Abstract
All organisms that produce fatty acids do so via a repeated cycle of reactions. In mammals and other animals, these reactions are catalyzed by a type I fatty acid synthase (FAS), a large multifunctional protein to which the growing chain is covalently attached. In contrast, most bacteria (and plants) contain a type II system in which each reaction is catalyzed by a discrete protein. The pathway of fatty acid biosynthesis in Escherichia coli is well established and has provided a foundation for elucidating the type II FAS pathways in other bacteria (White et al., 2005). However, fatty acid biosynthesis is more diverse in the phylum Actinobacteria: Mycobacterium, possess both FAS systems while Streptomyces species have only the multi-enzyme FAS II system and Corynebacterium species exclusively FAS I. In this review we present an overview of the genome organization, biochemical properties and physiological relevance of the two FAS systems in the three genera of actinomycetes mentioned above. We also address in detail the biochemical and structural properties of the acyl-CoA carboxylases (ACCases) that catalyzes the first committed step of fatty acid synthesis in actinomycetes, and discuss the molecular bases of their substrate specificity and the structure-based identification of new ACCase inhibitors with anti-mycobacterial properties.
Keywords: lipid biosynthesis, acyl-CoA carboxylase, Corynebacterium, Mycobacterium, Streptomyces, lipid homeostasis
Introduction
Within the Actinobacterial taxon (Gram-positive bacteria with high G + C content of DNA) three genera, Mycobacterium, Streptomyces and Corynebacterium, have been widely studied because of their remarkable impact on mankind, either as extremely dangerous pathogens, such as Mycobacterium tuberculosis, Mycobacterium leprae and Corynebacterium diphtheriae or, at the other extreme, as very useful sources of bioactive metabolites. In this sense streptomycetes are well known for their ability to produce a vast array of secondary metabolites widely used in human health (e.g. anticancer agents, immunossupressants, and antibiotics)(Hopwood, 1989) and corynebacteria for their ability to produce and secrete large amounts of L-amino acids, mainly used in the food and livestock industries (Kramer, 1994).
One motivation to present a comprehensive and comparative review of fatty acid biosynthesis in these three genera of actinomycetes was to focus on the remarkable differences between them in the content of their membrane and storage lipid components and consequently on the lipid biosynthetic machinery. Even though streptomycetes undergo a complex life cycle with distinctive developmental and morphological stages (Chater and Losick, 1997), the cell wall is similar to that of other Gram-positive bacteria, composed of a simple peptidoglycan mesh surrounding the cytoplasmic membrane. By contrast, the cell walls of Mycobacterium spp., as well as species of related genera including Corynebacterium, Gordonia, Nocardia, and Rhodococcus, are formed by a thick meso-diaminopimelic acid-containing peptidoglycan covalently linked to arabinogalactan, which in turn is esterified by long-chain α-alkyl, β-hydroxy fatty acids called mycolic acids (Brennan, 2003). While in mycobacteria these fatty acids have very long carbon chains (C60–90) and may contain various oxygen functions in addition to the β-hydroxyl group (Daffe and Draper, 1998), mycolic acids found in other actinomycetes consist of a mixture of saturated and unsaturated fatty acids with shorter carborn chains like C22–36 in corynomycolic acids (Collins et al., 1982; Minnikin and Goodfellow, 1980). Mycolic acids are believed to play a central role in the formation of a second permeability barrier functionally similar to the outer membrane in Gram-negative bacteria (Gebhardt et al., 2007; Liu et al., 1996). While the latter is a typical bilayer of phospholipid and lipopolysaccharide, in mycobacteria and corynebacteria it consists of a monolayer of mycoloyl residues covalently linked to the cell-wall arabinogalactan, to form mycoloyl arabinogalactan (MAG), or acylated to trehalose units fo form trehalose monomycolate (TMM) and trehalose dimycolate (TDM) (Brennan, 2003; Daffe and Reyrat, 2008) (Fig. 1). Besides mycolic acids, the outer membrane of mycobacteria also contains an array of very complex lipids relevant in the infection process of the pathogenic mycobacteria, such as dimycocerosate esters (DIMs), sulpholipids and mycoketides (Astarie-Dequeker et al., 2009; Camacho et al., 1999; Cox et al., 1999). In the last few years cell-free reconstitution studies demonstrated that several of these compounds are synthesized from acyl-CoA molecules, most probably derived from the fatty acid biosynthesis pathway, which are further elongated by specific FAS related enzymes known as polyketide synthases (PKS) (Onwueme et al., 2005; Sirakova et al., 2001; Trivedi et al., 2005).
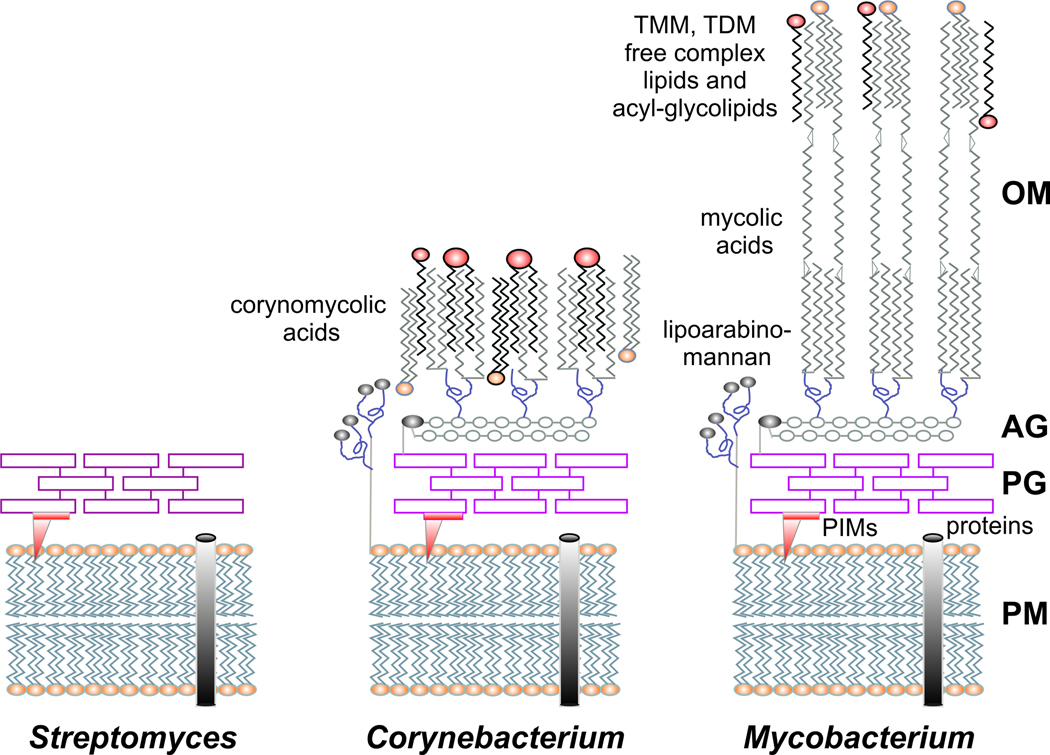
Fig. 1. Schematic representation of the cell envelope of actinomycetes.
Comparison of the cell wall structures from Streptomyces, Corynebacterium and Mycobacterium. In streptomycetes, as in other Gram-positive bacteria, the cell wall consists of a peptidoglycan layer (PG) that covers the cytoplasmic membrane (PM). Corynebacteria and mycobacteria contain a more complex cell wall including an additional layer of arabinogalactan (AG) and an outer membrane (OM) composed of mycolic acids, trehalose monomycolate (TMM), trehalose dimycolate (TDM) and free lipids. The mycobacterial cell wall has a high proportion of covalently attached mycolic acid residues, which constitute a permeability barrier contributing to antibiotic resistance and pathogenicity. PIMs: phosphatidylinositol-mannosides.
Another remarkable feature of mycobacteria and streptomycetes, but not of corynebacteria, is that de novo synthesized fatty acids are not only dedicated to building membrane phospholipids (PL) but are also incorporated into neutral lipid storage compounds, the triacylglycerides (TAG) (Arabolaza et al., 2008; Daniel et al., 2004). The synthesis of TAG and its accumulation within the cytoplasm as lipid droplets brings another layer of complexity in the lipid metabolism of these bacteria, and highliths another important difference between the three genera of actinomycetes discussed in this review.
General principles of bacterial fatty acid biosynthesis
The biosynthesis of fatty acids is the first step in the formation of membrane lipids and is essential for all cells, excepting the Archea in which the membranes are composed of glycerol-ether lipids instead of glycerol-ester lipids and are based on isoprenoid side chains (De Rosa et al., 1986). The basic features of fatty acid synthesis pathways, like the constituent chemical reactions and the identity of the component enzymes, were the focus of intensive studies more than four decades ago, especially in the laboratories of Vagelos (Alberts et al., 1969; Greenspan et al., 1969), Bloch (Odriozola et al., 1977) and Wakil (Pugh et al., 1966; Toomey and Wakil, 1966). Later on, the genes and enzymes of fatty acid and phospholipid metabolism were defined using E. coli as a model organism (Cronan and Rock, 1996; Rock and Cronan, 1996). However, although the E. coli paradigm provides a solid core of information that is shared by all bacteria, the biochemical details of the lipid metabolic pathways in other bacteria cannot be inferred only from this model. Some relevant differences between E. coli and other important non-actinomycetes human pathogens have been reviewed elsewhere (Rock, 2008).
The first committed step in fatty acid biosynthesis is the carboxylation of acetyl-CoA by acetyl-CoA carboxylase (ACC), to form the common extender unit malonyl-CoA, which is then made available to the enzymes of fatty acid biosynthesis by its conversion to malonyl-ACP by malonyl-CoA:ACP transacylase, usually called FabD. The chain elongation steps in fatty acid biosynthesis consist of an iterative series of reactions built on successive addition of a two-carbon unit to a nascent acyl group. Reaction intermediates (the growing acyl-chains) are covalently attached to the acyl carrier protein (ACP) through a thioester linkage to the terminal sulphydryl of the 4´-phosphopanteine prosthetic group. The first elongation step is initiated by the Claisen condensation of malonyl-ACP with an acyl-CoA, catalyzed by the first condensing enzyme, the β-ketoacyl-ACP synthase III or FabH, to form β-ketoacyl-ACP. In E. coli, two other mechanisms for the initiation of fatty acid biosynthesis have been described (Cronan and Rock, 2008). Additional cycles of elongation in the pathway are catalyzed by the FabF-FabB class of condensing enzymes. These proteins condense malonyl-ACP with acyl-ACP to extend the acyl chain by two carbons. This is the only irreversible step in the elongation cycle and so it is not surprising that the 3-ketoacyl-ACP synthases play key roles in regulating the product distribution of the pathway. Three enzymes complete the cycle by reducing the β keto group. First, the nicotinamide adenine dinucleotide phosphate (NADPH)-dependent β-ketoacyl-ACP reductase, or FabG, reduces the β keto group to a β-hydroxyl intermediate. Second, this is dehydrated by the β-hydroxyacyl-ACP dehydratase, known as FabZ or FabA, to a trans-2-enoyl-ACP. The third step involves reduction of the enoyl chain by the NADPH-dependent (FabI or FabL) or NADH-dependent (FabV) enoyl-ACP reductase. On completion of each cycle of condensation and chain reduction, the growing acyl chain is transferred back to the ketosynthase to initiate the next cycle.
Based on protein architecture, FASs are classified as type I or type II. The type II FAS consists of a dissociated system formed by a discrete and highly conserved group of enzymes encoded by a series of separate genes (Fig. 2). This system has been characterized from bacteria and parasites as well as mitochondria and chloroplasts (the site of fatty acid biosynthesis in plants), where it catalyses de novo fatty acid synthesis (except in mycolic acid producers), usually from acetyl-CoA (Rawlings, 1998; Rock and Cronan, 1996). The classical type I FAS consists of a multifunctional protein carrying all the catalytic domains on a single polypeptide chain (Fig. 2). These enzymes are typically found in eukaryotes (Schweizer and Hofmann, 2004) (though not in plants) and as a remarkable prokaryotic exception in the mycolic acid-producing actinomycetes (Bloch and Vance, 1977). FAS I enzymes can be subdivided according to their domain arrangements and oligomerization state. Microbial type I FASs consist of a single type of polypeptide organised as a hexamer (α6) (Bloch and Vance, 1977; Wood et al., 1978), with a different domain order compared to the vertebrate FAS (Schweizer and Hofmann, 2004). Mycobacteria and corynebacteria type I FASs are hexamers with a domain arrangement of: acyltransferase, enoyl reductase, dehydratase, malonyl/palmitoyl transferase, acyl carrier protein, 3-ketoacyl reductase and 3-ketoacyl synthase (AT-ER-DH-MPT-ACP-KR-KS), an organization that differs from that of the animal FAS (KS-AT-DH-ER-KR-ACP-TE) (Schweizer and Hofmann, 2004). Besides, animal FAS releases free fatty acids by hydrolytic cleavage catalyzed by a thioesterase (TE) domain (Schweizer and Hofmann, 2004), while the type I FASs of yeast, mycobacteria and corynebacteria use an integral malonyl/palmitoyl transferase activity (MPT) for transacylation of palmitate (or any other end acyl-enzyme product) from the enzyme to CoA (Bloch and Vance, 1977; Lynen, 1980).
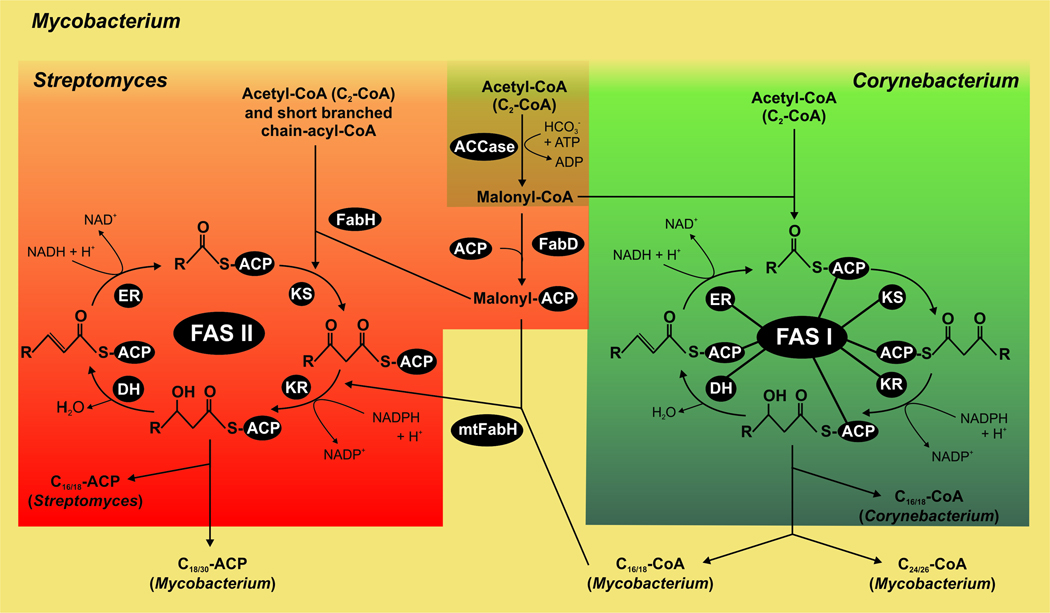
Fig. 2. Comparative model of fatty acid biosynthesis in actinomycetes.
Streptomyces Corynebacterium and Mycobacterium share the initiation step for fatty acid biosynthesis which is the carboxylation of acetyl-CoA to produce malonyl-CoA, catalyzed by the ACCase complex (brown). Both in Mycobacterium and Corynebacterium spp., malonyl-CoA and acetyl-CoA are condensed to be utilized by the multifunctional type I FAS for de novo biosynthesis of fatty acids (green). In Streptomyces, malonyl-CoA is converted to malonyl-ACP by FabD and then condensed with acetyl-CoA by FabH to form β-ketoacyl-ACP, which is used by the type II FAS dissociated system for de novo biosynthesis of fatty acids (red). Both in FAS I and FAS II systems, the chain elongation steps consist of an iterative series of reactions built on successive addition of a two-carbon unit to a nascent acyl group, and reaction intermediates are covalently attached to the acyl carrier protein (ACP). In Mycobacterium both systems are present, FAS I for de novo fatty acid biosynthesis and FAS II for the elongation of fatty acids for mycolic acid production. The differential features of Mycobacterium are shown in yellow.
