Abstract
Utilizing a genome-wide gene expression dataset generated from Affymetrix GeneChip® Human Exon 1.0ST array, we comprehensively surveyed the role of 322 X chromosome gene expression traits on cellular sensitivity to cisplatin and carboplatin. We identified 31 and 17 X chromosome genes whose expression levels are significantly correlated (after multiple testing correction) with sensitivity to carboplatin and cisplatin, respectively, in the combined HapMap CEU and YRI populations (false discovery rate, FDR<0.05). Of those, 14 overlap for both cisplatin and carboplatin. Employing an independent gene expression quantification method, the Illumina Sentrix Human-6 Expression BeadChip, measured on the same HapMap cell lines, we found that 4 and 2 of these genes are significantly associated with carboplatin and cisplatin sensitivity respectively in both analyses. Two genes, CTPS2 and DLG3, were identified by both genome-wide gene expression analyses as correlated with cellular sensitivity to both platinating agents. The expression of DLG3 gene was also found to correlate with cellular sensitivity to platinating agents in NCI60 cancer cell lines. In addition, we evaluated the role of X chromosome gene expression to the observed differences in sensitivity to the platinums between CEU and YRI derived cell lines. Of the 34 distinct genes significantly correlated with either carboplatin or cisplatin sensitivity, 14 are differentially expressed (defined as p<0.05) between CEU and YRI. Thus, sex chromosome genes play a role in cellular sensitivity to platinating agents and differences in the expression level of these genes are an important source of variation that should be included in comprehensive pharmacogenomic studies.
Keywords: platinum agents, X chromosome gene expression, population differences
Introduction
Despite the fact that gene expression has long been recognized to contribute to drug sensitivity (1), little is known regarding the expression of sex chromosome genes for their role in drug response. Furthermore, gender differences in drug sensitivity are observed both in cell lines (2) and in patients (3). Our previous studies have revealed in vitro inter-individual differences in platinum drug response, prompting us to evaluate whether sex chromosome genes may perhaps play a unique role in the transcriptome in governing inter-individual drug response. Indeed, genes on the sex chromosomes have been relatively understudied in relation to drug sensitivity (4, 5). In this study, we systematically evaluated the role of X chromosome genes in predicting platinum sensitivity.
Materials and Methods
In vitro cellular sensitivity to platinating agents
Fifty-seven unrelated HapMap CEU (Utah residents with ancestry from northern and western Europe), 59 unrelated YRI (Yoruba in Ibadan, Nigeria) and 90 unrelated ASN (Han Chinese in Beijing and Japanese in Tokyo) lymphoblastoid cell lines (LCLs) were purchased from Coriell Institute for Medical Research (Camden, NJ) (6) between 2001 and 2007. One advantage of the use of these cell lines is that extensive whole-genome genotype information is publicly available to the scientific community, which allows us to perform genotyping assays on randomly selected cell lines for randomly selected SNPs to authenticate the cell lines. This genotyping assay is routinely carried out in our Pharmacogenomic of Anticancer Agent Research (PAAR) cell line and genotyping core; furthermore, the cell lines have been the subject of extensive genetic studies (6). Cell line maintenance has been reported previously (2). Cells were treated with increasing concentrations of cisplatin or carboplatin for 48 and 72 h, respectively. Cell growth inhibition was assessed using the alamarBlueTM assay (BioSource International Inc., Camarillo, CA) (2). IC50 (the drug concentration required to inhibit 50% of cell growth) was determined by curve fitting of percent cell survival against drug concentration.
X chromosome gene expression
Our lab previously evaluated global baseline gene expression on 87 CEU and 89 YRI LCLs using the Affymetrix GeneChip® Human Exon 1.0 ST array (Affymetrix exon array) (7). We excluded from analysis all Affymetrix exon array probes that contain more than one known polymorphism to eliminate spurious associations due to hybridization artifacts (8). The Affymetrix gene expression data have been deposited into GEO (9) (Accession No: GSE9703). Three hundred and twenty two X chromosome transcript clusters/genes interrogated on the Affymetrix array and defined as expressed in LCLs (average normalized expression intensity greater than 5.34) were included in this study. In addition, baseline genome-wide gene expression data have been generated on 210 HapMap LCLs (including the CEU, YRI and ASN samples used in this study) using Illumina Sentrix Human-6 Expression BeadChip version 1 (Illumina array) (10). We downloaded these data from the Gene Expression Omnibus (GEO) database (Series Accession Number GSE6536). The probe intensities derived from 461 X chromosome genes (represented by 562 probes) were evaluated and compared to the Affymetrix exon array findings. From this comparison, we found 190 X chromosome genes are interrogated on both platforms.
Evaluating the relationship between X chromosome gene expression and platinum sensitivity
Linear regression was used to evaluate the role of X chromosome baseline gene expression levels in predicting sensitivity to carboplatin and cisplatin across populations. A schematic diagram of the study is illustrated in Figure 1. We utilized the Affymetrix exon array data for discovery and the Illumina array data to check robustness of the results. First, platinum IC50 values for all samples were log2 transformed to achieve approximate normality. Separate linear models were estimated within each population with transformed platinum IC50 values as the dependent variable and normalized expression intensities and an indicator of gender as independent variables. Subsequently, a meta-analysis was performed combining the individual p-values obtained for each X chromosome gene from the two populations to generate a cross-population p-value for each gene expression/platinum sensitivity correlation. Fisher’s method was used to combine p-values. Cisplatin and carboplatin data were analyzed separately. For multiple testing correction, FDRs were calculated using the q-value package in R (11). An FDR of < 0.05 was used as the statistical significance cutoff. The significant findings for cisplatin and carboplatin generated using the Affymetrix exon array data were compared and genes in the overlap were identified. We then evaluated, using Illumina array data, these gene candidates found in the overlap.
