited for Bmp pathway components displayed ectopic dorsal admp expression after 21 days of RNAi (yellow arrowheads, n > 9/9 for each condition). Weak dorsal expression of the ventral marker eye53 was detected in Smed-smad4(RNAi) animals (white arrowheads, n=5/11). Ventral and lateral marker expression was otherwise unaffected. (D) Non-amputated Smed-smad4(RNAi) animals displayed broad dorsal admp expression after 21 days of RNAi (yellow arrowheads, n=23/23) and ubiquitous admp expression after 82 days of RNAi (n=5/5). Non-amputated Smed-nog1(RNAi) animals displayed reduced ventral admp expression (white arrows, n=8/8) but normal netrin1 expression. Dotted lines: blastema boundary. (E) Top, phylogenetic diagram of bilaterians annotated with the existence of putative admp orthologs. Bottom, schematic of proposed conserved Bmp-Admp circuit in Xenopus embryos (left) and planarians (right). RNAi of smad1, smad4, and nog1 was shown to be effective (See Figure S4B). Bars: 100 microns for (A), (C); 200 microns for (B), (D). Anterior, up.
We next investigated whether the change in admp expression observed in bmp4(RNAi) animals reflected failure of specific regulation of admp or was the simple consequence of ventralization. Following Bmp pathway inhibition, we compared expression of admp to genes with similar ventral or lateral expression domains. Whereas regeneration occurs quickly (within days), intact non-amputated animals inhibited for bmp4 or the Bmp effectors smad1 or smad4 gradually become ventralized over a period of weeks [2, 3]. This slow transformation allows for greater temporal resolution in assessing the changes in gene expression that occur following Bmp signaling loss. After three weeks of RNAi, intact bmp4(RNAi), smad1(RNAi), and smad4(RNAi) animals all displayed dorsal expression of admp, despite little to no expansion in the expression of other tested genes (Figure 4C). Strikingly, smad4 inhibition resulted in broad dorsal expansion of admp expression after three weeks of RNAi and ubiquitous DV expression of admp after 82 days of RNAi (Figure 4D). In both of these cases, inhibition of smad4 resulted in more extensive dorsal expression of admp than did inhibition of either smad1 or bmp4 (Figure 4C and Figure S4C). Because Smad4 proteins are required for all forms of TGFβ superfamily signaling [21], these results suggest that admp expression may also be regulated by non-Bmp TGFβ signaling. In contrast to Bmp pathway RNAi, three weeks of nog1 RNAi in intact animals caused a potent reduction in the ventral admp expression domain without affecting other ventral markers (Figure 4D and Figure S4A). These data indicate that admp expression is negatively regulated by Bmp signaling during adult homeostasis and growth. Together with the observation that admp promotes bmp4 expression, we propose that inhibition of admp expression by Bmp4 produces a feedback circuit that buffers against fluctuations in Bmp signaling levels, conferring robustness in DV and ML patterning. This model is supported by the observation that admp depletion results in animals that are hypersensitive to bmp4 inhibition. Although planarians lack identified orthologs of the Bmp modulators chordin [22] and sizzled [23], the presence of a homolog of the Bmp pseudoreceptor bambi [24] (Figure S4D), as well as a greatly expanded family of noggin genes [25] and the ability of Admp to regulate nog1 expression (Figures S2D and S3D), suggests that additional regulatory mechanisms likely function to fine tune the activity of this central Bmp/Admp circuit.
Admp orthologs are widespread among protostomes
Because Smed-admp represents the first admp ortholog characterized in a protostome, we searched the genomes of other lophotrochozoans to determine whether admp orthologs are widespread in protostomes. Indeed, predicted admp orthologs were identified in the genomes of the snail Lottia gigantis, the leech Helobdella robusta, and the polychaete annelid Capitella teleta (Figure S1C). The presence of putative admp orthologs in these species, coupled with the expression pattern and functional properties of Smed-admp, suggest that a Bmp/Admp regulatory circuit is an ancestral and central feature of the DV axis (Figure 4E). This model predicts that an admp gene was present in the ancestor of C. elegans and Drosophila but subsequently lost in the evolution of these species; consequently the potential widespread significance of admp genes for the DV axis of Bilaterians was previously unknown.
Conclusions
Development proceeds in a remarkably reliable fashion despite the myriad forms that embryos assume and widely varying conditions they encounter [26]. Robust patterning of the DV axis is a crucial component of this process. The DV axis can scale during growth and, in some cases, is capable of restoration following surgical manipulation [9, 27-29]. How Bmp signaling is able to generate consistent DV pattern in diverse species and respond appropriately to perturbation has been a central mystery in developmental biology. Our data demonstrate that a molecular circuit of spatially opposed bmp4 and admp expression is crucial for the regeneration and maintenance of both DV and ML pattern in planarians. This circuit may function to buffer against changes in Bmp level that naturally arise from differences in patterned tissue size, the genotype of individuals, or environmental influences encountered. The requirement of a Bmp/Admp circuit for both planarian regeneration and deuterostome self-regulation [10, 11, 16] suggests that restoration of the DV axis in embryonic regulation and adult axial regeneration share mechanistic features. The presence of spatially opposed expression of bmp and admp in planarians (lophotrochozoans) and in several deuterostomes [15, 16, 30], suggests that a Bmp/Admp circuit is a widespread feature of the DV axis that emerged concurrent with the first bilaterally symmetric animals.
