Abstract
The radical S-adenosylmethionine (S-AdoMet) superfamily contains thousands of proteins that catalyze highly diverse conversions, most of which are poorly understood due to a lack of information regarding chemical products and radical-dependent transformations. We here report that NosL, involved in forming the indole side ring of the thiopeptide nosiheptide (NOS), is a radical S-AdoMet 3-methyl-2-indolic acid (MIA) synthase. NosL catalyzed an unprecedented carbon chain reconstitution of L-Trp to give MIA, showing removal of the Cα-N unit and shift of the carboxylate to the indole ring. Dissection of the enzymatic process upon the identification of products and a putative glycyl intermediate uncovered a radical-mediated, unusual fragmentation-recombination reaction. This finding unveiled a key step in radical S-AdoMet enzyme-catalyzed structural rearrangements during complex biotransformations. Additionally, NosL tolerated fluorinated L-Trps as the substrates, allowing for production of a regiospecifically halogenated thiopeptide that has not been found in over 80 entity-containing, naturally occurring thiopeptide family.
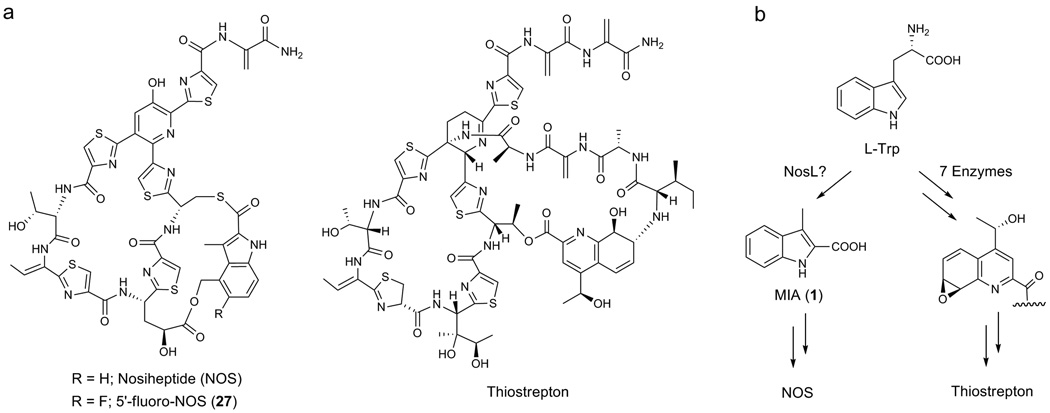
Thiopeptides are a class of clinically interesting and highly modified polythiazolyl antibiotics1–3. The potent activity of many members against various drug-resistant bacterial pathogens has promoted extensive investigations by chemical modification into new analogue development, towards overcoming their physical drawbacks such as water solubility for clinical use. Thiopeptides possess a characteristic macrocyclic core, consisting of a nitrogen-containing, 6-membered ring central to multiple thiazoles and dehydroamino acids, but vary in side chain (and/or ring) appending additional functionalities (Fig. 1a). In contrast to heroic efforts of chemical synthesis4, their newly established biosynthetic pathways for thiopeptide framework formation are remarkably concise5–11, showing conserved posttranslational modifications on a ribosomally synthesized precursor peptide. The reactions include cyclodehydrations and subsequent dehydrogenations to form aromatic thiazoles, dehydrations to generate dehydroamino acids, and an intramolecular cyclization to afford the nitrogen heterocycle.
Fig. 1.
Structures of polycyclic thiopeptides and their side ring formations. (a) NOS and 5’-fluoro-NOS, and thiostrepton. (b) Processing of L-Trp to afford the indolic acid moiety of NOS and the quinalic acid moiety of thiostrepton, respectively, via completely different routes.
For most polycyclic thiopeptides, the side ring formations are precursor peptide-independent12–16. They exclusively exploit L-Trp to furnish the variable functional groups, as exemplified by the indolic acid moiety of nosiheptide (NOS) and the quinalic acid moiety of thiostrepton (Fig. 1b). Whereas the quinalic acid formation may require seven enzymes to convert L-Trp in a process including methylation, desamination, oxidation, ring opening and recyclization, reduction, and epoxidation6,7, the indolic acid moiety formation in NOS biosynthesis involves two enzymes NosL and NosN, both of which encodes the putative radical S-adenosylmethionine (S-AdoMet) proteins, to process L-Trp via a completely different route9. Characterization of a side ring opened NOS analogue by inactivating nosN supported the methylation activity of NosN, which acts on the 3-methylindolyl group to furnish the 3,4-dimehtylindole moiety. This leaves NosL as a candidate to biosynthesize the key intermediate, 3-methyl-2-indolic acid (MIA, 1). We have confirmed the essentiality of NosL to MIA formation9,11: inactivation of nosL completely abolished NOS production, which can be restored either by feeding extraneous 1 or by heterologous complementation with its counterpart nocL from nocathiacin (a naturally occurring analogue of NOS) biosynthesis. However, whether NosL alone catalyzes such a complex conversion remains elusive, given that 1 biosynthesis from L-Trp requires multiple, chemically unusual reactions to process the carbon side chain (including removal of the Cα-N unit and shift of the carboxylate to the indole ring. Fig. 1b).
In this study, we characterized NosL indeed as a radical S-AdoMet protein for converting L-Trp to MIA, the process of which involves a radical-mediated, unusual fragmentation-recombination reaction. This characterization took advantage of efficient 1 production in the nosL-expressing Escherichia coli strain and in vivo and/or in vitro determination of substrate(s), products (including shunt products), and a putative glycyl intermediate in NosL-catalyzed conversion. In light of the substrate tolerance of NosL, regiospecific fluorination of NOS was achieved via a modified MIA intermediate, yielding a new halogenated thiopeptide that has not been found in the naturally occurring family.