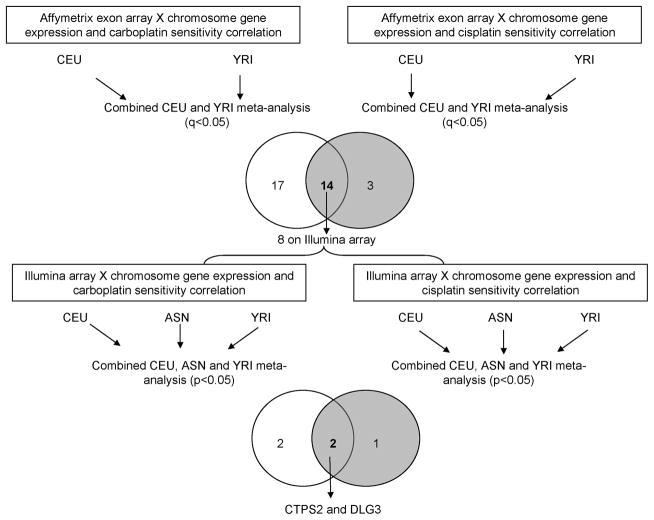
Figure 1.
Schematic diagram of the study. The top venn diagram shows the number of X chromosome genes whose expression significantly correlated with either carboplatin (left circle) or cisplatin (right circle) sensitivity (q<0.05). The number shown in the overlap region of the venn diagram represents the number of X chromosome genes whose expression significantly correlated with sensitivity to both platinums. When evaluating these 14 genes on Illumina array via meta-analysis, 4 (2+2) also correlated with carboplatin sensitivity (left circle) and 3 (1+2) correlated with cisplatin sensitivity (right circle). CEU = Caucasian; YRI = Yoruba people from Ibadan, Nigeria; ASN= Asian.
The same analysis was performed using gene expression measurements, assayed by the Illumina array. For this platform we used HapMap CEU, YRI and ASN populations. As in the Affymetrix exon array, we conducted a meta-analysis in the combined populations. P values less than 0.05 from the meta-analysis for cisplatin and carboplatin, evaluated separately, were identified.
In addition, to determine whether our results are confounded by X inactivation, we compared the expression levels of the identified genes between males and females within each population using Student’s t-tests.
Candidate gene expression evaluation in NCI-60 datasets
To explore the relevance of our findings in tumors, we examined the degree of correlation between candidate X chromosome gene expression and cellular susceptibility to cisplatin and carboplatin using NCI60 tumor cell lines. We downloaded the NCI-60 microarray expression and GI50 datasets (released in March, 2007) from the DTP/NCI Molecular Target Databases (12, 13). These datasets are comprised of gene expression data on untreated NCI-60 cell lines using different microarray platforms along with GI50 data. Linear regression was performed between the expression of a gene of interest and log10 carboplatin GI50. p<0.05 was considered statistically significant.
Regulatory variation and X chromosome gene expression
Previous studies in our laboratory have shown that platinum response-associated single nucleotide polymorphisms (SNPs) are enriched for expression quantitative trait loci (eQTLs) (14), suggesting the role of genetic variation in drug sensitivity and gene expression. Using the SCAN database (8), which hosts our genome-wide SNP and gene expression association data, we searched for evidence that eQTLs may be regulating these X chromosome genes that are significantly correlated with platinum sensitivity. The thirty-four genes (Table 1) whose expression levels are significantly correlated (q-value<0.05) with either carboplatin or cisplatin sensitivity were evaluated in the SCAN database. SNP-expression correlations that met the p<10−4 cutoff were reported. Of these, the associations between SNP genotype and platinum sensitivity were also evaluated to arrive at candidate SNPs whose genotypes met the p<10−4 threshold with both gene expression and cellular sensitivity to platinum.
Table 1.
X chromosome genes whose expression are signficantly correlated with either carboplatin or cisplatin sensitivity via a meta-analysis in the CEU and the CEU and YRI samples.