A number of functionally differentiated FAS variants, as well as a large family of FAS-related enzymes, have evolved by slight variations in the FAS pathways to produce a wide range of natural compounds in streptomycetes and complex lipids in mycobacteria (Gokhale et al., 2007a; Hertweck, 2009). For instance it is well known that many polyketides (PK) natural products, which have been a rich source of therapeutic compounds, are biosynthesized with comparable logic by multimodular PKSs, which are evolutionary descendant of type I FAS, or by type II PKS complexes highly homologue to FAS II enzymes. Moreover, the two systems could also share precursors, acetyl- and malonyl-CoA, which might establish a substrate competition effect; this can be the case for the biosynthesis of mycobacteria complex lipids but probably not for PK biosynthesis that generally occurs after growth arrest. The common mechanisms and the structural similarity of type I FAS and PKSs resulted in the proliferation of hibrid PKS-FAS pathways (Smith and Sherman, 2008). As an example, the very complex lipids of mycobacteria are made via a partnership between a FAS and a modular PKS (Gokhale et al., 2007b). All this functional and mechanistic overlapping brings over a very interesting question which is whether or not the fatty acid and PK synthesis share protein components.
Fatty acid synthase systems in three model actinomycetes
The complexity of lipid metabolism in actinomycetes, especially in the Corynebacterianeae suborder, became obvious soon after the complex lipids that form part of their cell wall started to be characterized and their biosynthetic pathways elucidated. The sequencing of the genomes of several species of Streptomyces, Mycobacterium and Corynebacterium soon provided very valuable information about lipid metabolism in these bacterial genera. This information, along with excellent biochemical studies, provided valuable information about the de novo synthesis of fatty acids as well as of complex lipids in actinomycetes.
Evolution of the fatty acid biosynthetic genes has followed a unique and very interesting path in actinomycetes. One of the most remarkable differences from the canonical E. coli type II fatty acid biosynthetic pathway is the type I megasynthase pathway found in the mycolic acid-producing actinomycetes. In corynebacteria, for example, none of the genes encoding FAS II components can be found, presumably because of the evolution of a FAS I system for de novo fatty acid biosynthesis. In mycobacteria, the FAS II system has adapted to elongate existing long-chain fatty acids to form the meromycolic chain of the mycolic acids, and this system coexists with the FAS I enzyme that is dedicated to de novo fatty acid biosynthesis. Finally, Streptomyces contains only the genes for the FAS II enzymes, as most bacteria do (Fig. 2).
Next we describe in detail the genetics and enzymology of the FAS systems used for fatty acid biosynthesis in the three model actinomycetes and their interaction with the PKS systems.
Streptomyces
Streptomyces utilizes a type II FAS to catalyze the de novo synthesis of fatty acids. The majority are made from branched starters such as isobutyryl, isovaleryl, and anteisovaleryl units to give odd- and even-numbered fatty acids with a methyl branch at the ω-terminus (80–90% of total fatty acid content); the rest are synthesized from unbranched starters such as acetyl and butyryl units (Kaneda, 1991; Wallace et al., 1995).
Characterization of the genes/enzymes involved in fatty acid biosynthesis in Streptomyces has received little attention compared to other bacteria, and most of the information available comes from predictive gene functions assigned by in silico genome analysis. Thus, the search for fatty acid biosynthetic genes in the most relevant streptomycetes genomes revealed various putative orthologues to several of the classical FAS II components. An extended analysis of all the annotated streptomycetes genomes revealed that some of the genes encoding the core enzymes involved in saturated fatty acid biosynthesis lie in a conserved fatty acid biosynthesis (fab) cluster that comprises the following genes: fabD, fabH, acpP and fabF. This cluster has been partially characterized in S. coelicolor and its relationship with the fatty acid biosynthetic pathway confirmed through genetic and biochemical experiments (Revill et al., 1995, , 1996; Revill et al., 2001). Further, the alignment of several actinomycete genomic regions containing this universal set of genes highlighted a remarkable synteny (Fig. 3), indicating a high degree of conservation in the evolution of this essential biosynthetic pathway.
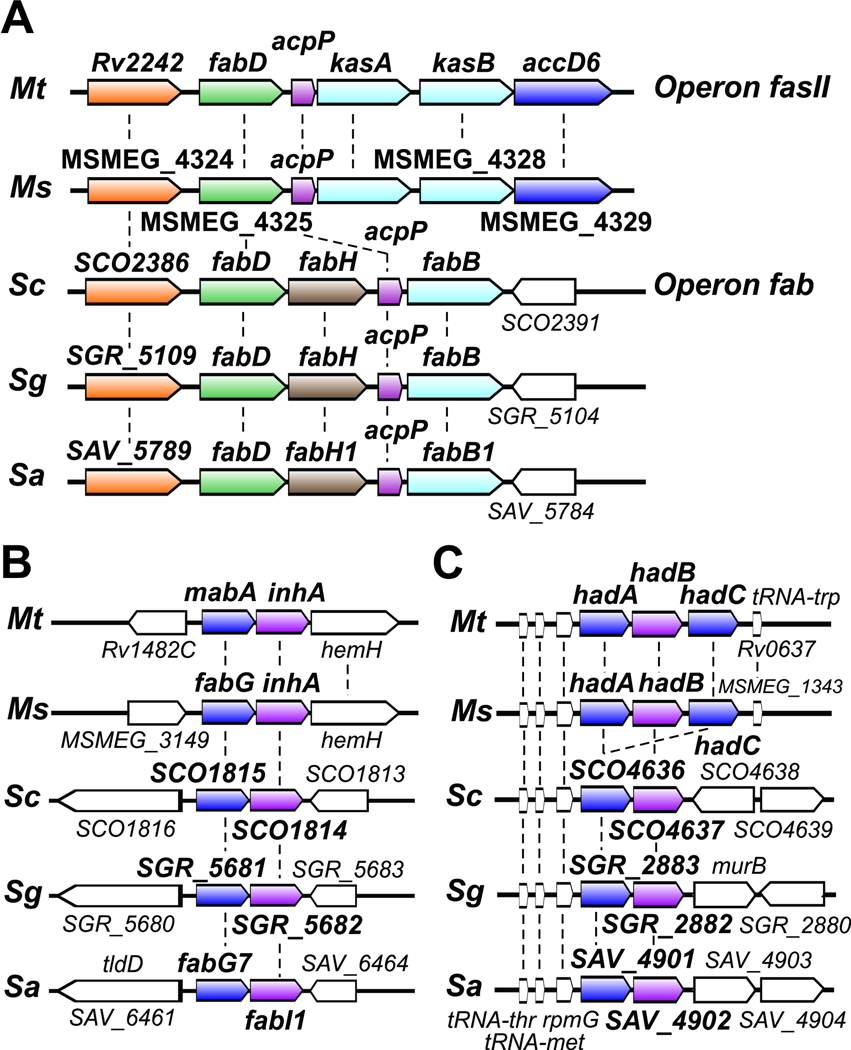
Fig. 3. Genetic organization of FAS II genes.
Synteny present in the fas II (fab) operons (A), the cluster of genes encoding for the FAS-II reductases (B) and the FAS II dehydratases (C) in some of the most relevant species of mycobacteria and streptomycetes: M. tuberculosis (Mt), M. smegmatis (Ms), S. coelicolor (Sc), S. griseous (Sg) and S. avermitilis (Sa).
fabD encodes the malonyl-CoA:ACP transacylase that provides the malonyl-ACP substrate to the condensing enzymes. FabD from S. glaucescens (sgFabD) could functionally replace the E. coli FabD protein when expressed in this microorganism (Summers et al., 1995). In vitro studies also demonstrated that sgFabD is active with several ACPs such as TcmM (the S. glaucescens tetracenomycin PKS ACP), AcpP (the E. coli FAS ACP) and FabC (the S. glaucescens FAS ACP) (Florova et al., 2002). The relaxed specificity of sgFabD for different ACPs suggested that the functional role of sgFabD is to provide malonyl-ACP precursors for both fatty acid and type II PK biosynthetic pathways (Florova et al., 2002). S. coelicolor has a single copy of fabD in the main fab cluster (Revill et al., 1995), suggesting that, as well as its essential role in fatty acid biosynthesis, scFabD is also shared with the type II PK biosynthetic pathway (Revill et al., 1995). On the other hand, S. griseus has three fabD homologues, (SGR_5108, SGR_6780, SGR_3283) suggesting the possibility of malonyl-CoA ACP transacylases dedicated to serve specific PKS ACPs.
In S. coelicolor fabH was proved to be essential, in accordance with its role as a gene encoding one of the FAS proteins (Revill et al., 2001). The generation of an S. coelicolor fabH mutant in the presence of a fabH gene from E. coli (with only 35% amino acid identity to the Streptomyces enzyme) resulted in a mutant strain that produced predominantly straight-chain fatty acids (86%) (Li et al., 2005), as expected for the presence of a FabH enzyme with a high specificity for acetyl-CoA like the ecFabH. These findings correlate with the biochemical properties of the S. glaucescens FabH protein, which shows broad substrate specificity for short-chain acyl-CoA substrates, consistent with its role in initiating branched- and straight-chain fatty acid biosynthesis and determining, at least in part, the fatty acid profile (Han et al., 1998).
The acpP gene is a confirmed member of the FAS complex in S coelicolor, as determined by its constitutive expression, its genomic clustering and its ability to function in E. coli fatty acid synthesis (Revill et al., 1996). The remaining two ACPs are developmentally regulated and required for the synthesis of the type II PKs synthesized by S. coelicolor, the blue-pigmented antibiotic actinorhodin and the grey pigment associated with the spore wall (Revill et al., 1996). When acpP was cloned in place of SCO5089 (encoding the actinorhodin ACP), pigmented PK production was severely compromised, implying that the FAS and PKS ACPs are biochemically incompatible (Revill et al., 1996).
The genes encoding the enoyl-ACP reductases, the β-ketoacyl-ACP reductase and the (3R)-hydroxyacyl-ACP dehydratase, have not yet been characterized in streptomycetes. However, based on the genetic and biochemical data available for these enzymes in related members of actinobacteria (see below) we can assign these functions to each of the putative genes found in streptomycetes. For example, in S. coelicolor, homologues to mabA-inhA from M. tuberculosis (Cole et al., 1998) (enoyl-ACP reductase and β-ketoacyl-ACP reductase, respectively) revealed a putative two-gene operon (SCO1814-SCO1815) with synteny not only in Streptomyces species but also among other actinomycetes (Fig. 3). SCO1814 is 47 % identical in a 263 aa overlap with InhA, and SCO1815 is 50.8% identical in a 455 aa overlap with MabA. Based on homology and synteny with the M. tuberculosis (3R)-hydroxyacyl-ACP dehydratase components (Sacco et al., 2007), we propose ORFs SCO4636-SCO4637 as the putative (3R)-hydroxyacyl-ACP dehydratase(s) of the FAS II system of S. coelicolor.
Mycobacterium
Mycobacteria are unusual in possessing both type I and type II FAS systems. Fatty acid biosynthesis in mycobacteria is initiated by a unique multifunctional FAS I enzyme, catalyzing the de novo synthesis of long-chain acyl-CoAs (C16:0- and C18:0) from acetyl-CoA and using malonyl-CoA as extender unit to yield butyryl-S-Enz. The C16:0- and C18:0-S-Enz, could either be converted to the CoA derivatives and used primarily for the synthesis of membrane phospholipids or further elongated by FAS I to produce C24:0-S-Enz in the fast-growing organism M. smegmatis (Bloch and Vance, 1977; Peterson and Bloch, 1977) and C26:0-S-Enz products in the slow-growing M. bovis and M. tuberculosis (Kikuchi et al., 1992), a characteristic behaviour of FAS I called “bimodal product distribution”. To understand further the bimodality of FAS I, Zimhony et al (Zimhony et al., 2004) generated a recombinant M. smegmatis strain in which the native fas1 gene was deleted and replaced with the M. tuberculosis fas1 gene (Zimhony et al., 2004). In the course of analyzing the recombinant M. smegmatis ΔfasI (attB::M. tuberculosis fas1) strain, it was found that the in vivo elongation of C16:0 did not follow the M. tuberculosis profile; rather, the new strain contained both fatty acids C24:0 and C26:0, challenging the concept that C16:0 elongation to produce C24:0 in M. smegmatis or to produce C26:0 in M. tuberculosis is exclusively a FAS I-dependent property, as described previously (Kikuchi et al., 1992; Peterson and Bloch, 1977), but most probably the result of a more complex interaction between FAS I and FAS II components. Furthermore, the bimodal product distribution shown by FAS I of mycobacteria not only depends on the intrinsic determinants of the FAS proteins but also on external factors that interfere with the elongation process. For instance, early studies carried out with FAS I from M. smegmatis demonstrated that acyl-CoA-binding substances like bovine serum albumin or certain mycobacterial polysaccharides could shift the FAS product pattern of mycobacterial type I FAS toward shorter-chain acids (Peterson and Bloch, 1977). More recently, Papaioannou et al (Papaioannou et al., 2007) studied the product-regulation mechanisms for fatty acid biosynthesis catalyzed by M. smegmatis FAS I. They found that the predominant formation of palmitic acid in the presence of synthetic O-alkyl polysaccharide is regulated by the removal of the end-product via a tight stoichiometric complex between the C16:0 fatty acid and the O-alkyl polysaccharide (gate-keeper mechanism) (Papaioannou et al., 2007). The bimodal pattern of product distribution has also been demonstrated with the yeast FAS, where the addition of acyl-CoA-binding protein to the assay mixture caused a dramatic decrease in the chain-length of acyl-CoA reaction products (Schjerling et al., 1996). In M. tuberculosis the C26:0 fatty acids synthesized by FAS I will become the substrate of a dedicated acyl-CoA carboxylase to generate the α–carboxy C26:0 fatty acid used as one of the substrate by the PKS 13 in the biosynthesis of mycolic acids.
The FAS I and FAS II systems are connected by a key condensing enzyme: the β-ketoacyl-ACP synthase III or FabH (Fig. 2). This enzyme catalyzes a decarboxylative condensation of malonyl-ACP with the acyl-CoA (C16:0 – C20:0) products of the type I FAS. The resulting 3-ketoacyl-ACP product is reduced to an acyl-ACP (extended by two carbons) and shuffled into the FAS II cycle. Enzymology studies carried out in the presence of malonyl-AcpM have shown that mtFabH can utilize a wide range of C12:0–C20:0 acyl-CoA substrates, consistent with structural studies (Scarsdale et al., 2001) and distinguishing it from FabH in other type II FAS systems that typically utilize C2–C6 acyl-CoA substrates (Qiu et al., 2005; Tsay et al., 1992). More recent studies have shown that in the acylation reaction performed with a range of acyl-CoAs (carried out in the absence of malonyl AcpM) mtFabH selectively forms a product with the dodecanoyl CoA (Sachdeva et al., 2008). These results support a previous hypothesis about the role of AcpM in modulating the substrate specificity of mtFabH (Brown et al., 2005). Structural and site-directed mutagenesis studies have helped to understand the molecular basis of the differences in substrate specificity between the E. coli and Mycobacterium FabH enzymes (Brown et al., 2005; Scarsdale et al., 2001). Both the E. coli and the M. tuberculosis FabH are homodimers with a CoA/malonyl-ACP binding channel leading from the enzyme surface to the buried active site cysteine residue. Unlike ecFabH, mtFabH contains a second hydrophobic channel leading from the active site. In the ecFabH structure this channel is blocked by a Phe87 residue, which constrains specificity to acetyl-CoA, while in mtFabH this residue is a Thr87, which permits binding of longer acyl chains. This same channel in mtFabH is capped by an α-helix, which limits bound acyl chain length to 16 carbons.