RESULTS
In vivo validation of NosL as a MIA synthase
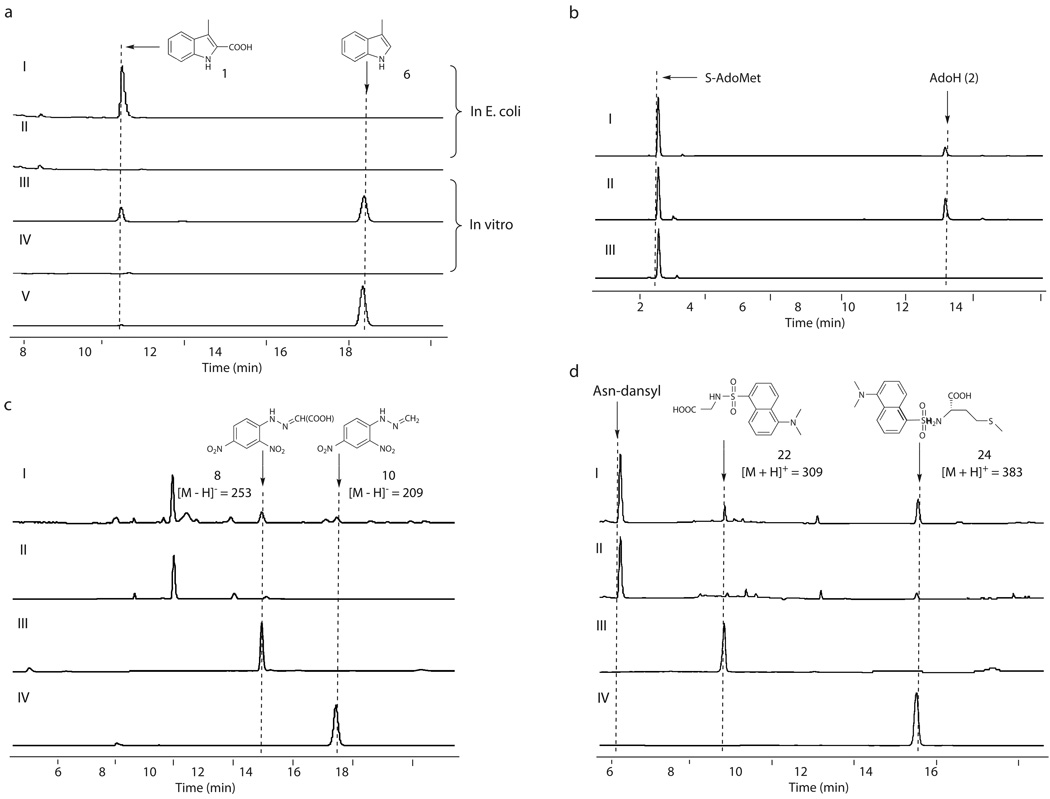
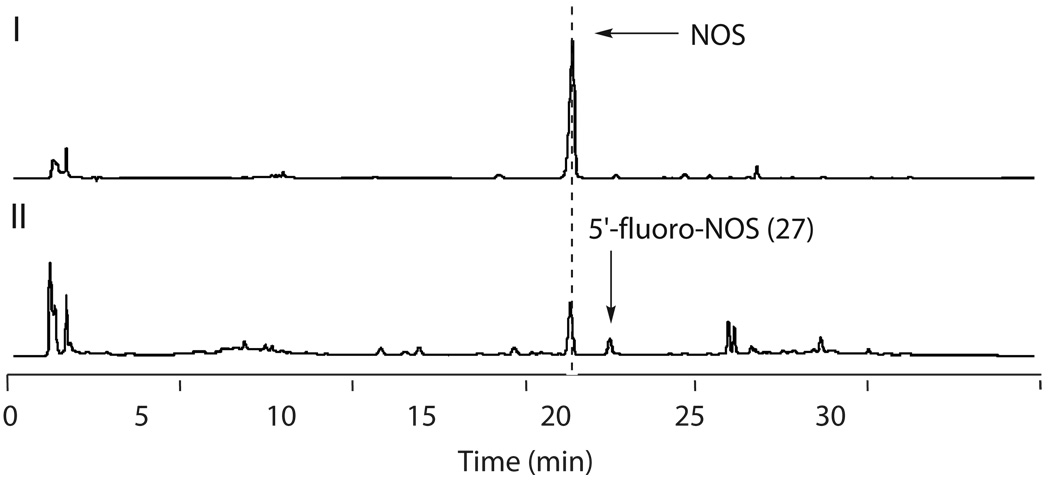
Considering that radical S-AdoMet proteins are sensitive to O2 for in vitro study in general17, we first set out to investigate the NosL function in vivo. nosL was introduced to and expressed in E. coli BL21(DE3), yielding the recombinant strain SL4101 to produce NosL in a N-terminally 6 × His-Tagged form. High performance liquid chromatography (HPLC) analysis revealed a compound produced by SL4101, which was absent in the control strain SL4100 carrying the vector alone (Fig. 2a). The molecular formula C10H9O2N for this compound was established by high-resolution (HR)-electron spray ionization (ESI)-Mass spectroscopy (MS), showing a [M – H]− ion at m/z = 174.05605 (174.05615 calculated). Additionally, 1H, 13C and selected 2D NMR analyses confirmed it to be 1, indicating that NosL is a novel MIA synthase. To probe the origin of 1, the nosL-expressing cells of SL4101 from Luria-Bertani (LB) broth were harvested, and inoculated into a nutrient-insufficient medium followed by further incubation with individual amino acids for a time course analysis of 1 production. Only the addition of L-Trp sped up 1 formation with the apparent rate of 150 µg/L per min in the initial 4 h, whereas the supplementation of other amino acid such as L-Gly or L-Ser had no significant effect on the 1-producing rate (Supplementary Fig. 1). This validated that L-Trp serves as the substrate for the production of 1. The relatively high yield of 1 (42 ± 5 mg/L over a 20-hour period) facilitated its purification and enabled further in vivo experiments to examine NosL catalysis.
Fig. 2.
Characterization of NosL-catalyzed reaction. All the examinations were performed at least in duplicate (for each having at least two parallel samples). (a) Validation of NosL as a MIA synthase. In vivo 1 production in the E. coli strains, including SL4101 harboring nosL (I) and SL4100 carrying the vector alone (II); and in vitro conversion of L-Trp to 1 and the shunt product 6 in the presence (III) and in the absence of S-AdoMet (IV), with the authentic 6 as a standard (V). (b) Consumption of S-AdoMet given the absence (I) and the presence of L-Trp (II), in contrast to that by omitting NosL from the reaction mixture (III). (c) Identification of the products with DNP derivatization by adding (I) and by omitting the substrate L-Trp (II), when authentic 7 (III) and 9 (IV) were subjected to DNP derivatization to generate the control derivatives. (d) Identification of the putative amino acid products with dansyl chloride derivatization, by using Asn as an internal standard for quantitative analysis. Production in the presence (I) and in the absence of L-Trp (II) while authentic 21 (III) and 23 (IV) were subjected to dansyl derivatization to generate the control derivatives for qualitative analysis.