| gene name | transcript cluster ID | Affymetrix |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| combined CEU and YRI | CEU | YRI | |||||||
| meta p values | meat q values | p values | q values | ||||||
| carboplatin | cisplatin | carboplatin | cisplatin | carboplatin | cisplatin | carboplatin | cisplatin | ||
| CD99L2 * | 4025771 | 1.70E-08 | 4.20E-09 | 2.73E-06 | 9.02E-07 | 6.23E-04 | 2.37E-06 | 1.24E-06 | 7.55E-05 |
| GPR174 * | 3982612 | 4.05E-07 | 7.01E-06 | 3.25E-05 | 7.54E-04 | 1.78E-05 | 1.15E-03 | 1.22E-03 | 3.91E-04 |
| STS * | 3967689 | 6.29E-06 | 2.85E-05 | 2.69E-04 | 1.53E-03 | 8.89E-06 | 8.16E-05 | 4.50E-02 | 2.48E-02 |
| DLG3 * | 3980643 | 6.70E-06 | 5.50E-04 | 2.69E-04 | 1.31E-02 | 1.11E-03 | 7.34E-02 | 3.85E-04 | 6.88E-04 |
| MID1IP1 * | 3974098 | 2.24E-05 | 1.12E-05 | 7.19E-04 | 8.00E-04 | 1.35E-01 | 1.36E-02 | 1.16E-05 | 5.42E-05 |
| CLIC2 | 4027769 | 3.40E-05 | 4.24E-03 | 9.11E-04 | 5.06E-02 | 2.60E-05 | 2.07E-03 | 9.39E-02 | 2.38E-01 |
| GRPR | 3970024 | 1.00E-04 | 8.04E-03 | 2.30E-03 | 6.66E-02 | 1.03E-02 | 1.26E-02 | 7.67E-04 | 8.10E-02 |
| TMEM164 * | 3987029 | 1.19E-04 | 1.75E-03 | 2.39E-03 | 2.35E-02 | 1.06E-03 | 2.29E-03 | 8.92E-03 | 7.96E-02 |
| ACSL4 * | 4017810 | 3.89E-04 | 2.64E-04 | 6.94E-03 | 7.43E-03 | 4.27E-04 | 6.12E-04 | 8.08E-02 | 3.69E-02 |
| MAP3K15 * | 4001850 | 5.44E-04 | 1.39E-04 | 7.35E-03 | 5.99E-03 | 1.97E-01 | 2.74E-02 | 2.53E-04 | 4.10E-04 |
| FAM3A * | 4027387 | 5.37E-04 | 9.10E-04 | 7.35E-03 | 1.78E-02 | 1.58E-01 | 6.52E-02 | 3.11E-04 | 1.35E-03 |
| WWC3 | 3968397 | 5.49E-04 | 3.28E-02 | 7.35E-03 | 1.43E-01 | 5.21E-04 | 8.38E-02 | 9.67E-02 | 6.26E-02 |
| CLCN5 * | 3977299 | 9.02E-04 | 8.41E-04 | 1.04E-02 | 1.78E-02 | 1.57E-02 | 6.82E-03 | 5.54E-03 | 1.18E-02 |
| CDKL5 | 3970642 | 8.49E-04 | 6.05E-02 | 1.04E-02 | 1.86E-01 | 2.55E-02 | 3.04E-01 | 3.19E-03 | 3.61E-02 |
| PJA1 | 4011464 | 1.18E-03 | 6.54E-03 | 1.26E-02 | 6.52E-02 | 2.60E-04 | 2.87E-02 | 4.50E-01 | 2.81E-02 |
| CTPS2 * | 4000839 | 1.73E-03 | 1.71E-04 | 1.40E-02 | 6.13E-03 | 9.71E-02 | 4.78E-02 | 1.86E-03 | 2.94E-04 |
| ATP6AP2 * | 3974556 | 1.46E-03 | 1.07E-03 | 1.40E-02 | 1.83E-02 | 2.18E-02 | 8.03E-03 | 6.83E-03 | 1.32E-02 |
| CNKSR2 | 3971219 | 1.72E-03 | 1.95E-02 | 1.40E-02 | 1.22E-01 | 5.34E-02 | 3.36E-01 | 3.34E-03 | 8.46E-03 |
| MED12 | 3980758 | 1.65E-03 | 3.64E-02 | 1.40E-02 | 1.51E-01 | 2.44E-04 | 1.18E-02 | 6.97E-01 | 5.04E-01 |
| GDI1 | 3996404 | 1.74E-03 | 1.38E-01 | 1.40E-02 | 2.90E-01 | 3.68E-03 | 8.71E-02 | 4.92E-02 | 3.52E-01 |
| SLC9A7 | 4006841 | 2.65E-03 | 4.92E-02 | 2.03E-02 | 1.75E-01 | 1.61E-01 | 8.77E-01 | 1.80E-03 | 9.74E-03 |
| MAGEH1 | 3978760 | 3.25E-03 | 6.15E-02 | 2.38E-02 | 1.86E-01 | 4.99E-01 | 3.98E-01 | 7.32E-04 | 2.81E-02 |
| GPC4 | 4022370 | 5.00E-03 | 8.06E-03 | 3.50E-02 | 6.66E-02 | 1.56E-02 | 1.35E-02 | 3.81E-02 | 7.59E-02 |
| TSC22D3 * | 4017381 | 5.84E-03 | 1.11E-03 | 3.87E-02 | 1.83E-02 | 2.54E-02 | 2.28E-01 | 2.79E-02 | 4.79E-04 |
| EDA2R * | 4011096 | 6.02E-03 | 3.03E-03 | 3.87E-02 | 3.83E-02 | 1.12E-03 | 7.24E-04 | 6.51E-01 | 4.64E-01 |
| PIM2 | 4007617 | 6.61E-03 | 2.86E-02 | 4.08E-02 | 1.43E-01 | 8.22E-01 | 1.41E-01 | 9.91E-04 | 3.16E-02 |
| PDZD11 | 4011637 | 7.11E-03 | 1.48E-01 | 4.23E-02 | 2.97E-01 | 6.13E-02 | 3.29E-01 | 1.44E-02 | 1.03E-01 |
| Cxorf21 | 4003895 | 7.65E-03 | 2.83E-01 | 4.39E-02 | 3.88E-01 | 2.63E-03 | 8.27E-02 | 3.66E-01 | 9.73E-01 |
| OFD1 | 3969455 | 8.62E-03 | 7.52E-03 | 4.53E-02 | 6.66E-02 | 2.42E-01 | 1.50E-01 | 4.56E-03 | 6.28E-03 |
| CFP | 4007164 | 8.26E-03 | 1.56E-01 | 4.53E-02 | 2.99E-01 | 1.75E-03 | 5.47E-02 | 6.01E-01 | 6.58E-01 |