In mycobacteria, the genes encoding the FAS II enzymes lie in different positions in the genome in three independent transcription units (Fig. 3). The first is formed by fabD-acpM-kasA-kasB-accD6 (Fig. 3A), the second contains two genes, mabA-inhA (Fig. 3B) (Cole et al., 1998), and the third consists of three genes, hadA, hadB and hadC (Fig. 3C). The first ORF of the five-gene operon encodes an enzyme with significant homology to FabD of other bacteria, which is a malonyl-CoA:ACP acyl transferase (MAT) (Fig. 3A). This enzyme catalyzes the formation of malonyl-ACP, which is then used by the condensing enzyme system for the extension of a growing fatty acid chain. mtFabD catalyzes in vitro the transacylation of malonate from malonyl-CoA to activated holo-AcpM (Kremer et al., 2001). A second enzyme with MAT activity was identified in M. tuberculosis and named FabD2 (Huang et al., 2006). This protein is 28% identical to FabD but its physiological role remains unknown. The second ORF of this operon encodes the acyl carrier protein AcpM, a C-terminally-extended homologue of bacterial ACPs, involved in carrying the growing acyl chain from one reactive centre to another until the complete cycle of fatty acid biosynthesis is finished. The elongation steps that follow the FabH-dependent condensation reaction of the FAS II system are conducted by the β-ketoacyl-AcpM enzymes encoded by kasA and kasB in the five-gene operon. KasA and KasB are 65% identical to each other and 46.8% and 45% identical to the S. coelicolor β-ketoacyl-ACP synthase II, FabF, respectively. Both KasA and KasB catalyze the condensation of acyl-AcpM with malonyl-AcpM, elongating the growing meromycolate chain by a further two carbons (Kremer et al., 2000) (Schaeffer et al., 2001). KasA and KasB have different chain-length specificities, suggesting a distinct role for each enzyme in the elongation of meromycolic acids. In fact, cell lysates of M. smegmatis overproducing KasA of M. tuberculosis could elongate fatty acids up to 40 carbons, while the overproduction of KasA and KasB resulted in the production of longer chain fatty acids up to 54 carbon atoms (Slayden and Barry, 2002). Based on these in vitro studies it was proposed that KasA is implicated in the initial elongation of mycolates which are extended to the full length molecules by KasB (Kremer et al., 2002; Slayden and Barry, 2002). This hypothesis was confirmed in vivo by generating kasB mutants both in Mycobacterium marinum (Gao et al., 2003) and in M. tuberculosis (Bhatt et al., 2007). Transposon mutagenesis in M. marinum gave rise to a kasB mutant strain which produced meromycolate chains two to four carbons shorter than the wild-type mycolates, with a significant reduction of the keto-mycolates (Gao et al., 2003). Similarly, a kasB mutant of M. tuberculosis also generated similar shortened mycolates (by up to six carbon atoms), but also showed the loss or reduction of the trans-cyclopropanated oxygenated mycolates. A KasA mutant could not be obtained (Bhatt et al., 2005), indicating that kasA is the only essential β-ketoacyl-ACP synthase II-encoding gene and that KasA is most probably the enzyme involved in initial extension of the mycolate chains, whereas KasB is primarily involved in the full-length extension of these molecules. Interestingly, the M. tuberculosis kasB mutant also lost its acid-fast staining and was unable to persist in immunocompetent infected mice, suggesting KasB as a putative new target for novel therapeutics.
The genes encoding the FAS-II reductases are found in the M. tuberculosis genome in a cluster containing mabA and inhA (Fig. 3B), which encode proteins that are functionally and structurally related. The inhA gene was identified as a putative target for INH (isoniazid) and ETH (ethionamide) in M. tuberculosis (Banerjee et al., 1994) and the InhA protein was demonstrated to catalyze the 2-trans-enoyl-ACP reduction, with a preference for long-chain substrates (Quemard et al., 1995). MabA encodes a β–ketoacyl-ACP reductase with preference for long-chain substrates and was shown to be involved in the elongation activity of FAS II (Marrakchi et al., 2002). In comparison with other bacterial reductases, MabA has unique functional and structural properties, such as a large hydrophobic substrate-binding pocket, which correlates with its preference for long-chain substrates consistent with its role in mycolic acid biosynthesis (Cohen-Gonsaud et al., 2002; Marrakchi et al., 2002). So far, there have been no successful reports of the generation of any mabA or inhA gene knockout, strongly suggesting their essentiality for mycobacterial viability.
Dehydration of the β–hydroxyacyl-ACP intermediate into trans-2-enoyl-ACP is catalyzed by the β–hydroxyacyl-ACP dehydratases in the M. tuberculosis cluster hadA-hadB-hadC (Sacco et al., 2007) (Fig. 3C). Heterologous expression of these proteins in E. coli led to the formation of two heterodimers, HadAB and HadBC, both with the properties expected for the mycobacterial FAS-II dehydratases: a marked specificity for both long-chain and ACP-bound substrates.
Corynebacterium
Corynebacteria also utilize the multifunctional type I FAS for de novo biosynthesis of fatty acids, which in these organisms are incorporated as part of the cell-membrane phospholipids or used as precursors for mycolic acid biosynthesis (Fig. 2). It is important to highlight that corynebacteria lack a type II FAS system that could elongate the C16:0 or C18:0 fatty acids generated by the type I FAS, so corynomycolic acids are synthesized through the direct condensation of the FAS I end products (Kalinowski et al., 2003).
The catalytic domain organization of the corynebacterial type I FAS follows the same pattern as in mycobacteria: AT-ER-DH-MPT-ACP-KR-KS (Schweizer and Hofmann, 2004). This, together with the high overall identity at the amino acid level (~ 50 %), suggests a conserved physiological function.
An in silico analysis of all the Corynebacterium genome sequences highlights the presence of two putative type I FASs in most species, with some exceptions in the pathogen Corynebacterium diphtheriae that only contains one type I FAS and in Corynebacterium jeikeium or Corynebacterium urealyticum that contain none, reflecting their strict dependence for growth on the presence of exogenous fatty acids (Tauch et al., 2005; Tauch et al., 2008). The presence of two FAS I enzymes in this genus was initially described in the former Brevibacterum ammoniagenes (Stuible et al., 1996), currently reclassified as Corynebacterium ammoniagenes and later in Corynebacterium glutamicum (Radmacher et al., 2005). In both cases it was shown through gene knockout experiments that one of these enzymes is essential for cell survival, FAS A and FAS-IA of C. ammoniagenes and C. glutamicum, respectively, while the second (FAS B or FAS-IB) is supplementary; however both enzymes are necessary to produce the characteristic lipid environment of this organism. Purification and further characterization of FAS A and FAS B from C. ammoniagenes showed that FAS A synthesizes principally palmitic (C16:0) and oleic (C18:1) acids while FAS B could only synthesize saturated C16:0 and C18:0 (stearic acid) fatty acids. In agreement with this biochemical characterization, mutational inactivation of the gene encoding FAS-IA in C. glutamicum results in an unsaturated fatty acid requirement (Radmacher et al., 2005). Supporting these data, both FAS A and FAS-1A contain a FabA-like domain which, in the type II FAS system, represents the enzymatic activity involved in the synthesis of unsaturated fatty acids. To date, our knowledge on the redundancy of type I FAS systems in the same organism is very limited.
Acyl-CoA carboxylases
Acetyl-CoA carboxylases
The first committed step in the biosynthesis of long-chain fatty acids in all animals, plants and bacteria is carried out by the acetyl-CoA carboxylase (ACC) (Wakil et al., 1983). ACC is a key enzyme that catalyzes the production of malonyl-CoA, which is further utilized by all the FAS systems for the elongation of the growing acyl chain. Malonyl-CoA is generated by the carboxylation of acetyl-CoA in a reaction that involves two separate hemi-reactions (Wakil et al., 1983) (Fig. 4A).
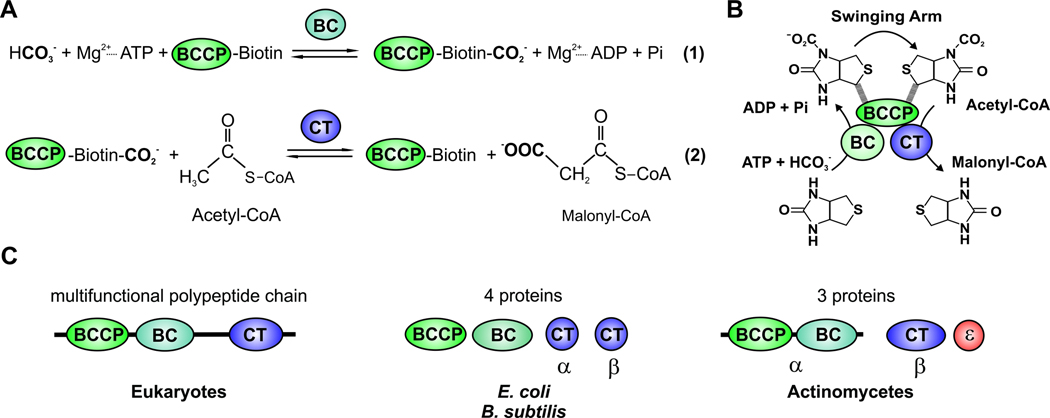
Fig. 4. Schematic diagram of ACC reaction and of domains and/or subunits of the biotin-dependent ACC.
(A) Stepwise enzymatic reactions of ACC. (B) ACC is composed of three different components, the biotin carboxylase component (BC), the biotin carboxyl carrier protein (BCCP) and carboxyltransferase (CT). The “Swinging Arm” of biotin-BCCP transports CO2 between the components BC and CT. Then the carboxyl group is transferred from biotin to acetyl-CoA to form malonyl-CoA. (C) The eukaryotic ACC is a multifunctional polypeptide chain that contains the three components (BC, BCCP and CT). Bacterial ACC is composed of multiple subunits. The E. coli and B. subtilis ACCs are formed by four proteins: the subunit BC, the subunit BCCP, and two proteins (α and β) that form the CT subunit. In actinomycetes the ACCase complex is made up of two main subunits: the α subunit, which contains the BC and BCCP components and the β subunit, which has the CT activity. In addition, actinomycete ACCases may include a third subunit, ε, the presence of which dramatically stimulates the specific activity of the enzyme complexes.
ACC is composed of three different components which allow it to carry out these two distinct reactions (Fig. 4B). The biotin carboxylase component (BC) catalyzes the first half-reaction, which involves the phosphorylation of bicarbonate by ATP to form a carboxyphosphate intermediate, followed by transfer of the carboxyl group to biotin to form carboxybiotin (Blanchard et al., 1999a; Blanchard et al., 1999b; Janiyani et al., 2001; Levert et al., 2000; Waldrop et al., 1994). In vivo, the cofactor biotin is attached to the biotin carboxyl carrier protein (BCCP) via an amide bond between the valeric acid side chain of biotin and the ε-amino group of a specific lysine residue (Athappilly and Hendrickson, 1995; Cronan, 2001; Reddy et al., 2000; Roberts et al., 1999; Yao et al., 1997). In the second reaction, catalyzed by the carboxyltransferase (CT) component, the carboxyl group is transferred from biotin to acetyl-CoA to form malonyl-CoA (Fig. 4A and B) (Blanchard and Waldrop, 1998; Lane and Lynen, 1963; Levert and Waldrop, 2002; Lynen, 1979). The “swinging arm” of biotin, consisting of a thumb-like loop region on BCCP, the lysine on the loop and biotin itself, is capable of moving alternatively between the BC and CT domains in order to transport the CO2 (Fig. 4B) (Perham, 2000).
Mammalian, yeast, and most other eukaryotic ACC complexes are large multifunctional enzymes containing the three components distributed in active domains on one polypeptide chain (Fig 4C) (Cronan and Waldrop, 2002). In contrast, in E. coli the three components are on four separate polypeptides that constitute the ACC complex (Fig 4C) (Cronan and Waldrop, 2002; Lane et al., 1974). This enzyme has long served as model system for biochemical and mechanistic studies of biotin-dependent carboxylases (Alberts et al., 1972; Blanchard et al., 1999b; Cronan and Waldrop, 2002).
Acyl-CoA carboxylases from actinomycetes
The first attempts to purify the ACC complex from actinomycetes resulted in the characterization of several enzymes with the ability to carboxylate not only acetyl-CoA but also other short chain acyl-CoA substrates, including propionyl- and butyryl-CoA (Erfle, 1973; Henrikson and Allen, 1979; Hunaiti and Kolattukudy, 1982). Consequently, these enzymes were referred to as acyl-CoA carboxylases (ACCase), and all of them consist of two subunits, a larger one (the α-chain) with the ability to carboxylate its covalently bound biotin group, and a smaller subunit (the β-chain) bearing the CT activity. More recently a third subunit, called ε, was also identified as part of several ACCase complexes (Diacovich et al., 2002; Gago et al., 2006). So far, this subunit has been identified only in ACCases from actinomycetes (Fig. 4C).
Genetic organization, substrate specificity and physiological role of acyl-CoA carboxylases from actinomycetes
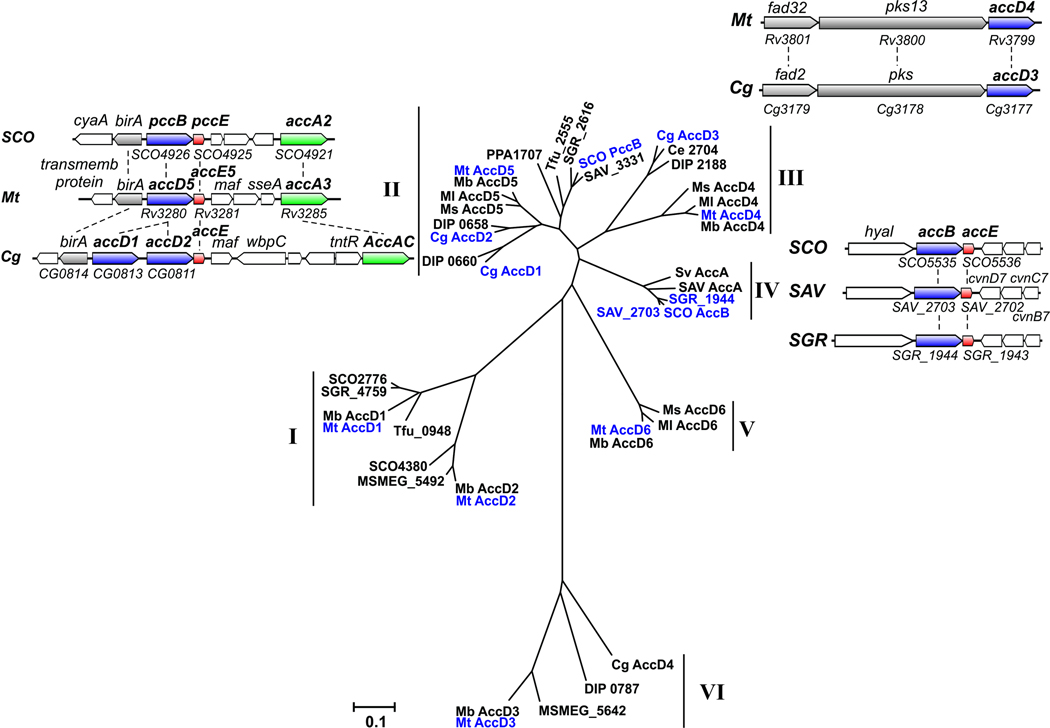
The increasing volume of sequence information revealed by the genome sequencing projects facilitated the studies of the ACCases from several actinomycetes, which rapidly revealed major aspects of their biochemical, physiological and structural properties (Daniel et al., 2007; Diacovich et al., 2002; Gago et al., 2006; Gande et al., 2004; Gande et al., 2007; Oh et al., 2006). In silico analysis of the S. coelicolor, M. tuberculosis and C. glutamicum genomes for the presence of putative ACCase components revealed a clear correlation between the number of putative ACCase genes and the diversity of lipid metabolism present in these microorganisms (Table 1). S. coelicolor, for example, has four genes encoding putative α subunits (accA1, accA2, pccA, SCO4381), four genes encoding β subunits (accB, pccB, SCO2776, SCO4380) and two genes encoding ε subunits (accE, pccE) (Bramwell et al., 1996; Diacovich et al., 2004; Rodriguez and Gramajo, 1999; Rodriguez et al., 2001). M. tuberculosis, as well as other free-living mycobacteria, also has an unusual number of genes related with putative ACCase complexes in their genomes; these genes encode three α subunits (accA1–3), six β subunits (accD1–6) (Cole et al., 1998) and one ε subunit (accE5) (Gago et al., 2006; Oh et al., 2006) (Table 1 and Fig. 5). Each of the six putative ACCases presumably serves a different physiological role, providing a vast variety of extender units for the biosynthesis of the rich diversity of mycobacterial lipids. The three genes (accA1, accA2, accA3) encoding the ACCase α subunits are highly conserved in mycobacteria, although in M. leprae only the ortholog of accA3 in M. tuberculosis, namely Ml0726, is found. This suggests that accA3 encodes the α subunit of the essential ACCase complexes. The focus in M. tuberculosis has been on the three putative essential ACCases (ACCase 4, ACCase 5 and ACCase 6) (see bellow), according to the transposon site hybridization (TraSH) analysis carried out by Sassetti et al (Sassetti et al., 2001; Sassetti and Rubin, 2003). The genome of C. glutamicum has four genes encoding the β subunits (accD1–4) of putative ACCases and only one gene encoding an α subunit (accBC) and one for an ε subunit (accE) (Table 1 and Fig. 5).