NosL as a Radical S-AdoMet protein for MIA formation
Based on sequence alignment NosL falls into the ThiH radical S-AdoMet subclass (around 20% identity, Supplementary Fig. 2), possessing a typical CxxxCxxC motif for [4Fe-4S] cluster nucleation and a Gly-containing segment involved in S-AdoMet binding. We therefore explored the nature of NosL for its cofactor binding and subsequent chemical transformations. In the CxxxCxxC motif, the Cys residues (corresponding to C95, C99 and C102) were systematically replaced to Ala. The constructs were introduced into E. coli BL21(DE3), yielding the recombinant strains SL4103 (for AxxxCxxC mutation of NosL), SL4105 (for CxxxAxxC mutation), and SL4107 (for CxxxCxxA mutation) for MIA (1) examination. These replacements completely abolished 1 production (Supplementary Fig. 3), demonstrating their indispensability to the NosL activity. The S-AdoMet binding potential was also evaluated in a similar way, by replacing G142 to Ala within the conserved GE/D motif. The recombinant strain SL4109 for expressing mutant NosL (G142A) lost the ability to produce 1 (Supplementary Fig. 3), supporting that NosL is a S-AdoMet-dependent protein.
Next, NosL was purified from SL4101 to homogeneity for assays in vitro. Whereas the aerobically isolated enzyme was inactive, the activity of the anaerobically isolated NosL was detectable but extremely low (Supplementary Fig. 4). NosL was then reconstituted under a strictly anaerobic condition, giving the purified sample (Supplementary Fig. 5a) in dark brownish color. This protein contained 5.1 ± 0.5 of Fe and 5.7 ± 0.3 of sulfide per molecule. The ultraviolet (UV)-visible absorptions of the reconstituted NosL are characteristic for [4Fe-4S] binding proteins (Supplementary Fig. 5b), displaying a A280/A400 ratio at 3.4 : 1 and an apparent shift by reducing with sodium dithionite. Further electron paramagnetic resonance (EPR) analysis at 13 K gave an axial spectrum (Supplementary Fig. 5c), showing approximate g values of 2.02 and 1.91. This supported the binding of a [4Fe-4S]+ cluster to NosL. Incubation of the reconstituted NosL with L-Trp and S-AdoMet was performed in the presence of dithionite, indeed showing MIA (1) production with the yield approximately 7-fold higher than that of the as-isolated enzyme without reconstitution (Fig. 2a and Supplementary Fig. 4a). In the reaction mixture the chemical reductant dithionite can be replaced with a natural reduction system containing flavodoxin, flavodoxin reductase, and NADPH, leading to ~100% improvement in 1 formation (Supplementary Fig. 6). NosL-catalyzed conversion cannot proceed in the absence of S-AdoMet (Fig. 2a), turn-over of which by NosL was further confirmed upon identification of the product 5’-deoxyadenosine (AdoH, 2, Fig. 2b). S-AdoMet consumption was independent of L-Trp, but significantly enhanced by adding L-Trp into the reaction mixture (Fig. 2b and Supplementary Fig. 4a). Together with the identification of methionine (discussed below) as the concomitant product originating from S-AdoMet, we conclude that NosL, whose activity is S-AdoMet-dependent, catalyzes 1 formation from L-Trp via an Ado•-initiated process.
NosL-catalyzed carbon chain reconstitution on L-Trp
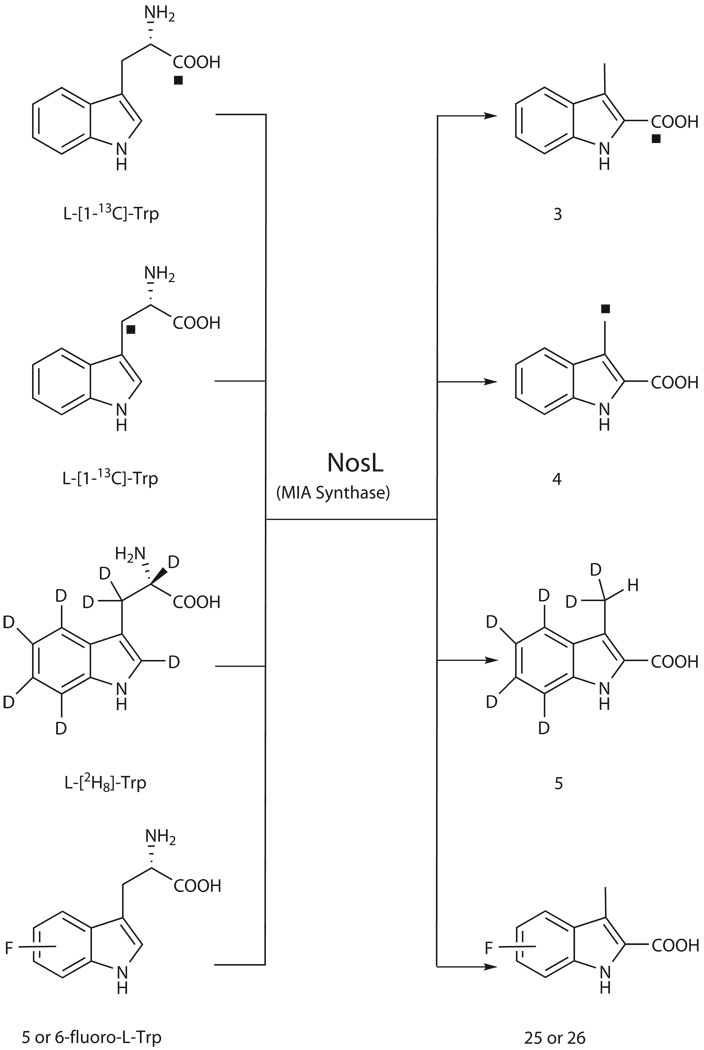
To investigate the mechanism of NosL-catalyzed conversion, we focused on the reconstitution of the carbon side chain of L-Trp. [1-13C] and [3-13C]-labeled L-Trps were fed individually into SL4101, resulting in enriched 13C resonances for the 2-carboxylate (at δ 164.0 for product 3) and 3-methyl group (at δ 8.83 for product 4) of MIA, respectively (Scheme 1 and Supplementary Fig. 7). Consistent with similar studies carried out in the NOS-producing strain Streptomyces actuosus12,13, these findings demonstrated that 1 biosynthesis involves an unprecedented carbon chain rearrangement of the L-Trp precursor, showing 1) the intramolecular migration of the carboxylate to C2 of the indole ring, 2) transformation of the methylene carbon to a methyl group, and 3) elimination of the α-carbon and its associated amino group. The feeding of L-[2H8]-Trp also supported this conclusion by allowing the production of [2H6]-MIA (5) (HR-ESI-MS m/z calcd. for C10D6H2O2N 180.09371, found 180.09343), and excluded the participation of the benzene ring in this conversion (given no change in deuterium substitutions, Scheme 1). A 0.04 ppm upfield-shift (from δ 2.48 relative to CH3) of the signal that corresponds to the CHD2 protons was found in the 1H NMR spectrum of 5 (Supplementary Fig. 8), ruling out adjacent hydrogen transfer, either from Cα or from C2 on the indole ring, to provide the third hydrogen atom of the methyl group. Further, using L-[2H8]-Trp as the substrate, in vitro assay of NosL-catalyzed reactions showed that the co-produced AdoH (2) (HR-ESI-MS m/z calcd. for C10H13N5O3 252.10967, found 252.10983) was not deuterium labeled. This clearly indicates that the Ado•-initiated rearrangement of the carbon chain does not start with a C-H bond cleavage on L-Trp to generate the substrate radical.