| LOC170082 | 3969396 | 8.75E-03 | 1.72E-01 | 4.53E-02 | 3.03E-01 | 6.34E-01 | 7.81E-01 | 1.77E-03 | 5.26E-02 |
| PIGA | 4000512 | 1.10E-02 | 2.76E-04 | 5.04E-02 | 7.43E-03 | 2.10E-02 | 1.79E-03 | 6.95E-02 | 1.33E-02 |
| GLA | 4015763 | 2.27E-02 | 1.52E-03 | 7.36E-02 | 2.34E-02 | 2.45E-02 | 1.67E-03 | 1.39E-01 | 9.33E-02 |
| PGK1 | 3982462 | 1.10E-01 | 1.67E-03 | 1.91E-01 | 2.35E-02 | 4.09E-02 | 2.50E-04 | 5.67E-01 | 6.89E-01 |
represents the 14 genes whose expression levels are significantly correlated with both carboplatin and cisplatin sensitivity (q-value<0.05) after a meta-analysis in the combined CEU and YRI samples, based on Affymetrix exon array data.
Bold: indicates that the gene expression level is correlated with sensitivity to both platinating agents (q-value<0.05) from a meta-analysis using the Affymetrix exon array data and is correlated with sensitivity to both platinating agents (p<0.05) from a meta-analysis (in the combined CEU, YRI and ASN data) using the Illumina array.
NA: not available.
CEU = Caucasian; YRI = Yoruba people from Ibadan, Nigeria; ASN= Asian.
Evaluating the contribution of X chromosome genes to the observed population differences in platinum sensitivity
We have previously reported significant in vitro differences among HapMap populations in cellular sensitivity to carboplatin and cisplatin (2, 15). In this study, we tested the 34 genes listed in Table 1 for differential expression between the CEU and YRI samples using a two tailed t-test. Raw p<0.05 was used as cutoff. In addition, we compared the minor allele frequency (MAF) in the two populations of any eQTLs for the differentially expressed X chromosome genes.
Results
Inter-individual cellular sensitivity differences to platinum agents
Large inter-individual variations in sensitivity to platinum agents were observed among unrelated samples obtained from the three HapMap populations. As shown previously, there were 32.5-, 17.4- and 19.9-fold variation for cisplatin IC50; and 8.8-, 12- and 5.2-fold variation for carboplatin IC50 in the CEU, YRI and ASN samples, respectively (2, 15).
X chromosome gene expression levels correlate with cellular sensitivity to platinum agents
Through a meta-analysis in the combined CEU and YRI samples (q-value<0.05) using the gene expression dataset from the Affymetrix exon array, we identified 31 and 17 X chromosome genes whose expression levels are significantly correlated with cellular sensitivity to carboplatin and cisplatin, respectively (Table 1). Fourteen of these genes are significantly correlated with both carboplatin and cisplatin sensitivity. For confirmation, using an independent gene expression quantification method (Illumina array), 8 of the 14 genes were found to be interrogated on the Illumina array. Four (CD99L2, STS, DLG3, and CTPS2) and three (DLG3, FAM3A, and CTPS2) of the 8 were found to correlate with carboplatin and cisplatin sensitivity, respectively, using a meta-analysis in the combined CEU, YRI and ASN samples (p<0.05). Two X chromosome genes, CTPS2 and DLG3, were found to correlate with both carboplatin and cisplatin sensitivity using both Affymetrix exon array and Illumina array data. Specifically, the increased expression of both genes is correlated with increased sensitivity to the two platinum agents studied (Figure 2).
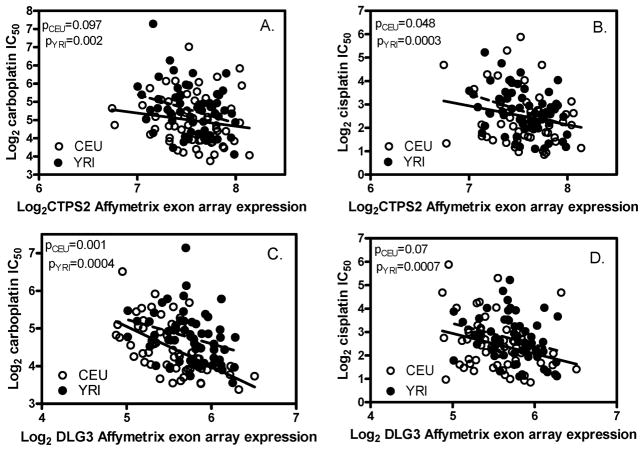
Figure 2.
The correlation between the expression of CTPS2 or DLG3 and cellular sensitivity to platinums in CEU and YRI. Affymetrix exon array expression data are presented. CTPS2 (A, B) and DLG3 (C, D) expression levels are significantly correlated with both carboplatin (A, C) and cisplatin (B, D) sensitivity.