Table 1.
Composition of acyl-CoA carboxylase complexes in actinomycetes.
| Complex/ Organism |
ACCase (ACC) |
ACCase (PCC) |
ACCase (long substrate) |
ACCase? | ACCase? | ACCase? |
|---|---|---|---|---|---|---|
| Streptomyces | ||||||
| α: accA2 (accA1) | α: accA2 (accA1) | α: pccA | α: SCO4381 | |||
| β: accB | β: pccB | - | β: SCO2776 | β: SCO4380 | - | |
| ε: accE | ε: pccE | |||||
| Mycobacterium | ||||||
| α: accA3 | α: accA3 | α: accA3 | α: accA1 | α: accA2 | ||
| β: accD6 | β: accD5 | β: accD4 | β: accD1 | β: accD2 | β: accD3 | |
| ε: accE5 | β: accD5 | |||||
| Corynebacterium | ||||||
| α: accBC | α: accBC | α: accBC | ||||
| β: accD1 | β: accD2 | β: accD3 | ||||
| ε: accE | β: accD2 | - | - | β: accD4 | ||
| ε: accE |
ACCase: acyl-CoA carboxylase, ACC: acetyl-CoA carboxylase, PCC: propionyl-CoA carboxylase, α: subunit containing BC and BCCP domains, β: subunit containing CT domain, ε: small subunit necessary for maximal ACCase activity.
Fig. 5. Phylogenetic analysis and genetic organization of CTs β subunits of ACCase complexes found in actinomycetes.
The unrooted phylogenetic tree was generated using the neighbor-joining method of the freely available Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Reliability of the tree was assessed by bootstrap analysis with 1,000 replications. Bootstrap values for the different families described in the text are all over 80. The tree is drawn to scale and the lengths of the branches are proportional to the inferred evolutionary distances. The bar indicates the relative number of substitutions per site. The organism key is as follows: Ce, Corynebacterium efficiens, Cg, Corynebacterium glutamicum; DIP, Corynebacterium diphtheriae; Mb, Mycobacterium bovis; Ml, Mycobacterium leprae; Mt, Mycobacterium tuberculosis; Ms, Mycobacterium smegmatis; PPA, Propionibacterium acnes; Sa, Streptomyces antibioticus; SAV, Streptomyces avermitilis; SCO, Streptomyces coelicolor; SGR, Streptomyces griseus; Sv, Streptomyces venezuelae; Tfu, Thermobifida fusca. The genetic organization and synteny are shown for CTs that have been characterized (Mt AccD4, Mt AccD5, Cg accD1, Cg AccD2, Cg AccD3, SCO PccB, and SCO AccB).
A phylogenetic analysis, including the information for most of the predicted β subunits (CT) associated with the carboxylase group of enzymes from several actinomycetes, defined six groups that can be clearly distinguished (I to VI) (Fig. 5). Group I includes the two CTs, AccD1 and AccD2, corresponding to the Mycobacterium ACCase complexes that have been implicated in the degradative metabolism of odd-numbered fatty acids (Cole et al., 1998). The accA1 and accD1 genes encode the α and β subunits of a putative carboxylase and are organized as an operon clustered in a locus next to fadD35 and fadE10, with a partial synteny with the locus containing pccA from S. coelicolor. In addition, the accA2 (α2) and accD2 (β2) genes are in a locus together with fadE12, fadE13, and echA7. The relationship of these two enzyme complexes with β-oxidation, as proposed by Cole et al (Cole et al., 1998), is possible but still remains an open question. In Streptomyces two of the β subunits included in Group I are also organized as operons together with their putative α subunits, suggesting that they form a discrete enzyme complex. This is the case for the α and β subunits pccA and SCO2776 and for SCO4381 and SCO4380, respectively. An enzymatic PCC complex was partially characterized in S. coelicolor (Bramwell et al., 1996) consisting of a biotinylated protein, PccA, of 88 kDa, and the CT of 66 kDa (product of SCO2776). No ε subunits have been associated to this enzyme complex. To date, the putative complex formed by the proteins encoded by SCO4381 and SCO4380 has not been studied. In this group, no CT orthologs were found for corynebacteria.
Group II is the only one where each organism analyzed is represented at least once, suggesting that members of this group might be involved in fundamental functions in actynobacteria. In M. tuberculosis, the main locus comprising accA3 (α3) (Rv3285) also contains birA (biotin ligase) (Rv3279), accD5 (β5) (Rv3280) and accE5 (ε) (Rv3281). This locus is syntenous with orthologous gene sets in S. coelicolor where the genes encoding the β and ε subunits, pccB (SCO4026) and pccE (SCO4025), also organized as an operon, are found in the same locus as accA2 (SCO4021) and the biotin ligase birA (SCO4027) (Fig. 5). In C. glutamicum the accD2 (CG0811), accE (CG0810), and accBC genes are also clustered, showing a partial synteny with their corresponding genes from streptomycetes and mycobacteria that includes the gene birA (CG0814). In addition to accD2 this cluster includes a second gene, named accD1 (CG0813), encoding a β subunit highly homologous to AccD2 (identity of 69 %), suggesting that one of these genes originated by a gene duplication event (Table 1 and Fig. 5).
The first studies that integrated the biochemical and genetic analysis of the actinomycete ACCases were initiated with the identification of the α and β subunits of two ACCases from S. coelicolor named PCC (Rodriguez and Gramajo, 1999) and ACC (see below, group IV) ((Rodriguez and Gramajo, 1999; Rodriguez et al., 2001). The PCC complex is composed by PccB (β subunit), PccE (ε subunit) and the α subunits AccA1 or AccA2, which are encoded by two identical genes named accA1 (SCO6271) and accA2 (SCO4021). Kinetic studies demonstrated that this enzyme efficiently carboxylates butyryl- and propionyl-CoA, with a clear preference for the latter (Diacovich et al., 2002), but not acetyl-CoA. The ε subunit resulted in a distinctive feature of the actinomycete ACCases (Diacovich et al., 2002) and its presence dramatically stimulates the specific activity of the enzyme complexes. The ε subunits are small basic proteins (pIs between 9 and 11) that, when present, seem to be essential for the formation of a stable α-β complex. In vivo, PCC may provide methylmalonyl- and ethylmalonyl-CoA for the biosynthesis of polyketides associated with S. coelicolor. In this sense the α subunit AccA1, which is identical to AccA2 but is found in a different locus and is expressed at the transition phase (Rodriguez et al., 2001), could be part of a stationary-phase PCC complex together with PccB and PccE, generating the substrates for a type III PKS (SCO7221) that utilizes methylmalonyl- and ethylmalonyl-CoA as extender units for the biosynthesis of germicidin (Song et al., 2006). Besides, the genes encoding a type I PKS (SCO6273, SCO6274 and SCO6275) are separated from accA1 by only 2 Kb, suggesting that this PKS could also be the receptor of the α-carboxylic acids synthesized by a stationary phase ACC or PCC.
The essential complex ACCase 5 from M. tuberculosis was also reconstituted from its purified components, the α biotinylated subunit AccA3, the β subunit AccD5, and the ε subunit AccE5 (Rv3281) (Gago et al., 2006). The close phylogenetic relationship between AccD5 and PccB from S. coelicolor (Fig. 5) helped to predict the biochemical properties of this enzyme, which was later confirmed by in vitro enzyme kinetics. ACCase 5 shows a clear substrate preference for propionyl-CoA compared with acetyl-CoA (specificity constant five fold higher), indicating that the main physiological role of this enzyme is to generate methylmalonyl-CoA for the biosynthesis of branched-chain fatty acids. Also, similarly to S. coelicolor, the addition of the ε subunit, which in this case binds tightly to the α-β subcomplex and not to the β subunit as it happens in streptomycetes, is essential for obtaining maximal enzyme activity (Gago et al., 2006). Controversial results about the size of the active ε subunit of this complex have been published (Oh et al., 2006). Significantly, in the paper published by Oh et al. the activity of the ACCase 5 complex, containing an ε subunit with extra amino acids in its N-terminal sequence, resulted in an enzyme with a specific activity 300-fold lower than the enzyme complex reconstituted by Gago et al. (Gago et al., 2006) with a shorter version of the ε subunit.
The putative ACC of C. glutamicum consists of the biotinylated α-subunit AccBC (63 kDa), the β-subunit AccD1 (58 kDa) included in group II, and the small peptide AccE of 8.9 kDa (Gande et al., 2007). This enzyme complex carboxylates acetyl- and propionyl-CoA, with higher affinity for the first one. AccD1 mutants grow very poorly; however, when the medium is supplemented with sodium oleate together with butter hydrolysate, growth of the accD1 mutant is restored to normal levels, confirming a direct relationship of accD1 with fatty acid biosynthesis (Gande et al., 2004).
In group III, we only found enzymes that belong to the mycolic acid-producing branch of actinomycetes, suggesting that they represent CTs with more specialized functions. In agreement with this finding, it has been recently demonstrated that members of this group are directly involved in mycolic acid biosynthesis (Gande et al., 2004; Gande et al., 2007; Portevin et al., 2005). So far, the ACCase 4 complex of M. tuberculosis has not been well characterized at the biochemical level. The fact that accD4 (Rv3799c) is clustered with and transcribed in the same orientation as the genes pks13 (Rv3800c) and fadD32 (Rv3801c) (Fig. 5), a condensase and an acyl-AMP ligase involved in the condensation reaction leading to mycolic acid formation (Trivedi et al., 2004), suggested that AccD4 should be the β component of a long-chain acyl-CoA carboxylase that generates the α-C26 carboxylic acid involved in the last condensation step of mycolic acid biosynthesis. Moreover, phylogenetic analysis shows that AccD4 ortologs are present only in the mycolic acid containing bacteria, confirming a specific role for this CT subunit in the biosynthesis of this complex lipid. Co-immunoprecipitation studies carried out in cell-free extracts of M. smegmatis demonstrated that AccD4 interacts with the α subunit AccA3 and the β subunit AccD5 (Portevin et al., 2005), suggesting that the ACCase 4 components would be AccA3, AccD4 and AccD5. Experiments carried out in our laboratory directed to reconstitute the AccD4-AccD5 hetero-oligomers in vitro and in vivo were unsuccessful (Kurth and Gago, unpublished), while the ACCase 5 containing AccD5 as the unique β subunit was unambiguously characterized as a PCC. Consequently, more sophisticated biochemical, structural and genetic studies will be needed to characterize in detail the enzyme dedicated to provide the α chain of mycolic acids in mycobacteria. In C. glutamicum the putative ACCase involved in corynomycolic acid biosynthesis was studied in more detail. Similarly to M. smegmatis, the AccD3 and AccD2 β subunits of corynebacteria (orthologous of the AccD4 and AccD5 subunits from M. smegmatis) co-immunoprecipitated with the α subunit AccBC (the orthologous of AccA3) and the ε subunit AccE when anti-His antibodies where used to treat cell-free extracts of C. glutamicum expressing either His6-AccD2 or His6-AccD3 (Gande et al., 2007). These experiments suggested that in this microorganism the ACCase involved in providing the substrates for corynomycolic acid synthesis is formed by all these subunits. The immunoprecipitation experiments were also supported by the construction of knockout mutants in the two β subunits. The accD2 and accD3 mutations resulted in the complete loss of free and extractable mycolates without affecting phospholipid synthesis and the possible accumulation of an unknown intermediate (Gande et al., 2004). In addition, in the accD2 and accD3 mutants, the cell wall-bound mycolic acids were absent, illustrating that both genes could be involved in mycolic acid synthesis. However, the biochemical studies performed on the cell-free extracts of these mutants or on cell-free extracts of cells overexpressing either AccD2 or AccD3 were less convincing (Gande et al., 2007). Furthermore, experiments carried out in vitro with the purified components, AccBC-AccD2-AccD3-AccE, show enzyme activities barely detectable either with acetyl-CoA or palmitoyl-CoA, and curiously the assays were not conducted using propionyl-CoA as a substrate which, based on the high identity (75%) found between AccD2 of C. glutamicum and AccD5 of M. tuberculosis, would be the preferred substrate of such complex.
Mycobacteria and corynebacteria lack group IV enzymes; this is particularly noteworthy as members of this group are probably essential ACC complexes that primarily recognize acetyl-CoA as a substrate, as has been demonstrated for the AccB subunit of S. coelicolor (Diacovich et al., 2002; Rodriguez et al., 2001). In this bacterium the genes encoding the β and ε subunits accB (SCO5535) and accE (SCO5536) are clustered as an operon (Table 1 and Fig. 5). As occurs with the PCC complex (see group II), the α subunit that conform the ACC complex in Streptomyces are either AccA1 or AccA2. Consequently, the α subunits are shared by the two complexes, although the β and ε subunits of the ACC complex are AccB and AccE while for PCC the β and ε subunits are PccB and PccE, respectively. Notably, in S. coelicolor the ε subunits interact with the corresponding β subunit and are not functionally interchangeable (Diacovich et al., 2002). ACC complex was reconstituted in vitro from their purified components, and their kinetic characterization was conducted with several acyl-CoA substrates (Diacovich et al., 2002; Rodriguez and Gramajo, 1999; Rodriguez et al., 2001). This study denoted a relaxed substrate specificity of this enzyme, with a similar specificity constant for acetyl-, propionyl-, and butyryl-CoA, to provide malonyl-, methylmalonyl-, and ethylmalonyl-CoA, respectively (Diacovich et al., 2002). The primary role of the S. coelicolor ACC appears to be the generation of malonyl-CoA for the biosynthesis of fatty acids during exponential phase, and for the biosynthesis of the polyketide actinorhodin during the stationary phase of growth (Rodriguez et al., 2001). Gene disruption experiments showed that AccB and AccA2 are essential for S. coelicolor viability (Rodriguez and Gramajo, 1999), which confirmed that these subunits are part of a dedicated ACC in this microorganism.
