Abstract
The binding affinity is determined by the Gibbs energy of binding (ΔG) which is the sum of enthalpic (ΔH) and entropic (-TΔS) contributions. Because the enthalpy and entropy contribute in an additive way to the binding energy, the same binding affinity can be achieved by many different combinations of enthalpic and entropic contributions; however, do compounds with similar binding affinities but different thermodynamic signatures (i.e. different ΔH, -TΔS combinations) exhibit the same functional effects? Are there characteristics of compounds that can be modulated by modifying their thermodynamic signatures? In this paper, we consider the minimization of unwanted conformational effects arising during the development of CD4/gp120 inhibitors, a new class of HIV-1 cell entry inhibitors. Competitive inhibitors of protein/protein interactions run the risk of triggering the very same signals that they are supposed to inhibit. Here, we show that for CD4/gp120 inhibitors the magnitude of those unwanted effects is related to the proportion in which the enthalpy and entropy changes contribute to the binding affinity. The thermodynamic optimization plot (TOP) previously proposed to optimize binding affinity can also be used to obtain appropriate enthalpy/entropy combinations for drug candidates.
Keywords: Binding Affinity, Enthalpy, Entropy, Thermodynamic Optimization, Isothermal Titration Calorimetry
The development of small molecules that are able to inhibit protein/protein interactions is a very active area of research (1-3). In many cases, the binding of a protein to another (e.g. protein ligand to cell surface receptor) generates a signal that is transmitted downstream to other proteins. In those situations, a major goal in drug design is the inhibition of the signal. Achieving that goal with a competitive inhibitor poses the risk that the inhibitor itself may act as a surrogate protein ligand and trigger the signal that needs to be inhibited. In fact, those unwanted effects have been reported for HIV-1 cell entry inhibitors (4).
The first event in HIV-1 infection is the binding of the virus envelope glycoprotein gp120 to the cell surface receptor CD4 (5, 6). The binding of CD4 triggers a conformational change in gp120 that renders the envelope glycoprotein able to bind to the chemokine coreceptor (CCR5 or CXCR4). This conformational change is characterized by a large scale structuring or folding event in gp120, which is reflected in a very large favorable enthalpy change and a very large unfavorable entropy change. In fact, the binding of CD4 structures the coreceptor binding site in gp120 allowing it to bind to the chemokine coreceptor (7). The binding of gp120 to the chemokine coreceptor triggers the sequence of events that leads to the fusion of viral and cell membranes and subsequent cell infection. Previously (4) we showed that a low molecular weight compound (NBD-556, shown in Figure 1) that competes with CD4 was able to activate the coreceptor site in gp120, allowing the virus to infect CD4-negative cells. We also found that this compound had a thermodynamic signature similar to the one exhibited by CD4, indicating that it triggered the same conformational changes in gp120 that lead to the activation of the coreceptor binding site. The question raised from those observations is whether it is possible to develop a CD4 competitive inhibitor that does not elicit the same unwanted conformational effects that lead to the activation of the coreceptor site.
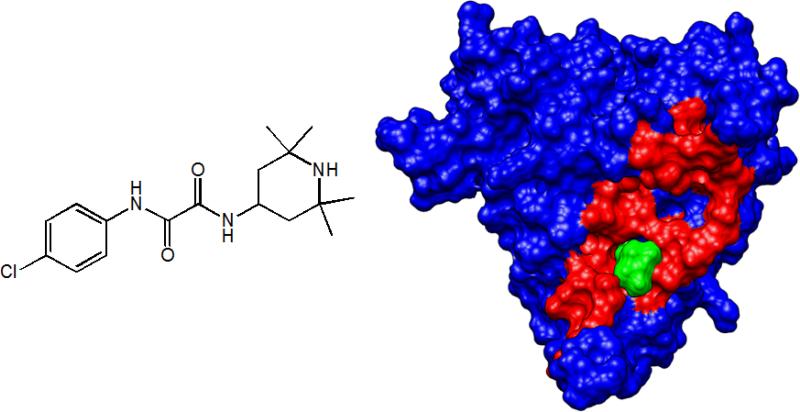
Figure 1.
NBD-556 is a low-molecular-weight compound (left panel) that binds to gp120, partially overlapping the footprint of CD4. The right panel shows gp120 (PDB entry 1G9N) with residues that are within 5 Å of CD4 in red. NBD-556 (green) interacts only with a few residues within the CD4 footprint but is still able to trigger a large conformational structuring of gp120 that results in enhancement of infection of CD4-negative cells (19).