Furthermore, we examined the correlation between the expression of CTPS2 and DLG3, and cisplatin and carboplatin GI50s in the NCI60 cancer cell lines. DLG3 expression was found to significantly correlate with carboplatin GI50 in all 60 cancer cell lines (p<0.05). This finding was confirmed by multiple gene expression arrays including Affymetrix U95 and U133.
Genetic variation may be regulating X chromosome genes whose expression correlates with platinum sensitivity
Using the SCAN database, we identified 36 and 57 eQTLs (at p<10−4) in CEU and YRI respectively, which predict the expression of the X chromosome genes that have been shown to be significantly associated (q-value<0.05) with either of the platinating agents (from Table 1). These SNPs, which are associated with baseline gene expression of 11 and 15 such X chromosome genes in CEU and YRI respectively, have genotypes that also associated with carboplatin or cisplatin sensitivity (meeting the p<10−4 cutoff). In CEU, 6 such eQTLs (rs11001716, rs11001720, rs7087332, rs7099295, rs4503472 and rs1865644), located in the intronic region of the C10orf11 gene on chromosome 10, are associated with carboplatin sensitivity and predict two X chromosome genes (GRPR and CDKL5) with expression levels that themselves show significant platinum associations (q-value<0.05). In YRI, 3 such eQTLs are associated with both carboplatin and cisplatin sensitivity and are targeting X chromosome genes (rs1595273 and CD99L2; rs9686286 and GLA and PIGA; rs9749439 and PIGA) with expression levels showing significant platinum associations (q-value<0.05). These findings provide strong evidence that the effect of the expression of X chromosome genes on platinum sensitivity may in part be mediated by genetic variants.
X chromosome gene expression patterns may in part explain population differences in response to platinating agents
Population differences, observed in vitro, in susceptibility to carboplatin and cisplatin were previously reported in HapMap YRI and ASN samples (15) as well as between CEU and YRI for carboplatin (2). After establishing the role of X chromosome gene expression in platinum sensitivity, we further evaluated whether sex chromosome gene expression patterns, especially on the X chromosome, contribute to the observed population differences in platinum sensitivity. Of the 34 genes (Table 1) whose expression levels are significantly correlated with sensitivity to either platinum (q-value<0.05), 14 genes showed evidence of differential expression between the CEU and YRI samples (p<0.05. Affymetrix exon array data). Nine (CD99L2, CLIC2, DLG3, CNKSR2, CXorf21, PIM2, ATP6AP2, GPR174, and MID1IP1) and seven (CD99L2, CLIC2, DLG3, PIM2, ATP6AP2, GPR174, and MID1IP1) of these 14 genes also meet the p<0.05 cutoff with carboplatin and cisplatin sensitivity, respectively, in the separate CEU and YRI populations with concordant effect direction.
Furthermore, we identified 24 eQTLs that predict (at the p<10−4 level) the expression of at least one of the differentially expressed X chromosome genes found to be important in platinum sensitivity (q-value<0.05). The majority of these SNPs show variation in allele frequencies in the CEU and YRI populations. As an example, one gene, CD99L2, is differentially expressed between the CEU and YRI samples (p=0.01). This gene’s expression is significantly correlated with both carboplatin and cisplatin sensitivity in both populations (p =0.002 and <0.0001 for carboplatin in CEU and YRI, respectively; p<0.0001 and =0.0003 for cisplatin in CEU and YRI, respectively; Figure 3A, 3B). Furthermore, an eQTL (rs1595273), whose genotype meets p<10−4 with the expression of CD99L2 is also associated with carboplatin and cisplatin sensitivity in YRI (p=7×10−5 and 1×10−4; Figure 3C, 3D). Interestingly, consistent with the observed difference in expression between the two populations, rs1595273 has MAF of 0.43 and 0.31 in CEU and YRI, respectively.
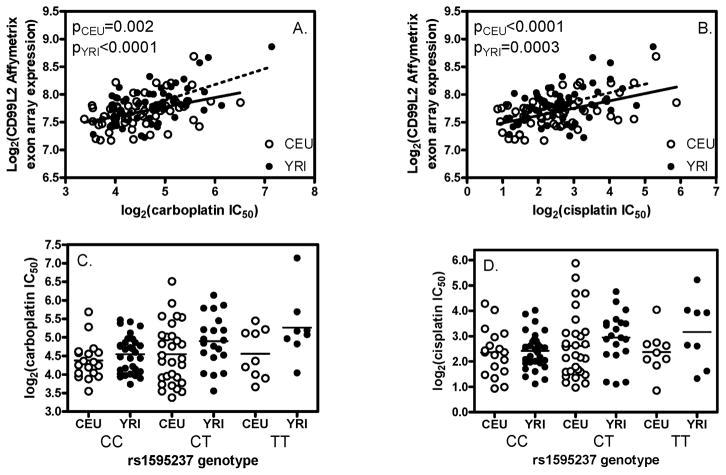
Figure 3.
The relationship between the expression of CD99L2, rs1595237 genotype and cellular sensitivity to platinums in CEU and YRI. Affymetrix exon array expression data are presented. CD99L2 expression level is significantly correlated with carboplatin (A) and cisplatin (B) in both CEU and YRI populations. rs1595237 genotype, which predicts the expression of CD99L2, meets the p<10−4 cutoff with carboplatin (C) and cisplatin (D) sensitivity in YRI (p=7×10−5 and 1×10−4).