Scheme 1.
NosL-catalyzed carbon chain reconstitution of L-Trp shown by labeling patterns and generation of fluorinated MIA analogues. The solid rectangle shows the 13C-labeled carbon atom. D, deuterium. F, fluorine.
Determination of the coproduct and shunt products
To probe the individual steps in MIA (1) formation, we carried out extensive survey of the product profile in NosL-catalyzed in vitro conversion. HPLC analysis of the reaction mixture revealed the first shunt product (Fig. 2a), which was deduced to be 3-methylindole (6) upon further GC-MS analysis with an authentic compound as the standard (Supplementary Fig. 9). Supplementation with 2,4-dinitrophenyl hydrazine (DNP) for derivatization led to identification of the second shunt product as glyoxylate (7) (corresponding to 8, the derivative 2-(2,4-dinitrophenyl) hydrazonoacetate. Fig. 2c and Supplementary Fig. 10). Thus, the first Cα-Cβ cleavage of L-Trp appears to provide the 3-methylindole part whereas the readdition-coupled, second cleavage may take place on the separated 2C-N unit to furnish the 2-carboxylate group. Consistent with this prediction, upon a similar derivatization we detected the production of formaldehyde (9) (corresponding to 10, the derivative 1-(2,4-dinitrophenyl)-2-methylene hydrazine. Fig. 2c and Supplementary Fig. 11), which most likely comes from the α-carbon of L-Trp.
Quantitative analysis of NosL-catalyzed in vitro reaction
We chose three representative products, including MIA (1) and 3-methylindole (6) from L-Trp, and AdoH (2) from S-AdoMet, to quantitatively evaluate NosL-catalyzed in vitro conversion (Supplementary Fig. 4). The yields of the products were enzyme dose-dependent, as the increase of the concentration of NosL accordingly improved the production of 1, 6, and 2 (Supplementary Fig. 4a). Their productions at 25°C were relatively constant in the presence of 1 mM dithionite, 500 µM L-Trp and 1 mM S-AdoMet, showing that about 39 ± 3 µM 1, 140 ± 20 µM 6 and 381 ± 30 µM 2, with a ratio around 1 : 3.6 : 9.8, were produced by 20 µM NosL catalysis over a two-hour period (Supplementary Fig. 4b). For conversion of the substrate L-Trp, under this condition one molecule of NosL produced approximate two molecules of 1 along with seven molecules of 6. The production of AdoH was consistently higher, showing a yield ratio around 2.1 : 1 for 2 corresponding to a sum of 1 and 6. For optimizing the reaction, we speculated that the reduction degree in the reaction mixture may impact the proportion of 1. We therefore measured the effect of the concentration of the chemical reductant dithionite on the yields of 1 and 6 (Supplementary Fig. 4c). Intriguingly, 6 production was highly dependent of dithionite concentration, whereas the yields of 1 were relative constant, around 30–42 µM in all assays. Particularly, when 100 µM dithionite was used, the ratio of 1 to 6 can be improved to around 3 : 1.
Prediction of the Cα-Cβ bond cleavage manner on L-Trp
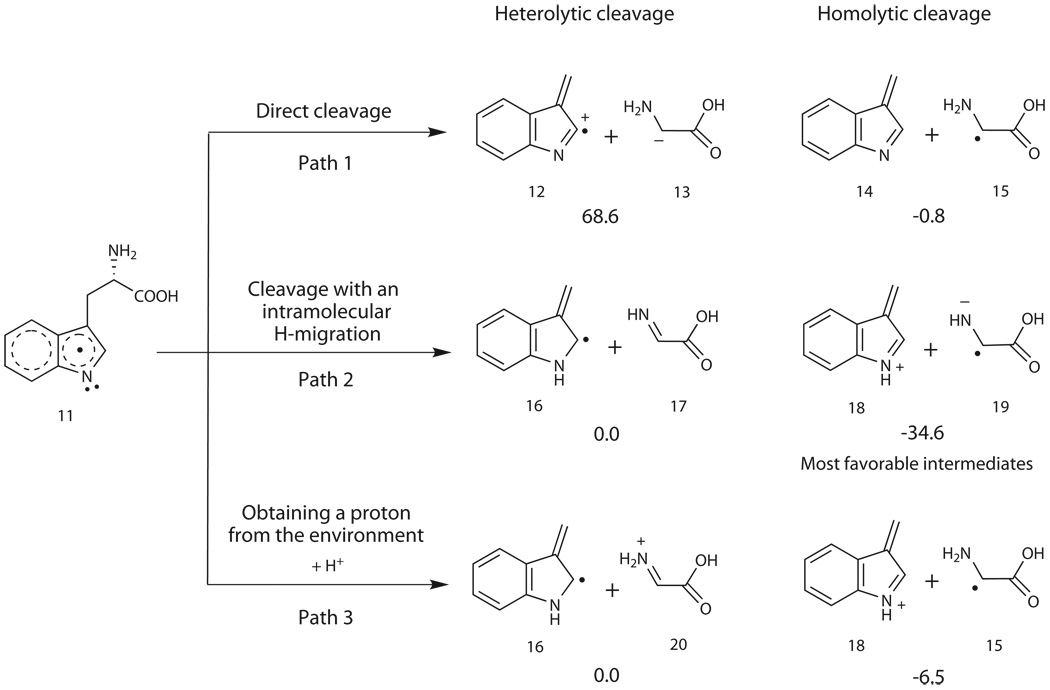
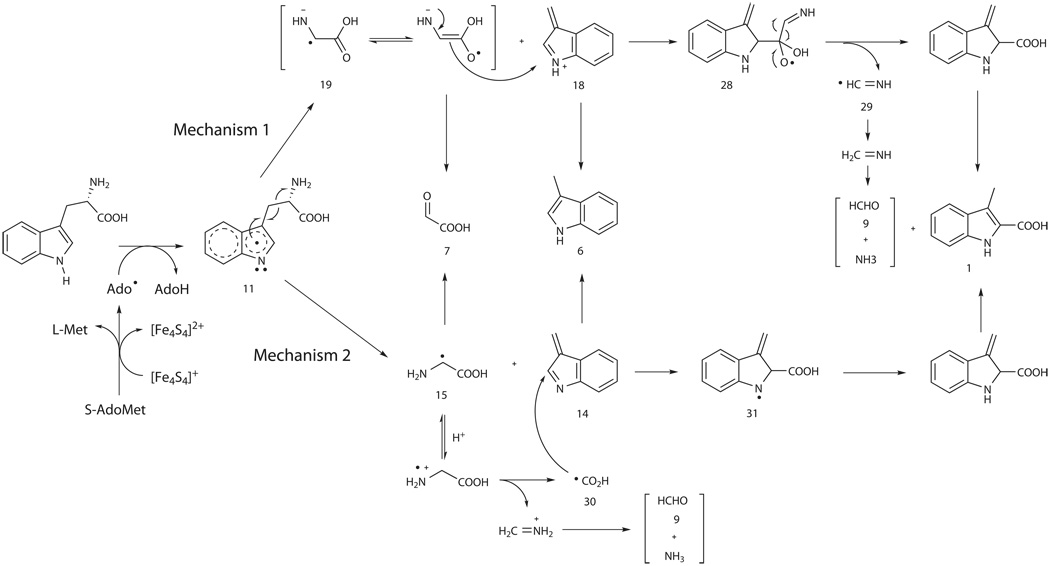
Inspired by identification of 3-methylindole (6) and glyoxylate (7) as the shunt products, we proposed that the Ado•-initiated carbon chain reconstitution of L-Trp may begin with a Cα-Cβ bond cleavage during NosL-catalyzed MIA (1) formation. According to the bond-breaking pattern, either via homolysis or via heterolysis, there are two sets of hypothetic intermediates that share a common fate to the shunt product pair (Fig. 3). Thus, the Density Functional Theory (DFT) calculation was performed to differentiate these two possibilities (Fig. 3, and Supplementary Figs. 12 and 13). While the direct cleavage of the L-Trp neutral radical (11) as outlined in Path 1 leads to the generation of the intermediates 12 and 13 (heterolytic) or 14 and 15 (homolytic), Path 2 indicates that the cleavage accompanied with an intramolecular hydrogen migration results in the production of the intermediates 16 and 17 (heterolytic) or 18 and 19 (homolytic). Given the intermediate 11 that may get a proton from the environment, the model shown in Path 3 is also considered, indicating the generation of the intermediates 16 and 20 (heterolytic) or 18 and 15 (homolytic). At all events the homolytic products are lower in energy than their corresponding heterolytic products, suggesting that the homolysis was thermodynamically favored to form 3-metheneindole and glycyl radical.