Group V contains proteins found only in mycobacteria (Fig. 5). The CT AccD6 that belongs to this group of CT subunits was recently characterized as the β component of an essential ACC from M. smegmatis (Kurth et al., 2009), suggesting that this group of proteins could represent the CTs of the essential ACCs of mycobacteria that have diverged from other known ACCs due to the intracellular lifestyle of this group of bacteria. A complex with acetyl-CoA carboxylase activity was reconstituted in vitro with the α subunit AccA3 and the CT β subunit AccD6 (Daniel et al., 2007); this enzyme shows similar specificity for both acetyl- or propionyl-CoA and does not require a ε subunit for full activity. The physiological role of this enzyme as the ACC in charge of providing malonyl-CoA for mycolic acid biosynthesis was first predicted based on the location of accD6 (Rv2247) as part of the fasII operon (fabD-acpM-kasA-kasB-accD6) (Fig. 3A) and later confirmed in M. smegmatis by the construction of a conditional mutant in accD6 (Kurth et al., 2009). Studies carried out on this mutant allowed AccD6 to be assigned as the essential CT component of the ACCase 6 enzyme complex, whose biological role is to generate malonyl-CoA for the biosynthesis of membrane and cell envelope fatty acids by the type I and type II FAS systems. The essentiality of accD6 was also demonstrated by genetic experiments (Kurth et al., 2009).
Recently it has been suggested that members of the most distantly related group VI, including the gene encoding the CT accD3 from mycobacteria, are not directly involved in lipid biosynthesis (Gande et al., 2004). It is hard to predict their functional role since none of the other members of this group have a known activity or a predicted function (Table 1 and Fig 5). In C. glutamicum, the accD4 mutant (the accD3 orthologue of M. tuberculosis) did not exhibit a phenotype in terms of total fatty acid or mycolic acid content. However, this mutant resulted in a uniform and significant decrease in ACCase activity (14C fixation) in C. glutamicum cell-free extracts, and overexpression of accD4 led to a uniform increase in activity, in both cases independently of the substrate used (acetyl-CoA, palmitoyl-CoA or palmitic acid). Therefore, the physiological role of this CT subunit remains unknown.
A remarkable feature that arises from this sequence alignment analysis is that all of the β subunits present in groups II and IV have a putative ε subunit genetically associated with the corresponding β-subunit-encoding gene, suggesting that the new group of ACCases originally described in streptomycetes is ubiquitously present in all the actinomycetes analyzed so far. This new class of ACCases could be considered as specific targets for the development of new antimycobacterial drugs based on their distinctive structure.
In summary, the ACCase complexes from actinomycetes not only catalyze the first committed step in fatty acid biosynthesis, providing malonyl-CoA for the iterative condensation steps that occurs during the synthesis of these macromolecules, but they also catalyze the carboxylation of different substrates, such as propionyl- butyryl- or long-chain acyl-CoAs, generating precursors for the synthesis of a variety of polyketides and complex lipids in these microorganisms.
Structural studies of acyl-CoA carboxylases: identification of inhibitors
The quaternary structure of ACCase complexes has been reported for M. smegmatis (α6β6) (Haase et al., 1982), M. tuberculosis (α6β6) (Gago et al., 2006), Sacharopolyspora erythraea (α4β4) (Hunaiti and Kolattukudy, 1982), Myxococcus xanthus (α6β6) (Kimura 1998) and S. coelicolor (Diacovich et al., 2002), and in all cases the stoichiometry of the complex subunits was 1:1. The individual analysis of each subunit shows that in most cases the β subunit adopts a stable hexameric conformation (Diacovich et al., 2004; Gago et al., 2006; Lin et al., 2006), while the α subunit is found both as a trimer and as a hexamer (Gago et al., 2006). As we mentioned above, some of these complexes need an ε subunit for full enzyme activity (Diacovich et al., 2002; Gago et al., 2006; Oh et al., 2006; Rodriguez et al., 2001), but the exact molar ratio between this subunit and the α and β subunits has not been determined.
Due to the instability of the bacterial ACCs, the structural details of the different subunits were studied separately. First, the crystal structure of the BC component of E. coli ACC was solved (Waldrop et al., 1994), and this was followed by the crystal structure of the biotinoyl domain of E. coli BCCP (Athappilly and Hendrickson, 1995) and the CT domain (Bilder et al., 2006). Other CT domains from biotin-dependent carboxylases were also solved by X-ray crystallography, including the 1.3S transcarboxylase subunit (Reddy et al., 2000) and the 12S carboxyltransferase (for pyruvate carboxylation) (Hall et al., 2003) of P. Shermanii, the tetrameric pyruvate carboxylase (Xiang and Tong, 2008; Yu et al., 2009), and the glutaconyl-CoA decarboxylase (for sodium ion pump) (Wendt et al., 2003).
The crystal structures of S. coelicolor AccB and PccB
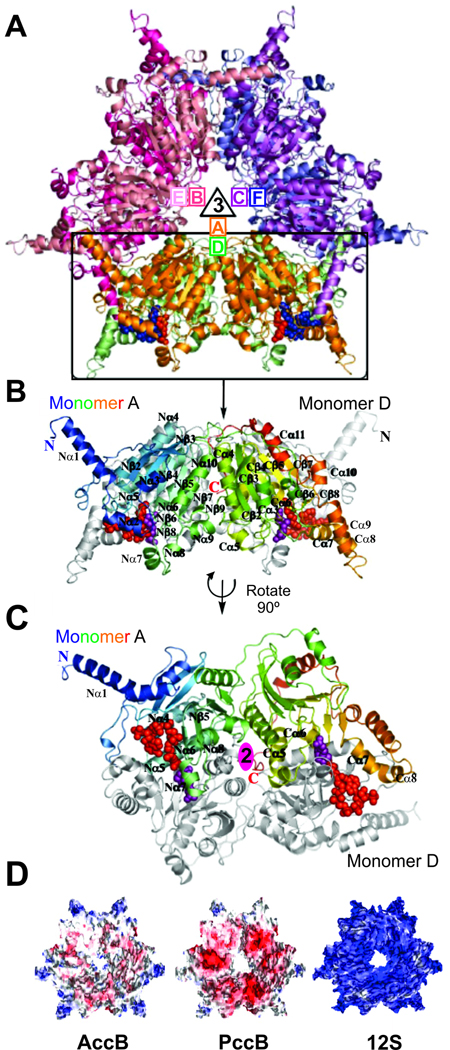
PccB is a 65 kDa protein that forms a hexamer in solution and in the crystal structure (Fig. 6A). The hexameric stoichiometry of PccB implies an overall stoichiometry of α6β6ε? for the holo complex, with a molecular weight of > 1 Mda. Similar to the Propionibacterium shermanii 12S transcarboxylase (Hall et al., 2003), the PccB hexamer has a 32-fold symmetry, where a 3-fold axis is perpendicular to a 2-fold axis (Fig. 6A). Specifically, A-B-C and D-E-F monomers form two tightly interacting trimer rings, while A–D, B–E, and C–F dimers interact extensively (Fig. 6A). Like 12S and yeast-CT (Hall et al., 2003; Zhang et al., 2003), PccB also reveals two domains (N- and C-domains) in each monomer, and both domains have a crotonase fold that consists of seven β-sheets and α-helices (Fig. 6B and C) (Holden et al., 2001).
Fig. 6. Crystal structure of PccB from S. coelicolor.
(A) The overall hexameric structure of PccB formed by two stacks of three monomers (A-B-C and D-E-F) related by the 3-fold axis. (B) Monomer A of PccB, colored from the N-terminus (blue) to the C-terminus (red), with two structurally similar domains (N and C). Monomer D is shown in black and white under monomer A. (C) The dimeric, di-domain interaction between monomer A and monomer D (after a 90° rotation) of PccB, related by the 2-fold axis. (D) Electronegative surfaces of AccB, PccB, and 12S (Diacovich et al., 2004; Hall et al., 2003) reveal key differences between these very similar CTs. Surfaces are colored according to their charges from red (−) to white to blue (+).
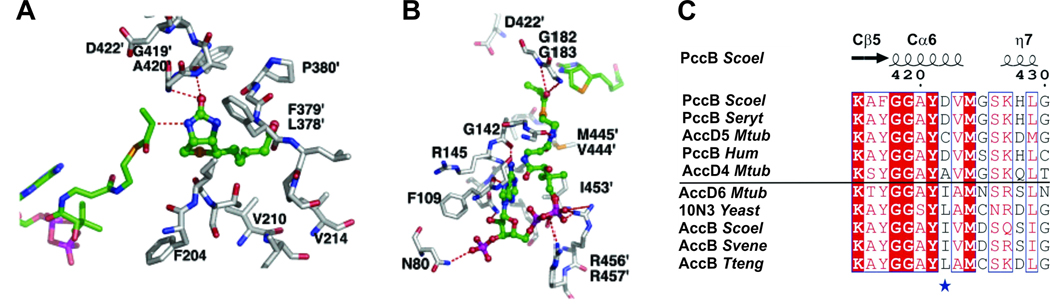
The biotin and acyl-CoA binding pockets of CTs are identified by the co-crystal structure of PccB bound with the substrate biotin and propionyl-CoA (Diacovich et al., 2004) (Fig. 7A). The binding pockets of acyl-coenzyme A and biotin are perpendicular to each other and meet at the junction of the L-shaped active site, near the intersection of Nβ5-Nα6 of one monomer and Cβ5-Cα6 of its dimeric partner (Fig. 7A and B), and shows that acyl-CoA binds predominantly from the N domain of the first monomer, while biotin binds from the C domain of its dimeric partner. Such a dimeric, di-domain active site arrangement is conserved across the biotin-dependent CTs, including PccB, 12S (a decarboxylase) (Hall et al., 2003), GCD (a sodium-transport decarboxylase) (Wendt et al., 2003) and yeast-CT (the β-subunit of ACC) (Zhang et al., 2003). Based on the biotin-propionyl-CoA co-crystal structure of PccB and its comparison with other CT structures, the binding motif of acyl-CoA and biotin to CT should be ubiquitous among biotin-based CTs, where dimeric, di-domain interactions are crucial for substrate binding. Although the overall structures of PccB, AccB (Tsai, unpublished results) and 12S are similar, there is a large difference in surface properties among these CTs (Fig. 6D). This may reflect a major difference in protein-protein interaction and in their corresponding biochemical roles.
Fig. 7. PccB active site.
(A) The binding pockets of acyl-CoA and biotin in PccB are perpendicular to each other and meet at the junction of the L-shaped active site. (B) The binding pocket of propionyl-CoA reveals that residue 422, at the end of the pocket, is the only residue different in AccB and PccB. (C) Sequence comparison of different CTs that accept propionyl-CoA as a substrate, like PccB, where position 422 (in PccB) is occupied by small residues (blue star), such as Asp or Cys, and CTs that accept acetyl-CoA as a substrate, such as AccB, where this residue is a larger, hydrophobic residue. Scoel (S. coelicolor), Seryt (S. erythraea), Mtub (M. tuberculosis), Hum (Human), Yeast, Svene (S. venezuelae), Tteng (T. tengcongensis).
Structure-based mutation: molecular basis of substrate specificity of S. coelicolor ACCases
The molecular basis of substrate specificity, i.e. how the CTs differentiate acetyl- from propionyl-CoA, has been addressed with the S. coelicolor β subunits AccB and PccB. Significantly, nearly all residues in the acyl-CoA binding pocket are conserved between AccB and PccB, except residue 422 of PccB (420 in AccB), which lies at the end of the acyl-CoA binding pocket (Fig. 7C). Residue 422 is an Asp in PccB and an Ile (420) in AccB. Sequence comparison indicates that for different CTs that accept propionyl-CoA as a substrate, from S. coelicolor to the human PccB, this position is occupied by small residues, such as Asp or Cys (Fig. 7C). On the other hand, for CTs that accept acetyl-CoA as a substrate, such as AccB, this residue is a larger hydrophobic residue. This led to the hypothesis that the amino acid at this position may help to define the shape of the acyl-CoA binding pocket. To test this hypothesis, the D422I:PccB and I420D:AccB mutants were compared. Wild type PccB does not accept acetyl-CoA as a substrate; however the D422I mutant of PccB should make the active site into a “mock AccB” to accept acetyl-CoA. The PccB D422I mutant not only accepted acetyl-CoA as its substrate, but also exhibited similar substrate specificity as wild type AccB for propionyl- and butyryl-CoA. Further, the AccB I420D mutant (the “mock PccB”), cannot accept acetyl-CoA as its substrate (Diacovich et al., 2004). In essence, by a single mutation in the active site, the substrate specificities of mutant ACC and PCC have been interchanged. Systematic mutations of PccB D422 and AccB I420 further confirmed the importance of this residue for substrate specificity (Arabolaza et al., 2010b). In summary, the above results confirmed that the CT domain is the major determinant for acyl-CoA specificity.
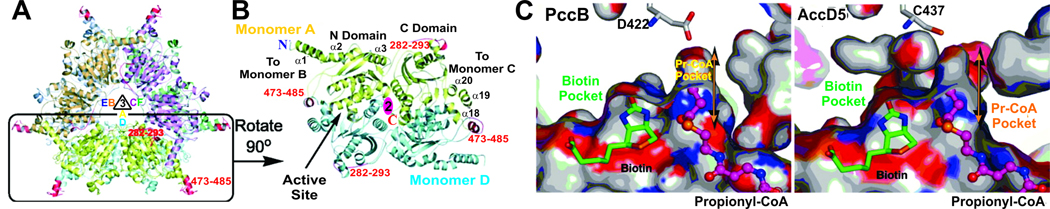
The crystal structure of M. tuberculosis AccD5
As mentioned above, M. tuberculosis contains six ACCase β subunits (AccD1–6) (Cole et al., 1998). In order to determine the molecular basis of substrate specificity for ACCase5 complex, a crystal structure of AccD5 was solved at 2.9 Å resolution (Lin et al., 2006). AccD5 consists of a 360-kDa hexamer (Fig. 8A), similar to the S. coelicolor PccB (Diacovich et al., 2004); however AccD5 differs from the dimeric yeast CT domains (Zhang et al., 2003). Similarly to PccB, the dimeric interface is also conserved in AccD5 (Fig. 8B). All of the biotin-binding residues are conserved between AccD5 and PccB. Significantly, residues in the acyl-CoA binding pocket are also highly conserved surrounding the acyl, pantetheine, ribose and adenine ring, while residues that interact with the bis-phosphate group, such as Q472, R471, and F468, are partially conserved among AccD4–6 and PccB (Lin et al., 2006) (Diacovich et al., 2004).
Fig. 8. Crystal structure of AccD5 from M. tuberculosis.
(A) The overall hexameric fold of AccD5 is similar to that of PccB. (B) The active site lies at the dimeric, di-domain interface. (C) A comparison of the active sites between PccB and AccD5 shows that AccD5 cannot accommodate a substrate larger than propionyl-CoA (Lin et al., 2006)
Significantly, the AccD5 crystal structure shows that the active site cannot accommodate an acyl chain with more than three carbons (Fig. 8C). Contrary to the previous proposal that the AccD4–5 complexes accept long-chain acyl-CoAs as their substrates (Gande et al., 2004), both crystal structure and kinetic assay consistently showed that AccD5 preferentially accepts propionyl-CoA as its substrate, producing methylmalonyl-CoA, the substrate for the biosynthesis of multimethyl-branched fatty acids such as mycocerosic, phthioceranic, hydroxyphthioceranic, mycosanoic and mycolipenic acids (Minnikin et al., 2002; Takayama et al., 2005), many of which are important factors for virulence, survival, multi-drug resistence, and latency development.
Structure-based inhibitor design of M. tuberculosis ACCases
Acetyl-CoA carboxylases have crucial roles in fatty acid metabolism and are attractive targets for drug discovery against a variety of human diseases, including diabetes, obesity, cancer and microbial infections. ACCs have also been the targets of herbicides that are currently used as commercial products. The reader can find excellent reviews on these topics elsewhere (Gronwald, 1994; Heath et al., 2001; Levert and Waldrop, 2002; Pacheco-Alvarez et al., 2002; Shen et al., 2004; Tong, 2005; Tong and Harwood, 2006).