A comparison of the expression levels between males and females in CTPS2, DLG3 and CD99L2 shows that the correlation between expression and drug sensitivity is not driven by the phenomenon of X inactivation (P>0.2 between females and males).
Discussion
Studies involving platinating agent sensitivity with X chromosome genes have been conducted on a single gene, or gene product. To date, there have been no comprehensive studies to explore the importance of X chromosome genes in predicting platinum sensitivity. Taking advantage of genome-wide gene expression array datasets, our study comprehensively explored the role of the expression of X chromosome genes in predicting platinum sensitivity. We identified 31 and 17 X chromosome genes whose expression levels are significantly correlated with carboplatin and cisplatin sensitivity in the combined HapMap CEU and YRI populations, 14 of which were found to be in the overlap between the two platinating agents evaluated in this study. Utilizing an independent expression quantification method (Illumina array), measured on the same HapMap cell lines, we found overlap with 4 and 2 of these genes, respectively. Two X chromosome genes were identified by both genome-wide gene expression analyses to be correlated with sensitivity to both platinating agents.
One interesting gene from our findings, CTPS2, encodes cytidine 5′-triphosphate (CTP) synthase II (16), the rate-limiting enzyme in the synthesis of cytosine nucleotides, which plays an important role in various metabolic processes and provide the precursors necessary for the synthesis of RNA and DNA. It has been shown that cancer cells that exhibit increased cell proliferation also exhibit an increased activity of CTPS2. Furthermore, our laboratory has reported a significant positive correlation between cellular sensitivity to platinating agents and cellular growth rate (17). Thus, it is plausible that increased CTPS2 gene expression leads to increased cellular proliferation, which in turn leads to increased platinum sensitivity.
Another novel gene, DLG3, encoding human neuroendocrine Dlg (NE-Dlg), is located within the 1.8-Mb dystonia-parkinsonism region at Xq13.1 (18). It also directly interacts with the colorectal tumor suppressor adenomatous polyposis coli (APC) and negatively regulates cell proliferation (19). In fact, overexpression of DLG3 in proliferating cells induces growth suppression and impairment of cell adhesive ability. Indeed, we observed significant correlation between DLG3 expression and platinum sensitivity in a set of 60 NCI60 cancer cell lines: the higher the expression of this gene, the less sensitivity a tumor cell shows to carboplatin treatment. As mentioned earlier, we have described a positive correlation between cellular proliferation and platinum sensitivity (17). This supports our finding in NCI60 cell lines that DLG3’s effect on platinum sensitivity may be mediated by its effects on cellular proliferation.. Further evaluation of the mechanism underlying the observed relationship between DLG3 expression and platinum sensitivity is warranted.
Previously, we observed in vitro population differences in sensitivity to platinum agents (9, 20, 21) that could be the result of transcription-level, epigenetic, posttranscriptional, and posttranslational effects; however, we hypothesized that differential gene expression may have a key role in the observed population differences. In this study, we chose to focus on X chromosome genes for their potential contribution to the observed population differences in cellular sensitivity to platinating agents. Of the 34 X chromosome genes whose expression levels are significantly correlated with platinum sensitivity, 14 are differentially expressed between the CEU and YRI samples. Furthermore, we also identified a set of eQTLs that predict the expression of the differentially expressed X chromosome genes and showed expected MAF variation between CEU and YRI. Whether these population differences in platinum sensitivity observed in vitro are recapitulated in clinical settings is not definitively established (22) but closer inspection of the available clinical data on ethnically-divergent chemotherapy outcomes may deserve increased attention based upon our findings.
There have been some reports of cisplatin effect on the expression of an X-linked inhibitor of apoptosis protein, XIAP, as a key determinant in chemosensitivity in various cancer cell lines (4, 5). Interestingly, XIAP, did not show significant correlation with either cisplatin or carboplatin sensitivity in our samples (p=0.17, 0.19 in CEU; 0.92 and 0.31 in YRI, for cisplatin and carboplatin, respectively). However, the majority of studies on XIAP and other platinum-implicated genes have been done after platinum therapy, suggesting that perhaps platinum-induced gene expression changes in XIAP are important in mediating alterations in platinum sensitivity (4, 5). Since our study evaluated baseline gene expression, it is not surprising that we did not find a correlation between XIAP expression and platinum sensitivity.
Our study has several recognized limitations. In comparing the independent gene expression quantification methods (the Illumina versus the Affymetrix array), it is well-known that the chromosomal coverage of these two platforms is different. For example, we evaluated 322 X chromosome genes using the Affymetrix exon array while 562 probes (representing 461 genes) were evaluated on the Illumina array with 190 overlap. Only 8 of the 14 genes identified using the Affymetrix exon array to be associated with sensitivity to cisplatin and carboplatin are interrogated on the Illumina array. Therefore, some important X chromosome genes may still be identified by one platform but not validated by the other. However, the X chromosome genes whose expression levels were shown to be important for platinum sensitivity across two different expression platforms constitute findings that are robust.
In summary, the current study demonstrates the importance of the expression of X chromosome genes in cellular sensitivity to platinum agents. This study raises a critical, but neglected, perspective on the relationship between the X chromosome and drug response. Our work shows that sex chromosome gene expression differences may be an important source of variation that should be included in comprehensive studies of pharmacogenomics.