Fig. 3.
Different patterns for cleaving the Cα-Cβ bond of L-Trp neutral radical 11. The DFT calculation-based, relative free energies (including solvent effect ΔGsol) are in kcal/mol. In Path 1 and Path 2 the energy of 16 + 17 is used as the reference zero energy, whereas in Path 3 the energy of 16 + 20 is used as the reference.
To experimentally support the hypothesis, we attempted to probe the putative intermediate in NosL-catalyzed reaction, by employing a derivatization associated, rapid quench method previously used for analyzing the glutamate mutase-catalyzed reaction. Glutamate mutase, a Ado•-producing, adenosylcobalamin (AdoCbl, coenzyme B12)-dependent protein, has been demonstrated to convert L-Glu into glycyl radical (trapped as glycine by using this quench technique) and acrylate during L-threo-3-methylaspartate formation18–20. The NosL-catalyzed reaction was quenched at 10 sec before derivatization with dansyl chloride. HPLC-MS analysis showed the production of glycine (21) (corresponding to the derivative Gly-dansyl, 22. Fig. 2d and Supplementary Fig. 14), as well as methionine (23) (corresponding to the derivative Met-dansyl, 24. Fig. 2d and Supplementary Fig. 15) deriving from S-AdoMet. Using Asn as an internal standard, the glycine species was observed in a yield of 2 ± 0.5 µM, equal to around 2.5% of the enzyme active site. Under the highly reductive condition this species is presumed to derive from the immediately uncoupled intermediate, glycyl radical, consistent with the predicted manner for homolytic cleavage of the Cα-Cβ bond of L-Trp during MIA formation.
Substrate flexibility of NosL
Unveiling of the NosL chemistry then promoted us to feed available analogues of L-Trp, including D-Trp and derivatives with different substitutions on the indole ring, into SL4101 to determine the substrate tolerance of NosL. Individual additions of most of these substrates had no effect on MIA (1) production, and failed to afford the expected 1 analogues (Supplementary Table 3). These indicate the stringent substrate specificity of NosL in stereochemistry and in functionalization of the indole ring. In contrast, feeding of 5- or 6-fluoro-DL-Trp (only the L-configuration isomer can be used as the substrate) led to generation of the distinct product with a yield approximately 20% of the concomitant 1 production (Supplementary Fig. 16). Spectroscopic analyses, including HR-ESI-MS, and 1H, 13C and 19F NMR (Supplementary Fig. 17), clearly established these compounds corresponding to 5-fluoro-MIA (25) and 6-fluoro-MIA (26), respectively (Scheme 1).
Generation of a fluorinated NOS analogue
To determine whether or not the fluorinated MIA (1) can be incorporated as an intermediate in vivo, we therefore fed 5-fluoro-DL-Trp into the NOS-producing strain S. actuosus. HPLC-MS analysis showed the dramatic decrease in NOS production (20%–25% relative to that of the wild type strain, Fig. 4), presumably due to competitive inhibition of biosynthetic enzymes in the substrates. A new product, with UV spectra similar to NOS, was clearly observed, showing a yield approximately 20% of the concomitant NOS production. According to the molecular formula as C51H42FN13O12 (HR-ESI-MS m/z calcd. 1238.13114, found 1238.12959), this compound was deduced to be a NOS analogue substituted by a fluorine atom at C5’ of the indole ring (Fig. 1a and Supplementary Fig. 18). For structural elucidation it was purified and then subjected to comparative spectroscopic analysis with NOS, showing the only difference in the indole moiety. 19F NMR, and 1D and 2D NMR spectra further supported this structure assignment of 5’-fluoro-NOS (27. Supplementary Fig. 19 and Table 4). The effect of the 5’-fluoro-substitution on antibacterial activity was consequently evaluated by bioassays against the test strain Bacillus subtillis, displaying the minimum inhibitory concentration (MIC) at 0.004 µM/mL for 27 to that at 0.008 µM/mL for NOS (Supplementary Table 5), Notably, there has yet to be a halogenated member of the naturally occurring thiopeptide antibiotics, which number over 80 compounds reported to date1.