The first antibacterial compounds that targeted the ACC were the pseudopeptidic pyrrolidinedione natural products moiramide B and andrimid that selectively inhibit the ACC-carboxyltransferase reaction (Freiberg et al., 2004; Pohlmann et al., 2005) and synthetic variations of each moiety of the modularly composed pyrrolidinediones have showed highly improved activities against gram-positive bacteria compared to those of previously reported variants. Unfortunately, no studies have been published addressing the efficacy of these compounds in actinobacteria. As an effort to find inhibitors for the ACCases of M. tuberculosis a structure-based in silico screening was conducted with UC Irvine ChemDB database (Chen et al., 2005) and the crystal structure of AccD5 (Lin et al., 2006). More than 100 putative AccD5 inhibitors were identified and assayed in vitro for inhibition of AccD5 and for the whole ACCase 5 enzyme complex. One of the inhibitors, NCI-65828, was found to be a competitive inhibitor of AccD5 with a KI of 13.1 µM (Lin et al., 2006). Based on the conserved structure of the AccD5 and AccD6 active sites, several inhibitors of AccD5 were also screened as potential inhibitors of AccD6 and it was found that the ligand NCI-172033 was capable of inhibiting AccD6 with a KI of 8 µM (Kurth et al., 2009). This compound also showed bactericidal activity against several pathogenic mycobacteria by causing a strong inhibition of both fatty acid and mycolic acid biosynthesis at minimal inhibitory concentrations. Overexpression of accD6 in M. smegmatis conferred resistance to NCI-172033, confirming AccD6 as the main target of the inhibitor and as a target for the development of new antimycobacterial agents (Kurth et al., 2009)
A key issue of structure-based inhibitor design is the versatility of the docking software and the lack of self-improving ability of the most commonly used docking softwares (Anderson, 2003). Building on the above promising results, the enzymatic assays of AccD5 and AccD6 were combined with virtual high-throughput screening, aiming to discover new AccD5 and AccD6 inhibitors as potential tuberculosis drugs (Swamidass et al., 2009). Using a new method (Influence Relevance Voter, IRV), the in silico screening program was turned into an artificial intelligence that self-improves by feeding the enzyme inhibition assay data back to the program. In this way, IRV achieves significantly more accurate prediction than other traditional screening programs. Further, IRV retrieves three times as many experimentally proven “hits” in its prediction-sorted list. This successfully developed “smart” program allows feedback of actual experimental data to improve its prediction power, which leads to improved inhibitors against AccD5 and AccD6. These results pave the way towards structure-based development of anti-tuberculosis therapeutics.
Controlling lipid homeostasis in actinomycetes
The de novo fatty acid biosynthetic pathway is a major focal point for the regulatory events that control lipid homeostasis. Regulation at the level of fatty acid biosynthesis is crucial not only because the biophysical properties of membranes are determined in large part by the composition of the fatty acids that are produced by de novo biosynthesis, but also for the metabolic energy that is expended in the formation of these molecules. Therefore, fatty acid production must be precisely controlled to support membrane biogenesis and prevent the wasteful expenditure of ATP.
Although significant progress has been made in recent years, most of the regulatory mechanisms that control fatty acid metabolic pathways have been elucidated in model bacteria, such as E. coli (Cronan and Subrahmanyam, 1998; DiRusso and Nystrom, 1998; Rock and Cronan, 1996; Zhang et al., 2006) and Bacillus subtilis (Matsuoka et al., 2007; Schujman et al., 2003; Schujman et al., 2006). These investigations revealed that this process involves both a biochemical control of key enzymes as well as a transcriptional regulation of the lipid genes. There is a rapid response of the integrated biochemical network, acting at the level of the activities of lipid biosynthetic enzymes and a central control at the level of gene expression involving specific transcription factors.
Regulation of expression of the genes for fatty acid synthesis is complex (Zhang and Rock, 2008). Currently, six transcriptional regulators have been identified, four of them - FadR (E. coli), DesR (B. subtilis), FabR (E. coli) and DesT (P. aeruginosa) - adjust the levels of unsaturated fatty acids in membrane phospholipids, while FapR (B. subtilis) and FabT (S. pneumonieae) are global transcriptional repressors that simultaneously control the expression of many genes involved in fatty acid and phospholipid metabolism (Cronan and Subrahmanyam, 1998; Lu and Rock, 2006; Schujman et al., 2003; Schujman et al., 2006; Schujman and de Mendoza, 2008; Zhang and Rock, 2009). An extensive bioinformatic analysis carried out in different actinomycetes genome databases demonstrated that these microorganisms lack FadR, FapR or FabR homologues. However, the existence of a transcriptional control of the genes required for fatty acid biosynthesis was recently reported both in S. coelicolor and in M. tuberculosis (Arabolaza et al., 2010a; Salzman et al., 2010). These studies describe the characterization of a novel type of transcription factor widely distributed among the actinobacteria (see below).
In most bacteria, the fatty acids produced by the biosynthetic pathway are incorporated exclusively into the membrane. The actinomycetes represent an exception since the product of microbial fatty acid biosynthesis, long chain acyl-ACP, can be directed to other cellular components such as complex structural lipids (e.g. mycolic acids or PDIM) or storage lipids (triacylglycerols). This feature adds a higher level of sophistication to the regulation of lipid homeostasis in these bacteria and the mechanisms involved in this global coordination are only recently coming to be understood.
Biochemical control of lipid biosynthesis in actinomycetes
While fatty acid biosynthesis has been reasonably well characterized in some genera of actinobacteria, the mechanisms that regulate the production of fatty acids and lipid molecules, either at the biochemical or at the genetic levels, are largely unknown. Only recent reports proposed a model for mycolic acid synthesis regulation based on the phosphorylation of KasA, KasB, FabH and MabA (Molle et al., 2006; Veyron-Churlet et al., 2009; Veyron-Churlet et al., 2010). Protein phosphorylation/dephosphorylation is a central mechanism for transduction of specific signals to various cellular processes, such as regulation of growth, differentiation, mobility and survival (Stock et al., 1989); in this sense these works represent the first evidence of this kind of regulation for the lipid metabolism. KasA and KasB are phosphorylated at high rates in vivo in Mycobacterium bovis BCG, mainly at two positions, and can be dephosphorylated by the mycobacterial Ser/Thr phosphatase PstP. In vitro experiments demonstrated that phosphorylation of KasA and KasB differentially modulates their condensing activities in the presence of either malonyl-ACP or C16-ACP (Molle et al., 2006). MtFabH is efficiently phosphorylated both in vitro and in vivo by several mycobacterial STPKs (serine, threonine and tyrosine kinases), particularly by PknF and PknA (Veyron-Churlet et al., 2009). A mutant variant of mtFabH, constructed to mimic constitutive phosphorylation, exhibited a marked decrease in transacylation, malonyl-ACP decarboxylation and condensing activities compared with the wild-type protein. MabA is also phosphorylated in vitro, as well as in vivo, by several M. tuberculosis STPKs (Veyron-Churlet et al., 2010). In this work, the authors provide a fundamental insight into the molecular mechanism(s) controlling mycolic acid biosynthesis through STPK-dependent phosphorylation of a FAS II enzyme. Thus the mycobacterial STPKs appear to play an important role in regulating the metabolism of mycolic acids, although whether phosphorylation of these proteins regulates the mycolic acid profile in vivo in response to environmental changes, and/or affects M. tuberculosis virulence in infected mice, has yet to be demonstrated.
On other hand, the Ser/Thr/Tyr phosphorylation phosphoproteome in C. glutamicum revealed that most of the phosphorylated proteins identified are enzymes acting on central metabolic pathways such as glycolysis, tricarboxylic cycle, fatty-acid metabolism and components of the protein synthesis machinery (Bendt et al., 2003). Particularly, an acyl-CoA synthetase (NgCl2774) and the acyl-CoA carboxylase α subunit (accBC) were found to be phosphorylated; however how this post-translational modification affects their enzyme activities and how this influences lipid metabolism is not known.
Transcriptional control of the fatty acid biosynthetic genes in actinomycetes
A major advance in the recognition of a transcriptional control of actinobacterial lipid synthesis occurred by the identification of FasR and MabR; two transcription factors that regulate the expression of the main fab operon in S. coelicolor and M. tuberculosis, respectively (Arabolaza et al., 2010a; Salzman et al., 2010). These novel regulators are highly conserved in most of the actinomycete genomes sequenced to date, including Arthrobacter, Frankia, Rhodococcus, Nocardia, Acidothermus, Bifidobacterium, Streptomyces and Mycobacterium, and the coding genes lie immediately upstream of their corresponding fab operons (Fig. 3A). An exception to this situation occurs in Corynebacterium, where no type II FAS exists and therefore no homologues to these transcription factors. The 21-bp imperfect palindromic motif (5´-TTTTGT-N9-ACAAAA-3´) identified as the regulator binding site was found to be highly conserved within actinomycetes and always associated with fab operons. Remarkably, although the binding motifs of S. coelicolor and M. tuberculosis lie at a comparable distance from the 5´ end of the first gene of the fab operon, at approximately −65 to −80 bp from the TSS (Fig. 3A), the binding of FasR to its operator sequence results in an activation of the fab genes in S. coelicolor (Arabolaza et al., 2010a) while the binding of MabR to its operator sequence results in a repression of these genes in M. tuberculosis (Salzman et al., 2010). Furthermore, these orthologues show another important difference. TraSH (transposon site hybridization) experiments carried out in M. tuberculosis suggested that mabR is an essential gene (Sassetti et al., 2001), and Salzman et al. (Salzman et al., 2010) demonstrated that the chromosomal copy of M. smegmatis mabR could only be disrupted in the presence of a wild-type copy of the gene provided in trans, giving a rigorous proof of the essentiality of this gene in mycobacteria. In sharp contrast, FasR has been shown to be non-essential for the survival of S. coelicolor (Arabolaza et al., 2010a). Thus, the evolutionary divergence found for this regulatory protein in S. coelicolor and in M. tuberculosis could be correlated with the different physiological roles of the FAS systems that each of them regulates.
Concluding Remarks
Actinomycetes include a very interesting group of bacteria that are ubiquitous in nature and well known for their distinct impacts on mankind. In actinomycetes, lipid metabolism is relevant not only for the generation of cell membrane phospholipids but also for the production of PK compounds with useful pharmaceutical properties, for the generation of storage compounds (TAG) with possible technological applications and for the synthesis of complex lipids, crucial for the permeability and pathogenicity of some pathogenic species. Therefore, the identification and further biochemical and structural characterization of each of the components of the fatty acid biosynthetic pathways, as well as understanding the regulation of lipid homeostasis, should have a major impact on the biology, biotechnology and disease control related with each of the organisms discussed in this review.
The genomic information available for several species of corynebacteria, mycobacteria and streptomycetes provides excellent opportunities for comparative genome analysis and for the extrapolation of gene functions between these genera. We have shown that several genes involved in fatty acid metabolism can be identified and analyzed in this way. This information when complemented with transcriptomic, proteomic and metabolomic data should result in a major step toward the understanding of this essential but poorly understood metabolic pathway in this group of organisms.
One of the novel aspects highlighted by this review in relation to lipid metabolism is a comparative analysis of the enzymes (ACCases) that catalyze the carboxylation of different acyl-CoAs and whose products are utilized not only in fatty acid biosynthesis but also in the synthesis of complex lipids and in the production of PK compounds. ACCases from actinomycetes present unique characteristics within this group of enzymes at both the biochemical and structural levels. The wide diversity of substrates recognized by the ACCase β subunits is an example of a remarkable divergent evolution of this class of enzymes, and provides great opportunities not only for understanding the molecular bases that dictate substrate specificity of the CTs but also for the use of these enzymes in protein engineering for the generation of new substrates for polyketide biosynthesis as well as the use of some of them as attractive target for the design of novel antibacterial drugs (Cronan and Waldrop, 2002; Heath et al., 2001). For instance, studies combining the structural data of some of the essential CTs subunits of M. tuberculosis with in silico screening of compounds available at the NCI database allowed the identification and characterization of two structurally different CT inhibitors, one of them with interesting bactericidal activity against MDR M. tuberculosis. In the future, investigation of fatty acid biosynthesis and its regulation in actinomycetes could open new avenues for therapeutics.
Acknowledgements
We are grateful to Prof. David Hopwood for helpful comments on the manuscript. Research support was provided by ANPCyT grant 01-13705 and NIH 1R03TW007982-02.