Table 2.
| Illumina | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| combined CEU, YRI and ASN | CEU | YRI | ASN | ||||||
| meta p values | meta q values | p values | p values | p values | |||||
| carboplatin | cisplatin | carboplatin | cisplatin | carboplatin | cisplatin | carboplatin | cisplatin | carboplatin | cisplatin |
| 1.91E-03 | 5.42E-02 | 4.65E-02 | 1.91E-01 | 9.03E-02 | 3.46E-01 | 2.69E-03 | 6.61E-02 | 1.19E-01 | 9.01E-02 |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 4.45E-03 | 7.84E-02 | 7.08E-02 | 2.04E-01 | 3.04E-04 | 3.94E-02 | 3.47E-01 | 2.48E-01 | 7.72E-01 | 3.53E-01 |
| 8.87E-04 | 9.31E-03 | 3.76E-02 | 8.65E-02 | 2.45E-02 | 2.72E-01 | 4.71E-03 | 1.47E-03 | 9.95E-02 | 5.11E-01 |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 6.09E-01 | 7.42E-01 | 5.96E-01 | 5.03E-01 | 1.47E-01 | 4.45E-01 | 9.69E-01 | 8.67E-01 | 7.39E-01 | 4.48E-01 |
| 5.58E-01 | 4.50E-01 | 5.74E-01 | 4.29E-01 | 3.90E-01 | 1.93E-01 | 4.38E-01 | 3.47E-01 | 5.08E-01 | 8.33E-01 |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 1.39E-01 | 1.78E-01 | 3.31E-01 | 3.13E-01 | 5.06E-02 | 2.80E-01 | 5.85E-01 | 3.51E-01 | 2.67E-01 | 1.17E-01 |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 1.13E-01 | 4.03E-03 | 3.07E-01 | 6.55E-02 | 7.78E-01 | 4.91E-01 | 7.64E-02 | 2.13E-01 | 9.81E-02 | 6.88E-04 |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 7.98E-02 | 4.84E-01 | 2.79E-01 | 4.46E-01 | 4.15E-02 | 9.86E-01 | 1.65E-01 | 1.33E-01 | 5.16E-01 | 4.91E-01 |
| 8.65E-01 | 6.64E-01 | 6.91E-01 | 4.90E-01 | 5.40E-01 | 6.43E-01 | 7.58E-01 | 5.67E-01 | 6.88E-01 | 3.54E-01 |
| 7.77E-02 | 7.46E-02 | 2.74E-01 | 2.04E-01 | 1.99E-01 | 4.82E-01 | 1.44E-01 | 5.49E-02 | 1.18E-01 | 1.21E-01 |
| 8.92E-03 | 5.76E-03 | 9.25E-02 | 7.03E-02 | 1.05E-01 | 3.15E-02 | 2.33E-02 | 3.63E-02 | 7.94E-02 | 9.80E-02 |
| 5.30E-01 | 6.64E-01 | 5.57E-01 | 4.90E-01 | 7.65E-01 | 4.97E-01 | 1.04E-01 | 2.99E-01 | 9.75E-01 | 8.69E-01 |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 9.85E-02 | 4.95E-02 | 2.87E-01 | 1.91E-01 | 1.24E-01 | 1.24E-01 | 7.71E-02 | 3.99E-01 | 4.99E-01 | 3.66E-02 |
| 5.79E-01 | 8.05E-01 | 5.77E-01 | 5.12E-01 | 9.77E-01 | 5.47E-01 | 1.14E-01 | 6.77E-01 | 8.41E-01 | 5.94E-01 |
| 1.14E-02 | 6.02E-02 | 1.06E-01 | 1.93E-01 | 2.11E-02 | 3.80E-01 | 4.27E-02 | 9.97E-02 | 2.94E-01 | 6.28E-02 |
| 2.69E-02 | 1.94E-01 | 1.70E-01 | 3.26E-01 | 3.28E-02 | 4.59E-01 | 6.49E-01 | 6.89E-02 | 3.76E-02 | 4.18E-01 |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 8.86E-02 | 2.27E-01 | 2.84E-01 | 3.43E-01 | 5.64E-01 | 5.91E-01 | 3.08E-02 | 3.09E-01 | 2.36E-01 | 9.28E-02 |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 1.05E-01 | 3.40E-01 | 2.99E-01 | 3.91E-01 | 7.80E-01 | 9.45E-01 | 2.19E-02 | 5.74E-02 | 3.08E-01 | 6.15E-01 |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 2.96E-01 | 7.84E-01 | 4.52E-01 | 5.07E-01 | 6.17E-02 | 4.32E-01 | 7.88E-01 | 6.86E-01 | 5.41E-01 | 6.83E-01 |
| 2.52E-01 | 1.08E-01 | 4.21E-01 | 2.40E-01 | 2.29E-01 | 8.02E-02 | 9.27E-02 | 1.53E-01 | 9.45E-01 | 4.46E-01 |
| 3.71E-01 | 1.70E-01 | 4.97E-01 | 3.03E-01 | 4.73E-02 | 1.73E-02 | 8.55E-01 | 8.27E-01 | 9.67E-01 | 7.53E-01 |
Acknowledgments
This study is supported by NIH/NIGMS Pharmacogenomics of Anticancer Agents grant U01GM61393, and by the University of Chicago Breast Cancer SPORE grant P50 CA125183. RSH received support from NIH/NIGMS grant K08GM089941, University of Chicago Cancer Center Support Grant (#P30 CA14599), and Breast Cancer SPORE Career Development Award.