Fig. 4.
Production of NOS and fluorinated thiopeptide by S. actuosus without the supplementation (I) and with the supplementation of 5-fluoro-DL-Trp (II). All the examinations were performed in duplicate (for each having two parallel samples).
DISCUSSION
The radical S-AdoMet superfamily currently comprises thousands of proteins that participate in numerous biochemical processes in animals, plants, and microorganisms17,21–24. These proteins share a common mechanism to generate a powerful oxidant Ado•, thereby initiating highly diverse transformations relevant to DNA repair and in the biosynthesis of vitamins, coenzymes, and antibiotics. Biochemical mechanisms of most of such conversions have not been characterized, due to a lack of information about the enzyme functions and radical-mediated transformations. Focusing on the characterization of NosL and NosL-catalyzed reaction, we herein unveiled a radical-mediated, unusual fragmentation-recombination process, which represents a key step in understanding radical S-AdoMet enzyme-catalyzed complex structure rearrangements.
The mechanism for NosL-catalyzed reaction was proposed in Scheme 2. Distinct from the widely found, C-H bond cleavage in the reactions controlled by known radical S-AdoMet enzymes17, Ado• in NosL catalysis may abstract the hydrogen atom from N1 on the indole ring of L-Trp to generate 11, the neutral substrate radical. The homolysis is thermodynamically favorable, leading to the Cα-Cβ bond cleavage for the first fragmentation to give the transient units 3-metheneindole and glycine radical. Taking a simultaneous proton transfer into account, the resulting cationic 3-metheneindole (18) and glycine radical (19) are more stable, facilitating their subsequent re-addition to generate the radical intermediate 28 (Mechanism 1). Elimination of the Cα-N unit (giving the methanimine radical 29) from 28 can proceed to furnish the 2-carboxylate group to yield MIA (1), and the hydrogen rebound to 29 results in the production of CH2=NH, which is hydrolyzed in the aqueous medium to form formaldehyde (9) and ammonia. Alternatively, according to a glycine decarboxylation-based free radical mechanism proposed previously25, the second fragmentation on the separated 2C-N unit (glycyl radical 15) could precede the re-addition, and results in a direct reposition of the carboxylate at C-2 of the indole ring (Mechanism 2): 1) the protonated radical 15 may undergo a C-C bond cleavage to generate carboxyl radical 30; and 2) the addition of 30 to 3-mtheneindole 14 can yield radical 31, hydrogen rebound to which eventually leads to the production of 1.
Scheme 2.
Mechanistic hypothesis for NosL-catalyzed conversion. For Mechanism 1, the recombination may take place between cationic 18 and radical 19 followed by elimination of the Cα-N unit from the resulting radical 28; and for Mechanism 2, the first separated radical 15 could undergo the second fragmentation to generate carboxyl radical 30 before the addition onto 14.
NosL-catalyzed in vitro conversion produced 1 along with two shunt products 3-methylindole (6) and glyoxylate (7), which may originate from the intermediate pair 3-metheneindole and glycyl radical, respectively. We proposed that the Cα-Cβ bond cleavage-post biotransformation, such as the inefficient recombination, may serve as a limitation step in NosL-catalyzed in vitro process. The over-produced intermediates could be released from the active site of NosL into the aqueous medium: while 3-metheneindole is aromatized by reduction to 6, glycyl radical is rapidly degraded to 7 and ammonia. The one electron-unpaired organic radicals have been known to be highly active and unstable. For instance the substrate-based, proposed glycyl radical (Scheme 2), if it fails to be further processed (e.g. addition to the intermediate 3-metheneindole), may lose an electron to generate dehydroglycine followed by hydrolysis for glyoxylate production. The similar result has been found in the in vitro reaction catalyzed by ThiH26. Without the associated components for turning over gylcyl radical or dehydroglycine in thiazole formation, ThiH catalyzes the Cα-Cβ bond cleavage of L-Tyr and gives p-cresol and glyoxylate as the products.
Substrate fragmentation is known in Ado•-induced biotransformations, as exemplified by ThiH and by newly characterized HydG (24% identity to NosL)27, which catalyzes the L-Tyr cleavage to generate p-cresol and cyanide. NosL, ThiH and HydG may share a common paradigm to process the radical-based, aromatic amino acid substrates for the Cα-Cβ bond cleavage, despite their difference in the fate of the resulting gylcyl radical (or dehydroglycine) intermediate. However, the fragmentation-recombination found in NosL chemistry is rare. To our knowledge, the only known example is glutamate mutase, the radical AdoCbl-dependent protein that converts L-Glu to the β-methyl branched product (but without fragment elimination)20,24. This strategy might be employed in certain Ado•-mediated structural rearrangements, such as the ThiC-catalyzed complex conversion of 5-aminoimidazole ribonucleotide to 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate in thiamine biosynthesis28.
In conclusion, we have uncovered NosL as a MIA synthase that catalyzes an unprecedented carbon side chain reconstitution of L-Trp. The radical-mediated, unusual fragmentation-recombination process may be general for radical S-AdoMet protein-catalyzed structural rearrangements in certain uncharacterized biotransformations. Taking advantage of the substrate tolerance of NosL, regiospecific fluorination of NOS could be achieved via a modified MIA intermediate. The NOS analogue produced in this fashion and bearing fluorine at the 5' position showed the improved pharmaceutical property. This application of combinatorial biosynthesis, enabled by our elucidation of NosL chemistry, compliments recent advances in understanding sequence permutations of the precursor peptide29,30.
METHODS
Bacterial strains, plasmids and primers
Please see Supplementary Tables 1 and 2
Materials
Please see Supplementary Methods.
Production of MIA in vivo
Construction of the recombinant strain SL4101 for expressing nosL was described in Supplementary Methods. SL4101 was cultured in LB medium. Once the cell density reached 0.6–0.8 at OD600 nm, the Fe(NH4)2SO4 solution was added into the culture broth to a concentration of 50 µM, followed by incubation on ice for 10 min. After addition of isopropyl-β-D-thiogalactopyranoside (IPTG, 100 µM in final), nosL expression had been induced at 25°C for 6–8 h. While the cells were harvested by centrifugation for following experiments (see below), the supernatant was directly subjected to HPLC analysisfor product examination. SL4100 carrring the vector pET28a alone was utilized as the control in the parallel analytic process. Site-specific mutagenesis for exploring the cofactor binding nature of NosL was described in Supplementary Methods.