References
- Alberts AW, Nervi AM, Vagelos PR. Acetyl CoA carboxylase, II. Demonstration of biotin-protein and biotin carboxylase subunits. Proc Natl Acad Sci U S A. 1969;63:1319–1326. doi: 10.1073/pnas.63.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts AW, Bell RM, Vagelos PR. Acyl carrier protein. XV. Studies of - ketoacyl-acyl carrier protein synthetase. J Biol Chem. 1972;247:3190–3198. [PubMed] [Google Scholar]
- Anderson AC. The process of structure-based drug design. Chem Biol. 2003;10:787–797. doi: 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Arabolaza A, Rodriguez E, Altabe S, Alvarez H, Gramajo H. Multiple pathways for triacylglycerol biosynthesis in Streptomyces coelicolor. Appl Environ Microbiol. 2008;74:2573–2582. doi: 10.1128/AEM.02638-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabolaza A, D'Angelo M, Comba S, Gramajo H. FasR, a novel class of transcriptional regulator, governs the activation of fatty acid biosynthesis genes in Streptomyces coelicolor. Mol Microbiol. 2010a;78:47–63. doi: 10.1111/j.1365-2958.2010.07274.x. [DOI] [PubMed] [Google Scholar]
- Arabolaza A, Shillito ME, Lin TW, Diacovich L, Melgar M, Pham H, Amick D, Gramajo H, Tsai SC. Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor. Biochemistry. 2010b;49:7367–7376. doi: 10.1021/bi1005305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarie-Dequeker C, Le Guyader L, Malaga W, Seaphanh FK, Chalut C, Lopez A, Guilhot C. Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog. 2009;5:e1000289. doi: 10.1371/journal.ppat.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athappilly FK, Hendrickson WA. Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing. Structure. 1995;3:1407–1419. doi: 10.1016/s0969-2126(01)00277-5. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR., Jr inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- Bendt AK, Burkovski A, Schaffer S, Bott M, Farwick M, Hermann T. Towards a phosphoproteome map of Corynebacterium glutamicum. Proteomics. 2003;3:1637–1646. doi: 10.1002/pmic.200300494. [DOI] [PubMed] [Google Scholar]
- Bhatt A, Kremer L, Dai AZ, Sacchettini JC, Jacobs WR., Jr Conditional depletion of KasA, a key enzyme of mycolic acid biosynthesis, leads to mycobacterial cell lysis. J Bacteriol. 2005;187:7596–7606. doi: 10.1128/JB.187.22.7596-7606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A, Fujiwara N, Bhatt K, et al. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc Natl Acad Sci U S A. 2007;104:5157–5162. doi: 10.1073/pnas.0608654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder P, Lightle S, Bainbridge G, et al. The structure of the carboxyltransferase component of acetyl-coA carboxylase reveals a zinc-binding motif unique to the bacterial enzyme. Biochemistry. 2006;45:1712–1722. doi: 10.1021/bi0520479. [DOI] [PubMed] [Google Scholar]
- Blanchard CZ, Waldrop GL. Overexpression and kinetic characterization of the carboxyltransferase component of acetyl-CoA carboxylase. J Biol Chem. 1998;273:19140–19145. doi: 10.1074/jbc.273.30.19140. [DOI] [PubMed] [Google Scholar]
- Blanchard CZ, Amspacher D, Strongin R, Waldrop GL. Inhibition of biotin carboxylase by a reaction intermediate analog: implications for the kinetic mechanism. Biochem Biophys Res Commun. 1999a;266:466–471. doi: 10.1006/bbrc.1999.1844. [DOI] [PubMed] [Google Scholar]
- Blanchard CZ, Chapman-Smith A, Wallace JC, Waldrop GL. The biotin domain peptide from the biotin carboxyl carrier protein of Escherichia coli acetyl-CoA carboxylase causes a marked increase in the catalytic efficiency of biotin carboxylase and carboxyltransferase relative to free biotin. J Biol Chem. 1999b;274:31767–31769. doi: 10.1074/jbc.274.45.31767. [DOI] [PubMed] [Google Scholar]
- Bloch K, Vance D. Control mechanisms in the synthesis of saturated fatty acids. Annu Rev Biochem. 1977;46:263–298. doi: 10.1146/annurev.bi.46.070177.001403. [DOI] [PubMed] [Google Scholar]
- Bramwell H, Hunter IS, Coggins JR, Nimmo HG. Propionyl-CoA carboxylase from Streptomyces coelicolor A3(2): cloning of the gene encoding the biotin-containing subunit. Microbiology. 1996;142:649–655. doi: 10.1099/13500872-142-3-649. [DOI] [PubMed] [Google Scholar]
- Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2003;83:91–97. doi: 10.1016/s1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- Brown AK, Sridharan S, Kremer L, Lindenberg S, Dover LG, Sacchettini JC, Besra GS. Probing the mechanism of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III mtFabH: factors influencing catalysis and substrate specificity. J Biol Chem. 2005;280:32539–32547. doi: 10.1074/jbc.M413216200. [DOI] [PubMed] [Google Scholar]
- Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- Cohen-Gonsaud M, Ducasse S, Hoh F, Zerbib D, Labesse G, Quemard A. Crystal structure of MabA from Mycobacterium tuberculosis, a reductase involved in long-chain fatty acid biosynthesis. J Mol Biol. 2002;320:249–261. doi: 10.1016/S0022-2836(02)00463-1. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Collins MD, Goodfellow M, Minnikin DE. Fatty acid composition of some mycolic acid-containing coryneform bacteria. J Gen Microbiol. 1982;128:2503–2509. doi: 10.1099/00221287-128-11-2503. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- Cronan JE, Rock CO. Biosynthesis of membrane lipids. In: Neidhardt F, editor. Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 612–636. [Google Scholar]
- Cronan JE, Rock CO. Biosynthesis of membrane lipids. In: Böck A, Curtiss R 3rd, Kaper J, Karp P, Neidhardt F, Nyström T, Slauch J, Squires C, Ussery D, editors. EcoSal, Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press; 2008. Chapter 3.6.4. [Google Scholar]
- Cronan JE, Jr, Subrahmanyam S. FadR, transcriptional co-ordination of metabolic expediency. Mol Microbiol. 1998;29:937–943. doi: 10.1046/j.1365-2958.1998.00917.x. [DOI] [PubMed] [Google Scholar]
- Cronan JE., Jr The biotinyl domain of Escherichia coli acetyl-CoA carboxylase. Evidence that the "thumb" structure id essential and that the domain functions as a dimer. J Biol Chem. 2001;276:37355–37364. doi: 10.1074/jbc.M106353200. [DOI] [PubMed] [Google Scholar]
- Cronan JE, Jr, Waldrop GL. Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- Chater K, Losick R. The mycelial life-style of Streptomyces coelicolor A3(2) and its relatives. In: Shapiro J, Dworkin M, editors. Bacteria as Multicellular Organisms. Oxford University Press; 1997. pp. 149–182. [Google Scholar]
- Chen J, Swamidass SJ, Dou Y, Bruand J, Baldi P. ChemDB: a public database of small molecules and related chemoinformatics resources. Bioinformatics. 2005;21:4133–4139. doi: 10.1093/bioinformatics/bti683. [DOI] [PubMed] [Google Scholar]
- Daffe M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- Daffe M, Reyrat JM. In: The Mycobacterial Cell Envelope. Daffe M, Reyrat JM, editors. Washington, D.C: ASM Press; 2008. [Google Scholar]
- Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Oh TJ, Lee CM, Kolattukudy PE. AccD6, a member of the Fas II locus, is a functional carboxyltransferase subunit of the acyl-coenzyme A carboxylase in Mycobacterium tuberculosis. J Bacteriol. 2007;189:911–917. doi: 10.1128/JB.01019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa M, Gambacorta A, Gliozzi A. Structure, biosynthesis, and physicochemical properties of archaebacterial lipids. Microbiol Rev. 1986;50:70–80. doi: 10.1128/mr.50.1.70-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacovich L, Peiru S, Kurth D, Rodriguez E, Podesta F, Khosla C, Gramajo H. Kinetic and structural analysis of a new group of Acyl-CoA carboxylases found in Streptomyces coelicolor A3(2) J Biol Chem. 2002;277:31228–31236. doi: 10.1074/jbc.M203263200. [DOI] [PubMed] [Google Scholar]
- Diacovich L, Mitchell DL, Pham H, Gago G, Melgar MM, Khosla C, Gramajo H, Tsai SC. Crystal structure of the beta-subunit of acyl-CoA carboxylase: structure-based engineering of substrate specificity. Biochemistry. 2004;43:14027–14036. doi: 10.1021/bi049065v. [DOI] [PubMed] [Google Scholar]
- DiRusso CC, Nystrom T. The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol Microbiol. 1998;27:1–8. doi: 10.1046/j.1365-2958.1998.00645.x. [DOI] [PubMed] [Google Scholar]
- Erfle JD. Acetyl-CoA and propionyl-CoA carboxylation by Mycobacterium phlei. Partial purification and some properties of the enzyme. Biochim Biophys Acta. 1973;316:143–155. doi: 10.1016/0005-2760(73)90004-0. [DOI] [PubMed] [Google Scholar]
- Florova G, Kazanina G, Reynolds KA. Enzymes involved in fatty acid and polyketide biosynthesis in Streptomyces glaucescens: role of FabH and FabD and their acyl carrier protein specificity. Biochemistry. 2002;41:10462–10471. doi: 10.1021/bi0258804. [DOI] [PubMed] [Google Scholar]
- Freiberg C, Brunner NA, Schiffer G, Lampe T, Pohlmann J, Brands M, Raabe M, Habich D, Ziegelbauer K. Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors with antibacterial activity. J Biol Chem. 2004;279:26066–26073. doi: 10.1074/jbc.M402989200. [DOI] [PubMed] [Google Scholar]
- Gago G, Kurth D, Diacovich L, Tsai SC, Gramajo H. Biochemical and structural characterization of an essential acyl coenzyme A carboxylase from Mycobacterium tuberculosis. J Bacteriol. 2006;188:477–486. doi: 10.1128/JB.188.2.477-486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gande R, Gibson KJ, Brown AK, Krumbach K, Dover LG, Sahm H, Shioyama S, Oikawa T, Besra GS, Eggeling L. Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in Corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J Biol Chem. 2004;279:44847–44857. doi: 10.1074/jbc.M408648200. [DOI] [PubMed] [Google Scholar]
- Gande R, Dover LG, Krumbach K, Besra GS, Sahm H, Oikawa T, Eggeling L. The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. J Bacteriol. 2007;189:5257–5264. doi: 10.1128/JB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao LY, Laval F, Lawson EH, Groger RK, Woodruff A, Morisaki JH, Cox JS, Daffe M, Brown EJ. Requirement for kasB in Mycobacterium mycolic acid biosynthesis, cell wall impermeability and intracellular survival: implications for therapy. Mol Microbiol. 2003;49:1547–1563. doi: 10.1046/j.1365-2958.2003.03667.x. [DOI] [PubMed] [Google Scholar]
- Gebhardt H, Meniche X, Tropis M, Kramer R, Daffe M, Morbach S. The key role of the mycolic acid content in the functionality of the cell wall permeability barrier in Corynebacterineae. Microbiology. 2007;153:1424–1434. doi: 10.1099/mic.0.2006/003541-0. [DOI] [PubMed] [Google Scholar]
- Gokhale RS, Sankaranarayanan R, Mohanty D. Versatility of polyketide synthases in generating metabolic diversity. Curr Opin Struct Biol. 2007a;17:736–743. doi: 10.1016/j.sbi.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Gokhale RS, Saxena P, Chopra T, Mohanty D. Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat Prod Rep. 2007b;24:267–277. doi: 10.1039/b616817p. [DOI] [PubMed] [Google Scholar]
- Greenspan MD, Alberts AW, Vagelos PR. Acyl carrier protein. 13. Beta-ketoacyl acyl carrier protein synthetase from Escherichia coli. J Biol Chem. 1969;244:6477–6485. [PubMed] [Google Scholar]
- Gronwald JW. Herbicides inhibiting acetyl-CoA carboxylase. Biochem Soc Trans. 1994;22:616–621. doi: 10.1042/bst0220616. [DOI] [PubMed] [Google Scholar]
- Haase FC, Henrikson KP, Treble DH, Allen SH. The subunit structure and function of the propionyl coenzyme A carboxylase of Mycobacterium smegmatis. J Biol Chem. 1982;257:11994–11999. [PubMed] [Google Scholar]
- Hall PR, Wang YF, Rivera-Hainaj RE, Zheng X, Pustai-Carey M, Carey PR, Yee VC. Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core. EMBO J. 2003;22:2334–2347. doi: 10.1093/emboj/cdg244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Lobo S, Reynolds KA. Characterization of beta-ketoacyl-acyl carrier protein synthase III from Streptomyces glaucescens and its role in initiation of fatty acid biosynthesis. J Bacteriol. 1998;180:4481–4486. doi: 10.1128/jb.180.17.4481-4486.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RJ, White SW, Rock CO. Lipid biosynthesis as a target for antibacterial agents. Prog Lipid Res. 2001;40:467–497. doi: 10.1016/s0163-7827(01)00012-1. [DOI] [PubMed] [Google Scholar]
- Henrikson KP, Allen SH. Purification and subunit structure of propionyl coenzyme A carboxylase of Mycobacterium smegmatis. J Biol Chem. 1979;254:5888–5891. [PubMed] [Google Scholar]
- Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- Holden HM, Benning MM, Haller T, Gerlt JA. The crotonase superfamily: divergently related enzymes that catalyze different reactions involving acyl coenzyme a thioesters. Acc Chem Res. 2001;34:145–157. doi: 10.1021/ar000053l. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. Antibiotics: opportunities for genetic manipulation. Philos Trans R Soc Lond B Biol Sci. 1989;324:549–562. doi: 10.1098/rstb.1989.0067. [DOI] [PubMed] [Google Scholar]
- Huang YS, Ge J, Zhang HM, Lei JQ, Zhang XL, Wang HH. Purification and characterization of the Mycobacterium tuberculosis FabD2, a novel malonyl-CoA:AcpM transacylase of fatty acid synthase. Protein Expr Purif. 2006;45:393–399. doi: 10.1016/j.pep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Hunaiti AR, Kolattukudy PE. Isolation and characterization of an acyl-coenzyme A carboxylase from an erythromycin-producing Streptomyces erythreus. Arch Biochem Biophys. 1982;216:362–371. doi: 10.1016/0003-9861(82)90222-3. [DOI] [PubMed] [Google Scholar]
- Janiyani K, Bordelon T, Waldrop GL, Cronan JE., Jr Function of Escherichia coli biotin carboxylase requires catalytic activity of both subunits of the homodimer. J Biol Chem. 2001;276:29864–29870. doi: 10.1074/jbc.M104102200. [DOI] [PubMed] [Google Scholar]
- Kalinowski J, Bathe B, Bartels D, et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Rainwater DL, Kolattukudy PE. Purification and characterization of an unusually large fatty acid synthase from Mycobacterium tuberculosis var. bovis BCG. Arch Biochem Biophys. 1992;295:318–326. doi: 10.1016/0003-9861(92)90524-z. [DOI] [PubMed] [Google Scholar]
- Kramer R. Systems and mechanisms of amino acid uptake and excretion in prokaryotes. Arch Microbiol. 1994;162:1–13. doi: 10.1007/BF00264366. [DOI] [PubMed] [Google Scholar]
- Kremer L, Douglas JD, Baulard AR, et al. Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J Biol Chem. 2000;275:16857–16864. doi: 10.1074/jbc.M000569200. [DOI] [PubMed] [Google Scholar]
- Kremer L, Nampoothiri KM, Lesjean S, Dover LG, Graham S, Betts J, Brennan PJ, Minnikin DE, Locht C, Besra GS. Biochemical characterization of acyl carrier protein (AcpM) and malonyl-CoA:AcpM transacylase (mtFabD), two major components of Mycobacterium tuberculosis fatty acid synthase II. J Biol Chem. 2001;276:27967–27974. doi: 10.1074/jbc.M103687200. [DOI] [PubMed] [Google Scholar]
- Kremer L, Dover LG, Carrere S, Nampoothiri KM, Lesjean S, Brown AK, Brennan PJ, Minnikin DE, Locht C, Besra GS. Mycolic acid biosynthesis and enzymic characterization of the beta-ketoacyl-ACP synthase A-condensing enzyme from Mycobacterium tuberculosis. Biochem J. 2002;364:423–430. doi: 10.1042/BJ20011628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth DG, Gago GM, de la Iglesia A, Bazet Lyonnet B, Lin TW, Morbidoni HR, Tsai SC, Gramajo H. Accase 6 is the essential acetyl-CoA carboxylase involved in fatty acid and mycolic acid biosynthesis in mycobacteria. Microbiology. 2009;155:2664–2675. doi: 10.1099/mic.0.027714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane MD, Lynen F. The biochemical function of biotin, VI. Chemical structure of the carboxylated active site of propionyl carboxylase. Proc Natl Acad Sci U S A. 1963;49:379–385. doi: 10.1073/pnas.49.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane MD, Moss J, Polakis SE. Acetyl coenzyme A carboxylase. Curr Top Cell Regul. 1974;8:139–195. [PubMed] [Google Scholar]
- Levert KL, Lloyd RB, Waldrop GL. Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin? Biochemistry. 2000;39:4122–4128. doi: 10.1021/bi992662a. [DOI] [PubMed] [Google Scholar]
- Levert KL, Waldrop GL. A bisubstrate analog inhibitor of the carboxyltransferase component of acetyl-CoA carboxylase. Biochem Biophys Res Commun. 2002;291:1213–1217. doi: 10.1006/bbrc.2002.6576. [DOI] [PubMed] [Google Scholar]
- Li Y, Florova G, Reynolds KA. Alteration of the fatty acid profile of Streptomyces coelicolor by replacement of the initiation enzyme 3-ketoacyl acyl carrier protein synthase III (FabH) J Bacteriol. 2005;187:3795–3799. doi: 10.1128/JB.187.11.3795-3799.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TW, Melgar MM, Kurth D, Swamidass SJ, Purdon J, Tseng T, Gago G, Baldi P, Gramajo H, Tsai SC. Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:3072–3077. doi: 10.1073/pnas.0510580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Barry CE, 3rd, Besra GS, Nikaido H. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J Biol Chem. 1996;271:29545–29551. doi: 10.1074/jbc.271.47.29545. [DOI] [PubMed] [Google Scholar]