We are grateful for excellent technical support provided by Mr. Ken Hecht in maintaining the cell lines.
The abbreviations used are
- EBV
Epstein-Barr Virus
- IC50
the concentration required to inhibit 50% of cell growth
- CEU
Centre d’Etude du Polymorphisme Humain (CEPH) Utah residents with northern and western European ancestry
- YRI
Yoruba people from Ibadan, Nigeria
- ASN
Han Chinese in Beijing and Japanese in Tokyo
References
- 1.Ramaswamy S, Golub TR. DNA Microarrays in Clinical Oncology. J Clin Oncol. 2002;20:1932–41. doi: 10.1200/JCO.2002.20.7.1932. [DOI] [PubMed] [Google Scholar]
- 2.Huang RS, Kistner EO, Bleibel WK, Shukla SJ, Dolan ME. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Molecular Cancer Therapeutics. 2007;6:31–6. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser J. Gender in the Pharmacy: Does It Matter? Science. 2005;308:1572. doi: 10.1126/science.308.5728.1572. [DOI] [PubMed] [Google Scholar]
- 4.Kim S, Lee S, Cho J, Son S, Choi S, Yun Y, et al. Combination of ginsenoside Rg3 with docetaxel enhances the susceptibility of prostate cancer cells via inhibition of NF-kappaB. Eur J Pharmacol. 2010;631:1–9. doi: 10.1016/j.ejphar.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 5.Roa W, Chen H, Fulton D, Gulavita S, Shaw A, Th’ng J, et al. X-linked inhibitor regulating TRAIL-induced apoptosis in chemoresistant human primary glioblastoma cells. Clin Invest Med. 2003;26:231–42. [PubMed] [Google Scholar]
- 6.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang RS, Duan S, Shukla SJ, Kistner EO, Clark TA, Chen TX, et al. Identification of Genetic Variants Contributing to Cisplatin-Induced Cytotoxicity using a Genome-wide Approach. Am J Hum Genet. 2007;81:427–37. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, et al. SCAN: SNP and copy number annotation. Bioinformatics. 26:259–62. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Duan S, Kistner E, Bleibel W, Huang R, Clark T, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–40. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stranger B, Nica A, Forrest M, Dimas A, Bird C, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci USA. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd MR, Paull KD. Some practical consideration and applications of the National Cancer Institute in vitro anticancer drug discovery screen. Drug Dev Des. 1995;34:91–109. [Google Scholar]
- 13.Weinstein JN. ‘Omic’ and hypothesis-driven research in the molecular pharmacology of cancer. Curr Opin Pharmacol. 2002;2:361–5. doi: 10.1016/s1471-4892(02)00185-6. [DOI] [PubMed] [Google Scholar]
- 14.Gamazon E, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic Drug Susceptibility Associated SNPs are Enriched in Expression Quantitative Trait Loci. Proc Natl Acad Sci USA. 2010;107:9287–92. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Donnell PH, Gamazon E, Zhang W, Stark AL, Kistner-Griffin EO, Huang RS, et al. Population differences in platinum toxicity as a means to identify novel genetic susceptibility variants. Pharmacogenet Genomics. 2010;20:327–37. doi: 10.1097/FPC.0b013e3283396c4e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Kuilenburg A, Meinsma R, Vreken P, Waterham H, van Gennip A. Identification of a cDNA encoding an isoform of human CTP synthetase. Biochim Biophys Acta. 2000;1492:548–52. doi: 10.1016/s0167-4781(00)00141-x. [DOI] [PubMed] [Google Scholar]
- 17.Stark AL, Zhang W, Mi S, Duan S, O’Donnell PH, Huang RS, et al. Heritable and non-genetic factors as variables of pharmacologic phenotypes in lymphoblastoid cell lines. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.3. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stathakis D, Lee D, Bryant P. DLG3, the gene encoding human neuroendocrine Dlg (NE-Dlg), is located within the 1.8-Mb dystonia-parkinsonism region at Xq13.1. Genomics. 1998;49:310–3. doi: 10.1006/geno.1998.5243. [DOI] [PubMed] [Google Scholar]
- 19.Makino K, Kuwahara H, Masuko N, Nishiyama Y, Morisaki T, Sasaki J, et al. Cloning and characterization of NE-dlg: a novel human homolog of the Drosophila discs large (dlg) tumor suppressor protein interacts with the APC protein. Oncogene. 1997;14:2425–33. doi: 10.1038/sj.onc.1201087. [DOI] [PubMed] [Google Scholar]
- 20.Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nature Genetics. 2007;39:226–31. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storey J, Madeoy J, Strout J, Murfel M, Ronald J, Akey J. Gene-expression variation within and among human populations. Am J Hum Genet. 2007;80:502–9. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millward MJ, Boyer MJ, Lehnert M, Clarke S, Rischin D, Goh BC, et al. Docetaxel and carboplatin is an active regimen in advanced non-small-cell lung cancer: a phase II study in Caucasian and Asian patients. Ann Oncol. 2003;14:449–54. doi: 10.1093/annonc/mdg118. [DOI] [PubMed] [Google Scholar]