Time Course-Based Analysis of MIA Production in vivo
The harvested SL4101 cells described above were resuspended in a nutrient-insufficient medium (yeast extract 0.1%, tryptone 0.5%, glucose 0.4%, NaCl 0.5%, glutamine 0.05%, K2HPO4 0.2%, MgSO4 0.01%, and FeSO4 0.001%), with OD600 nm of 0.6–0.8. Variable amino acids, including L-Trp, L-Ser and L-Gly, were individually supplemented to the broth (5 mM in final) before further incubation at 28°C. MIA production was measured by HPLC at 0.5, 1, 2, 3, 4, 10, or 20 h. The MIA production is approximately linear with respect to time in initial 4 hr, and the apparent rate was accordingly estimated.
Feeding of Isotope Labeled L-Trps and Variable L-Trp Analogues
The procedure is similar to that of the time course-based analysis for MIA production, except the individual supplementation of isotope labeled substrates (i.e. L-[2H8]-Trp, L-[1-13C]-Trp and L-[3-13C]-Trp), D-Trp, and derivatives with different substitution on the indole ring (i.e. 1-methyl-L-Trp, 2-methyl-DL-Trp, 4-methyl-DL-Trp, 6-methyl-DL-Trp, 5-hydroxyl-L-Trp, 5-methoxy-DL-Trp, 5-bromo-DL-Trp, 5-fluoro-DL-Trp, and 6-fluoro-DL-Trp) to a concentration of 1 mM before further incubation at 28°C for 8 h. The production of labeled or functionally substituted MIAs was monitored by HPLC-MS.
Structural characterization of MIA and its analogues, and a new NOS analogue by feeding 5-fluoro-DL-Trp into the NOS-producing strain was summarized in Supplementary Methods.
Reconstitution of NosL
Production and anaerobic purification of NosL were summarized in Supplementary Methods. A previously reported procedure31 was modified to reconstitute the active enzyme. Dithiothreitol was added to the purified protein (10 mM in final). Under this reductive condition, Fe(NH4)2SO4 solution (50 mM) was added carefully into the suspension to a final concentration of 500 µM. After 10 min, Na2S solution (50 mM) was added in the same way to a concentration of 500 µM. The solution had been incubated on ice for 5–7 h. The resulting dark brown suspension was then subjected to desalting on a column (10-DG, Bio-Rad, USA) that was pre-equilibrated with the elution buffer (50 mM Tris-HCl, 25 mM NaCl, 10 mM dithiothreitol, and 10% glycerol, pH 8.0). The colored fraction was collected, and concentrated to 100 µM for in vitro assay. Protein characterization was described in Supplementary Methods.
Assay of the NosL activity in vitro
The 100 µL of reaction mixture contained 10 mM dithiothreitol, 500 µM dithionite, 200 µM S-AdoMet, 200 µM L-Trp (or L-[2H8]Trp), and 20 µM reconstituted NosL in 50 mM Tris-HCl buffer (pH 8.0). Reactions were initiated by adding of S-AdoMet to the 10 min pre-incubated mixture (as a negative control that contains the components except S-AdoMet), and then incubated at 25°C for 2 h. To terminate the reaction, trifluoroacetic acid (TFA) was added to a final concentration of 10% (v/v) to inactivate the enzyme. After removal of the precipitates by centrifugation, the supernatant was subjected to HPLC-MS analysis. For evaluation of a natural reduction system, the chemical reductant dithionite was replaced by 0.5 mM NADPH, 50 µM flavodoxin, 20 µM flavodoxin reductase.
For GC-MS analysis, the supernatant was further extracted by ethyl acetate. For derivatization, 2,4-dinitrophenylhydrazine (DNP) was added to the supernatant (10 mM in final), which was further incubated at 37°C for 30 min.
Quantitative analysis of product formation in vitro
The 100 µL of reaction mixture contained 10 mM dithiothreitol, 1 mM dithionite, 1 mM S-AdoMet, and 500 µM L-Trp, in 50 mM Tris-HCl buffer (pH 8.0). The reconstituted NosL varying in concentration (10 µM, 20 µM, 40 µM, or 80 µM) was added into each reaction mixture. The 20 µM NosL catalysis was used for a time course analysis (by terminating at 10 min, 30 min, 60 min, 90 min, or 120 min). To measure the effect of dithionite concentration on the yields of MIA and 3-methylindole, each 100 µM of reaction mixture contained 20 µM reconstituted NosL, 10 mM dithiothreitol, 1 mM SAM, 500 µM L-Trp, and dithionite varying in concentration (100 µM, 200 µM, 500 µM, 1 mM, 2 mM, or 4 mM) in 50 mM Tris-HCl buffer (pH 8.0). The workups for the initiation, incubation and termination of reactions, and for the analysis of products were identical to those described above.
Probing of the putative intermediate in NosL-Catalyzed Conversion
Trapping of glycine species in NosL-catalyzed conversion was performed according to a modified procedure18. The 100 µL of reaction mixture contained 20 mM dithiothreitol, 2 mM dithionite, 1 mM S-AdoMet, 1 mM L-Trp, and 80 µM reduced NosL in 50 mM Tris-HCl buffer (pH 8.0). Reactions were initiated by adding of S-AdoMet to the 10 min pre-incubated mixture (as a negative control that contains the components except S-AdoMet) followed by immediately mixing, and then terminated at 10 sec by addition of TFA to a final concentration of 15% (v/v) to precipitate the enzyme. After removal of the precipitate by centrifugation, Na2CO3 (1.5 M in final) and dansyl chloride (1 mM in final) were supplemented to the supernatant, which was further incubated at 50°C for 60 min. 20 µL of TFA was then added to the solution, and the dansyl derivatives were subjected to HPLC-MS analysis. For quantifying the yield of glycine species, the amino acid Asn (20 µM) was used as an internal standard.
Supplementary Material
Acknowledgements
We thank Prof. H. G. Floss, University of Washington, for providing of S. actuosus ATCC25421 and his pioneer work on NOS biosynthesis, and Prof. Yuheng Zhang and Dr. Wei Tong, High Magnetic Field Laboratory, Chinese Academy of Sciences, for the assistance of EPR analysis. This work was supported in part by grants from National Institutes of Health of U.S (CA094426 to B.S.), National Natural Science Foundation (20832009, 30525001 and 20921091), Ministry of Science and Technology (2009ZX09501-008), National Basic Research Program (“973 program”, 2010CB833200), Chinese Academy of Sciences (KJCX2-YW-H08 and KSCX2-YW-G-06), and Science and Technology Commission of Shanghai Municipality (09QH1402700) of China (all to W.L).