- Lu YJ, Rock CO. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol Microbiol. 2006;59:551–566. doi: 10.1111/j.1365-2958.2005.04951.x. [DOI] [PubMed] [Google Scholar]
- Lynen F. New experiments of biotin enzymes. CRC Crit Rev Biochem. 1979;7:103–119. doi: 10.3109/10409237909105428. [DOI] [PubMed] [Google Scholar]
- Lynen F. On the structure of fatty acid synthetase of yeast. Eur J Biochem. 1980;112:431–442. doi: 10.1111/j.1432-1033.1980.tb06105.x. [DOI] [PubMed] [Google Scholar]
- Marrakchi H, Ducasse S, Labesse G, Montrozier H, Margeat E, Emorine L, Charpentier X, Daffe M, Quemard A. MabA (FabG1), a Mycobacterium tuberculosis protein involved in the long-chain fatty acid elongation system FAS-II. Microbiology. 2002;148:951–960. doi: 10.1099/00221287-148-4-951. [DOI] [PubMed] [Google Scholar]
- Matsuoka H, Hirooka K, Fujita Y. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J Biol Chem. 2007;282:5180–5194. doi: 10.1074/jbc.M606831200. [DOI] [PubMed] [Google Scholar]
- Minnikin DE, Goodfellow M. Lipid composition in the classification and identification of acid-fast bacteria. Soc Appl Bacteriol Symp Ser. 1980;8:189–256. [PubMed] [Google Scholar]
- Minnikin DE, Kremer L, Dover LG, Besra GS. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem Biol. 2002;9:545–553. doi: 10.1016/s1074-5521(02)00142-4. [DOI] [PubMed] [Google Scholar]
- Molle V, Brown AK, Besra GS, Cozzone AJ, Kremer L. The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J Biol Chem. 2006;281:30094–30103. doi: 10.1074/jbc.M601691200. [DOI] [PubMed] [Google Scholar]
- Odriozola JM, Ramos JA, Bloch K. Fatty acid synthetase activity in Mycobacterium smegmatis. Characterization of the acyl carrier protein-dependent elongating system. Biochim Biophys Acta. 1977;488:207–217. doi: 10.1016/0005-2760(77)90178-3. [DOI] [PubMed] [Google Scholar]
- Oh TJ, Daniel J, Kim HJ, Sirakova TD, Kolattukudy PE. Identification and characterization of Rv3281 as a novel subunit of a biotin-dependent acyl-CoA Carboxylase in Mycobacterium tuberculosis H37Rv. J Biol Chem. 2006;281:3899–3908. doi: 10.1074/jbc.M511761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwueme KC, Vos CJ, Zurita J, Ferreras JA, Quadri LE. The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog Lipid Res. 2005;44:259–302. doi: 10.1016/j.plipres.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Pacheco-Alvarez D, Solorzano-Vargas RS, Del Rio AL. Biotin in metabolism and its relationship to human disease. Arch Med Res. 2002;33:439–447. doi: 10.1016/s0188-4409(02)00399-5. [DOI] [PubMed] [Google Scholar]
- Papaioannou N, Cheon HS, Lian Y, Kishi Y. Product-regulation mechanisms for fatty acid biosynthesis catalyzed by Mycobacterium smegmatis FAS I. Chembiochem. 2007;8:1775–1780. doi: 10.1002/cbic.200700380. [DOI] [PubMed] [Google Scholar]
- Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- Peterson DO, Bloch K. Mycobacterium smegmatis fatty acid synthetase. Long chain transacylase chain length specificity. J Biol Chem. 1977;252:5735–5739. [PubMed] [Google Scholar]
- Pohlmann J, Lampe T, Shimada M, Nell PG, Pernerstorfer J, Svenstrup N, Brunner NA, Schiffer G, Freiberg C. Pyrrolidinedione derivatives as antibacterial agents with a novel mode of action. Bioorg Med Chem Lett. 2005;15:1189–1192. doi: 10.1016/j.bmcl.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Portevin D, Sousa-D'Auria C, Montrozier H, Houssin C, Stella A, Laneelle MA, Bardou F, Guilhot C, Daffe M. The acyl-AMP ligase FadD32 and AccD4-containing acyl-CoA carboxylase are required for the synthesis of mycolic acids and essential for mycobacterial growth: identification of the carboxylation product and determination of the acyl-CoA carboxylase components. J Biol Chem. 2005;280:8862–8874. doi: 10.1074/jbc.M408578200. [DOI] [PubMed] [Google Scholar]
- Pugh EL, Sauer F, Waite M, Toomey RE, Wakil SJ. Studies on the mechanism of fatty acid synthesis. 13. The role of beta-hydroxy acids in the synthesis of palmitate and cis vaccenate by the Escherichia coli enzyme system. J Biol Chem. 1966;241:2635–2643. [PubMed] [Google Scholar]
- Qiu X, Choudhry AE, Janson CA, Grooms M, Daines RA, Lonsdale JT, Khandekar SS. Crystal structure and substrate specificity of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus. Protein Sci. 2005;14:2087–2094. doi: 10.1110/ps.051501605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemard A, Sacchettini JC, Dessen A, Vilcheze C, Bittman R, Jacobs WR, Jr, Blanchard JS. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- Radmacher E, Alderwick LJ, Besra GS, Brown AK, Gibson KJ, Sahm H, Eggeling L. Two functional FAS-I type fatty acid synthases in Corynebacterium glutamicum. Microbiology. 2005;151:2421–2427. doi: 10.1099/mic.0.28012-0. [DOI] [PubMed] [Google Scholar]
- Rawlings BJ. Biosynthesis of fatty acids and related metabolites. Nat Prod Rep. 1998;15:275–308. doi: 10.1039/a815275y. [DOI] [PubMed] [Google Scholar]
- Reddy DV, Shenoy BC, Carey PR, Sonnichsen FD. High resolution solution structure of the 1.3S subunit of transcarboxylase from Propionibacterium shermanii. Biochemistry. 2000;39:2509–2516. doi: 10.1021/bi9925367. [DOI] [PubMed] [Google Scholar]
- Revill WP, Bibb MJ, Hopwood DA. Purification of a malonyltransferase from Streptomyces coelicolor A3(2) and analysis of its genetic determinant. J Bacteriol. 1995;177:3946–3952. doi: 10.1128/jb.177.14.3946-3952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revill WP, Bibb MJ, Hopwood DA. Relationships between fatty acid and polyketide synthases from Streptomyces coelicolor A3(2): characterization of the fatty acid synthase acyl carrier protein. J Bacteriol. 1996;178:5660–5667. doi: 10.1128/jb.178.19.5660-5667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revill WP, Bibb MJ, Scheu AK, Kieser HJ, Hopwood DA. Beta-ketoacyl acyl carrier protein synthase III (FabH) is essential for fatty acid biosynthesis in Streptomyces coelicolor A3(2) J Bacteriol. 2001;183:3526–3530. doi: 10.1128/JB.183.11.3526-3530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EL, Shu N, Howard MJ, Broadhurst RW, Chapman-Smith A, Wallace JC, Wallace JC, Morris T, Cronan JE, Jr, Perham RN. Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry. 1999;38:5045–5053. doi: 10.1021/bi982466o. [DOI] [PubMed] [Google Scholar]
- Rock CO, Cronan JE. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- Rock CO. Fatty acid and phospholipid metabolism in prokaryotes. In: Vance DennisE, Vance JeanE., editors. Biochemistry of Lipids, Lipoproteins and Membranes. Elsevier; 2008. pp. 59–96. [Google Scholar]
- Rodriguez E, Gramajo H. Genetic and biochemical characterization of the alpha and beta components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2) Microbiology. 1999;145(145):3109–3119. doi: 10.1099/00221287-145-11-3109. [DOI] [PubMed] [Google Scholar]
- Rodriguez E, Banchio C, Diacovich L, Bibb MJ, Gramajo H. Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor A3(2) Appl Environ Microbiol. 2001;67:4166–4176. doi: 10.1128/AEM.67.9.4166-4176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco E, Covarrubias AS, O'Hare HM, Carroll P, Eynard N, Jones TA, Parish T, Daffe M, Backbro K, Quemard A. The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2007;104:14628–14633. doi: 10.1073/pnas.0704132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva S, Musayev F, Alhamadsheh MM, Neel Scarsdale J, Tonie Wright H, Reynolds KA. Probing reactivity and substrate specificity of both subunits of the dimeric Mycobacterium tuberculosis FabH using alkyl-CoA disulfide inhibitors and acyl-CoA substrates. Bioorg Chem. 2008;36:85–90. doi: 10.1016/j.bioorg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman V, Mondino S, Sala C, Cole ST, Gago G, Gramajo H. Transcriptional regulation of lipid homeostasis in mycobacteria. Mol Microbiol. 2010;78:64–77. doi: 10.1111/j.1365-2958.2010.07313.x. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarsdale JN, Kazanina G, He X, Reynolds KA, Wright HT. Crystal structure of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III. J Biol Chem. 2001;276:20516–20522. doi: 10.1074/jbc.M010762200. [DOI] [PubMed] [Google Scholar]
- Schaeffer ML, Agnihotri G, Volker C, Kallender H, Brennan PJ, Lonsdale JT. Purification and biochemical characterization of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthases KasA and KasB. J Biol Chem. 2001;276:47029–47037. doi: 10.1074/jbc.M108903200. [DOI] [PubMed] [Google Scholar]
- Schjerling CK, Hummel R, Hansen JK, Borsting C, Mikkelsen JM, Kristiansen K, Knudsen J. Disruption of the gene encoding the acyl-CoA-binding protein (ACB1) perturbs acyl-CoA metabolism in Saccharomyces cerevisiae. J Biol Chem. 1996;271:22514–22521. doi: 10.1074/jbc.271.37.22514. [DOI] [PubMed] [Google Scholar]
- Schujman GE, Paoletti L, Grossman AD, de Mendoza D. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev Cell. 2003;4:663–672. doi: 10.1016/s1534-5807(03)00123-0. [DOI] [PubMed] [Google Scholar]
- Schujman GE, Guerin M, Buschiazzo A, Schaeffer F, Llarrull LI, Reh G, Vila AJ, Alzari PM, de Mendoza D. Structural basis of lipid biosynthesis regulation in Gram-positive bacteria. EMBO J. 2006;25:4074–4083. doi: 10.1038/sj.emboj.7601284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schujman GE, de Mendoza D. Regulation of type II fatty acid synthase in Gram-positive bacteria. Curr Opin Microbiol. 2008;11:148–152. doi: 10.1016/j.mib.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Schweizer E, Hofmann J. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol Mol Biol Rev. 2004;68:501–517. doi: 10.1128/MMBR.68.3.501-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Volrath SL, Weatherly SC, Elich TD, Tong L. A mechanism for the potent inhibition of eukaryotic acetyl-coenzyme A carboxylase by soraphen A, a macrocyclic polyketide natural product. Mol Cell. 2004;16:881–891. doi: 10.1016/j.molcel.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Sirakova TD, Thirumala AK, Dubey VS, Sprecher H, Kolattukudy PE. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J Biol Chem. 2001;276:16833–16839. doi: 10.1074/jbc.M011468200. [DOI] [PubMed] [Google Scholar]
- Slayden RA, Barry CE., III The role of KasA and KasB in the biosynthesis of meromycolic acids and isoniazid resistance in Mycobacterium tuberculosis. Tuberculosis (Edinb.) 2002;82:149–160. doi: 10.1054/tube.2002.0333. [DOI] [PubMed] [Google Scholar]
- Smith JL, Sherman DH. Biochemistry. An enzyme assembly line. Science. 2008;321:1304–1305. doi: 10.1126/science.1163785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Barona-Gomez F, Corre C, Xiang L, Udwary DW, Austin MB, Noel JP, Moore BS, Challis GL. Type III polyketide synthase beta-ketoacyl-ACP starter unit and ethylmalonyl-CoA extender unit selectivity discovered by Streptomyces coelicolor genome mining. J Am Chem Soc. 2006;128:14754–14755. doi: 10.1021/ja065247w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JB, Ninfa AJ, Stock AM. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuible HP, Wagner C, Andreou I, Huter G, Haselmann J, Schweizer E. Identification and functional differentiation of two type I fatty acid synthases in Brevibacterium ammoniagenes. J Bacteriol. 1996;178:4787–4793. doi: 10.1128/jb.178.16.4787-4793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers RG, Ali A, Shen B, Wessel WA, Hutchinson CR. Malonyl-coenzyme A: acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry. 1995;34:9389–9402. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- Swamidass SJ, Azencott CA, Lin TW, Gramajo H, Tsai SC, Baldi P. Influence relevance voting: an accurate and interpretable virtual high throughput screening method. J Chem Inf Model. 2009;49:756–766. doi: 10.1021/ci8004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Wang C, Besra GS. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev. 2005;18:81–101. doi: 10.1128/CMR.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A, Kaiser O, Hain T, et al. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J Bacteriol. 2005;187:4671–4682. doi: 10.1128/JB.187.13.4671-4682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A, Trost E, Tilker A, et al. The lifestyle of Corynebacterium urealyticum derived from its complete genome sequence established by pyrosequencing. J Biotechnol. 2008;136:11–21. doi: 10.1016/j.jbiotec.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L, Harwood HJ., Jr Acetyl-coenzyme A carboxylases: versatile targets for drug discovery. J Cell Biochem. 2006;99:1476–1488. doi: 10.1002/jcb.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomey RE, Wakil SJ. Studies on the mechanism of fatty acid synthesis. XVI. Preparation and general properties of acyl-malonyl acyl carrier protein-condensing enzyme from Escherichia coli. J Biol Chem. 1966;241:1159–1165. [PubMed] [Google Scholar]
- Trivedi OA, Arora P, Sridharan V, Tickoo R, Mohanty D, Gokhale RS. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, Mohanty D, Gokhale RS. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol Cell. 2005;17:631–643. doi: 10.1016/j.molcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Tsay JT, Oh W, Larson TJ, Jackowski S, Rock CO. Isolation and characterization of the beta-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J Biol Chem. 1992;267:6807–6814. [PubMed] [Google Scholar]
- Veyron-Churlet R, Molle V, Taylor RC, Brown AK, Besra GS, Zanella-Cleon I, Futterer K, Kremer L. The Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J Biol Chem. 2009;284:6414–6424. doi: 10.1074/jbc.M806537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyron-Churlet R, Zanella-Cleon I, Cohen-Gonsaud M, Molle V, Kremer L. Phosphorylation of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein reductase MabA regulates mycolic acid biosynthesis. J Biol Chem. 2010;285:12714–12725. doi: 10.1074/jbc.M110.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- Waldrop GL, Rayment I, Holden HM. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochemistry. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- Wallace KK, Zhao B, McArthur HA, Reynolds KA. In vivo analysis of straight-chain and branched-chain fatty acid biosynthesis in three actinomycetes. FEMS Microbiol Lett. 1995;131:227–234. doi: 10.1111/j.1574-6968.1995.tb07781.x. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Schall I, Huber R, Buckel W, Jacob U. Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase. EMBO J. 2003;22:3493–3502. doi: 10.1093/emboj/cdg358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SW, Zheng J, Zhang YM, Rock The structural biology of type II fatty acid biosynthesis. Annu.Rev.Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- Wood WI, Peterson DO, Bloch K. Subunit structure of Mycobacterium smegmatis fatty acid synthetase. Evidence for identical multifunctional polypeptide chains. J Biol Chem. 1978;253:2650–2656. [PubMed] [Google Scholar]
- Xiang S, Tong L. Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat Struct Mol Biol. 2008;15:295–302. doi: 10.1038/nsmb.1393. [DOI] [PubMed] [Google Scholar]
- Yao X, Wei D, Soden C, Jr, Summers MF, Beckett D. Structure of the carboxy-terminal fragment of the apo-biotin carboxyl carrier subunit of Escherichia coli acetyl-CoA carboxylase. Biochemistry. 1997;36:15089–15100. doi: 10.1021/bi971485f. [DOI] [PubMed] [Google Scholar]
- Yu LP, Xiang S, Lasso G, Gil D, Valle M, Tong L. A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure. 2009;17:823–832. doi: 10.1016/j.str.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang Z, Shen Y, Tong L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science. 2003;299:2064–2067. doi: 10.1126/science.1081366. [DOI] [PubMed] [Google Scholar]
- Zhang YM, White SW, Rock CO. Inhibiting bacterial fatty acid synthesis. J.Biol.Chem. 2006;281:17541–17544. doi: 10.1074/jbc.R600004200. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Rock CO. Transcriptional regulation in bacterial membrane lipid synthesis. J Lipid Res. 2009;50 Suppl:S115–S119. doi: 10.1194/jlr.R800046-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimhony O, Vilcheze C, Jacobs WR., Jr Characterization of Mycobacterium smegmatis expressing the Mycobacterium tuberculosis fatty acid synthase I (fas1) gene. J Bacteriol. 2004;186:4051–4055. doi: 10.1128/JB.186.13.4051-4055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]