Supplementary Material
Acknowledgements
We thank the Reddien Lab for discussions and support and Dr. Irving Wang for his artistic contributions. P.W.R. is an early career scientist of the Howard Hughes Medical Institute. We acknowledge support by NIH R01GM080639, ACS RSG-07-180-01-DDC, the Keck Foundation, and a Thomas D. and Virginia W. Cabot career development professorship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information includes Supplemental Experimental Procedures, four figures and three movies.
References
- 1.Lowe CJ, Terasaki M, Wu M, Freeman RM, Jr., Runft L, Kwan K, Haigo S, Aronowicz J, Lander E, Gruber C, et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddien PW, Bermange AL, Kicza AM, Sánchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- 3.Molina MD, Saló E, Cebrià F. The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev Biol. 2007;311:79–94. doi: 10.1016/j.ydbio.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Orii H, Watanabe K. Bone morphogenetic protein is required for dorso-ventral patterning in the planarian Dugesia japonica. Dev Growth Differ. 2007;49:345–349. doi: 10.1111/j.1440-169X.2007.00931.x. [DOI] [PubMed] [Google Scholar]
- 5.De Robertis EM, Sasai Y. A common plan for dorsoventral patterning in Bilateria. Nature. 1996;380:37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson EL, Anderson KV. Decapentaplegic acts as a morphogen to organize dorsal-ventral pattern in the Drosophila embryo. Cell. 1992;71:451–461. doi: 10.1016/0092-8674(92)90514-d. [DOI] [PubMed] [Google Scholar]
- 7.Hammerschmidt M, Serbedzija GN, McMahon AP. Genetic analysis of dorsoventral pattern formation in the zebrafish: requirement of a BMP-like ventralizing activity and its dorsal repressor. Genes Dev. 1996;10:2452–2461. doi: 10.1101/gad.10.19.2452. [DOI] [PubMed] [Google Scholar]
- 8.Dale L, Howes G, Price BM, Smith JC. Bone morphogenetic protein 4: a ventralizing factor in early Xenopus development. Development. 1992;115:573–585. doi: 10.1242/dev.115.2.573. [DOI] [PubMed] [Google Scholar]
- 9.Spemann H. Embryonic Development and Induction. 1938 [Google Scholar]
- 10.Reversade B, De Robertis EM. Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogenetic field. Cell. 2005;123:1147–1160. doi: 10.1016/j.cell.2005.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Zvi D, Shilo BZ, Fainsod A, Barkai N. Scaling of the BMP activation gradient in Xenopus embryos. Nature. 2008;453:1205–1211. doi: 10.1038/nature07059. [DOI] [PubMed] [Google Scholar]
- 12.Niehrs C. On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development. 137:845–857. doi: 10.1242/dev.039651. [DOI] [PubMed] [Google Scholar]
- 13.Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 14.Moos M, Jr., Wang S, Krinks M. Anti-dorsalizing morphogenetic protein is a novel TGF-beta homolog expressed in the Spemann organizer. Development. 1995;121:4293–4301. doi: 10.1242/dev.121.12.4293. [DOI] [PubMed] [Google Scholar]
- 15.Lele Z, Nowak M, Hammerschmidt M. Zebrafish admp is required to restrict the size of the organizer and to promote posterior and ventral development. Dev Dyn. 2001;222:681–687. doi: 10.1002/dvdy.1222. [DOI] [PubMed] [Google Scholar]
- 16.Joubin K, Stern CD. Molecular interactions continuously define the organizer during the cell movements of gastrulation. Cell. 1999;98:559–571. doi: 10.1016/s0092-8674(00)80044-6. [DOI] [PubMed] [Google Scholar]
- 17.Petersen CP, Reddien PW. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- 18.Gurley KA, Rink JC, Sánchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–1221. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- 20.Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 22.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM. Xenopus chordin: a novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HX, Ambrosio AL, Reversade B, De Robertis EM. Embryonic dorsal-ventral signaling: secreted frizzled-related proteins as inhibitors of tolloid proteinases. Cell. 2006;124:147–159. doi: 10.1016/j.cell.2005.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massague J, Niehrs C. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- 25.Molina MD, Saló E, Cebrià F. Expression pattern of the expanded noggin gene family in the planarian Schmidtea mediterranea. Gene Expr Patterns. 2009;9:246–253. doi: 10.1016/j.gep.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Waddington CH. Canalization of Development and the Inheritance of Acquired Characters. Nature. 1942;150:563–565. [Google Scholar]
- 27.Cooke J. Scale of body pattern adjusts to available cell number in amphibian embryos. Nature. 1981;290:775–778. doi: 10.1038/290775a0. [DOI] [PubMed] [Google Scholar]
- 28.De Robertis EM. Spemann's organizer and self-regulation in amphibian embryos. Nat Rev Mol Cell Biol. 2006;7:296–302. doi: 10.1038/nrm1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spratt NT, Haas H. Integrative mechanism in development of the early chick blastoderm. I. Regulative potentiality of separate parts. J. Exp. Zool. 1960:97–137. [Google Scholar]
- 30.Dosch R, Niehrs C. Requirement for anti-dorsalizing morphogenetic protein in organizer patterning. Mech Dev. 2000;90:195–203. doi: 10.1016/s0925-4773(99)00245-2. [DOI] [PubMed] [Google Scholar]
- 31.Cebrià F, Guo T, Jopek J, Newmark PA. Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev Biol. 2007;307:394–406. doi: 10.1016/j.ydbio.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.