Footnotes
Author contributions
Q.Z., D.C., Y.Y. and L.D. provided the experimental evidences; Y.L. performed the theoretical calculations; Q.Z., B.S. and W.L. analyzed the data and wrote the paper; and W.L. designed the research. All authors discussed results and approved the final manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available online at http://xxxx
References
- 1.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem. Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 2.Arndt H-D, Schoof S, Lu J-Y. Thiopeptide antibiotic biosynthesis. Angew. Chem. Int. Ed. 2009;48:6770–6773. doi: 10.1002/anie.200901808. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Kelly WL. Recent advances in thiopeptide antibiotic biosynthesis. Nat. Prod. Rep. 2010;27:153–164. doi: 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 4.Nicolaou KC, Chen JS, Edmonds DJ, Estrada AA. Recent advances in the chemistry and biology of naturally occurring antibiotics. Angew. Chem. Int. Ed. 2009;48:660–719. doi: 10.1002/anie.200801695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown LCW, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc. Natl. Acad. Sci. USA. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao R, et al. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J. Am. Chem. Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 8.Morris RP, et al. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J. Am. Chem. Soc. 2009;131:5946–4955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 9.Yu Y, et al. Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem. Biol. 2009;4:855–864. doi: 10.1021/cb900133x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, et al. Identification and analysis of the biosynthetic gene cluster encoding the thiopeptide antibiotic cyclothiazomycin in Streptomyces hygroscopicus 10–22. Appl. Environ. Microbiol. 2010;76:2335–2344. doi: 10.1128/AEM.01790-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Y, et al. Moving posttranslational modifications forward to biosynthesize the glycosylated thiopeptide nocathiacin I in Nocardia sp. ATCC202099. Mol. BioSysts. 2010;6:1180–1185. doi: 10.1039/c005121g. [DOI] [PubMed] [Google Scholar]
- 12.Houck DR, Chen L-C, Keller PJ, Beale JM, Floss HG. Biosynthesis of the modified peptide antibiotic nosiheptide in Streptomyces actuosus. J. Am. Chem. Soc. 1988;110:5800–5806. [Google Scholar]
- 13.Mocek U, et al. Biosynthesis of the modified peptide antibiotic nosiheptide in Streptomyces actuosus. J. Am. Chem. Soc. 1993;115:7557–7568. [Google Scholar]
- 14.Mocek U, et al. Biosynthesis of the modified peptide antibiotic thiostrepton in Streptomyces azureus and Streptomyces laurentii. J. Am. Chem. Soc. 1993;115:7992–8001. [Google Scholar]
- 15.Smith TM, Priestley ND, Knaggs AR, Nguyen T, Floss HG. 3,4-Dimethylindole-2-carboxylate and 4-(1-Hydroxyethyl)-2-quinolinecarboxylate activating enzymes from the nosiheptide and thiostrepton producers, Streptomyces actuosus and Streptomyces laurentii. J. Chem. Soc. Chem. Commun. 1993;21:1612–1614. [Google Scholar]
- 16.Priestley ND, et al. Studies on the biosynthesis of thiostrepton: 4-(1-hydroxyethyl)-quinoline-2-carboxylate as a free intermediate on the pathway to the quinaldic acid moiety. Bioorg. Med. Chem. 1996;4:1135–1147. doi: 10.1016/0968-0896(96)00126-5. [DOI] [PubMed] [Google Scholar]
- 17.Frey PA, Hegeman AD, Ruzika FJ. The radical SAM superfamily. Critical Rev. Biochem. Mol. Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 18.Chih HW, Marsh ENG. Pre-steady-state kinetic investigation of intermediates in the reaction catalyzed by adenosylcobalamin-dependent glutamate mutase. Biochemistry. 1999;38:13684–13691. doi: 10.1021/bi991064t. [DOI] [PubMed] [Google Scholar]
- 19.Chih H-W, March NG. Mechanism of glutamate mutase: Identification and kinetic competence of acrylate and glycyl radical as intermediates in the rearrangment of glutamate to methylaspartate. J. Am. Chem. Soc. 2000;122:10732–10733. [Google Scholar]
- 20.Banerjee R. Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem. Rev. 2003;103:2083–2094. doi: 10.1021/cr0204395. [DOI] [PubMed] [Google Scholar]
- 21.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet Y, Drennan CL. AdoMet radical proteins--from structure to evolution--alignment of divergent protein sequences reveals strong secondary structure element conservation. Nucleic Acids Res. 2004;32:4015–4025. doi: 10.1093/nar/gkh728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SC, Frey PA. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem. Sci. 2007;32:101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Marsh ENG, Patterson DP, Li L. Adenosyl radical: reagent and catalyst in enzyme reactions. Chem Bio Chem. 2010;11:604–621. doi: 10.1002/cbic.200900777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonifacic M, Stefanic I, Hug GI, Armstrong DA, Asmus K-D. Glycine decarboxylation: the free radical mechanism. J. Am. Chem. Soc. 1998;120:9930–9940. [Google Scholar]
- 26.Kriek M, Martins F, Challand MR, Croft A, Roach PL. Thiamine biosynthesis in Escherichia coli: identification of the intermediate and by-product derived from tyrosine. Angew. Chem. Ind. Ed. 2007;46:9223–9226. doi: 10.1002/anie.200702554. [DOI] [PubMed] [Google Scholar]
- 27.Driesener RC, et al. [FeFe]-hydrogenase cyanide ligands derived from S-adenosylmethionine-dependent cleavage of tyrosine. Angew. Chem. Int. Ed. 2010;49:1687–1690. doi: 10.1002/anie.200907047. [DOI] [PubMed] [Google Scholar]
- 28.Chatterjee A, et al. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat. Chem. Biol. 2008;4:758–765. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acker MG, Bowers AA, Walsh CT. Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J. Am. Chem. Soc. 2009;131:17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowers AA, Acker MG, Koglin A, Walsh CT. Thiazolyl peptide antibiotic biosynthesis: a cascade of posttranslational modifications on ribosomal nascent proteins. J. Am. Chem. Soc. 2010;132:7519–7527. doi: 10.1074/jbc.R110.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kriek M, et al. Thiazole synthase from Escherichia coli - an investigation of the substrates and purified proteins required for activity in vitro. J. Biol. Chem. 2007;282:17413–17423. doi: 10.1074/jbc.M700782200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.