The binding affinity is a function of the Gibbs energy, , which itself is a function of two quantities, the enthalpy (ΔH) and entropy (ΔS) changes, ΔG = ΔH - TΔS. Because of this additive character, many ΔH, -TΔS combinations can give rise to the same binding affinity. Since the enthalpy and entropy changes reflect different types of interactions, it can reasonably be hypothesized that compounds with similar affinities but different thermodynamic signatures may exhibit different characteristics. In fact, previous studies with HIV-1 protease inhibitors have indicated that inhibitors with similar affinities but different enthalpy/entropy profiles display different susceptibilities to drug resistance mutations (8). In this paper, we explore that hypothesis within the context of the optimization of CD4/gp120 inhibitors and the suppression of the unwanted conformational structuring effect.
RESULTS AND DISCUSSION
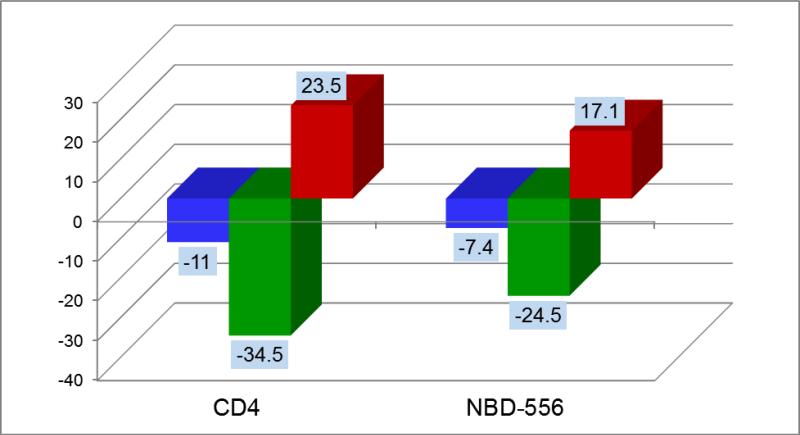
The extracellular portion of CD4 (MW = 44 kDa) binds to gp120 with an affinity close to 8 nM in a process characterized by a large favorable enthalpy and large unfavorable entropy (ΔH = -34.5; -TΔS = 23.5 at 25°C) (9, 10). These values are much larger than those usually found in protein/protein associations, indicating that an additional process takes place during CD4/gp120 binding. In fact, those large enthalpy/entropy values have been associated with a large conformational structuring or folding in gp120 (9, 10). From the available crystallographic structures of the gp120-CD4 complex, always obtained with the additional presence of an antibody that mimics the coreceptor (11-13), it is possible to evaluate the interactions between CD4 and gp120. CD4 forms 12 hydrogen bonds and 219 van der Waals contacts with 26 residues in gp120 (13). The contact surface between the two proteins buries from the solvent 1956 Å2 of which 63% is hydrophobic (surface areas were calculated according to Lee and Richards (14)). If CD4 and gp120 had the same conformation in solution as in the complex, a small favorable enthalpy and a favorable entropy would have been expected (15). In fact, antibodies that do not induce a structuring in gp120 (e.g. b12) exhibit that pattern (16, 17). The favorable entropy would have originated primarily from the burial of hydrophobic surface upon binding and the favorable enthalpy from the interactions established between the two proteins (8, 18). Calculations based on the burial of the interacting surface from the solvent (15) show that CD4 binding to gp120 would theoretically be associated with a negative change in heat capacity on the order of -0.4 kcal/(K × mol) which is much smaller in magnitude than the measured value of -1.8 kcal/(K × mol) indicating that a much larger surface is buried from the solvent upon binding than just the gp120-CD4 interface. A large negative heat capacity change would usually be associated with a favorable entropy change due to the large associated desolvation. However, the experimental entropy change is large and unfavorable, indicating that the large structuring effect more than compensates for the favorable change in desolvation entropy.
NBD-556 (MW = 337.84) shown in Figure 1 is a small non-peptidic molecule that binds to the gp120 cavity where Phe-43 in CD4 binds (4, 19, 20). NBD-556 is a competitive inhibitor of CD4 characterized by a binding affinity of 3.7 μM. Despite the small size, NBD-556 binds with a thermodynamic signature that resembles that of CD4 (ΔH = -24.5 kcal/mol, -TΔS = 17.1 kcal/mol at 25 °C) (Figure 2) that causes a similar structuring in gp120 and in turn triggers the infection of CD4-negative cells (4); i.e. NBD-556 acts as a surrogate CD4, an unwanted effect. The interactions between NBD-556 and gp120 have been evaluated by docking NBD-556 into the Phe-43 cavity of gp120 (PDB entry 1G9N) (19). The unwanted conformational structuring triggered by NBD-556 is also reflected in the observed changes in the CD spectrum of gp120 upon binding. Compared to CD4, the complex of NBD-556 with gp120 only buries from the solvent only 668 Å2 of which 69% are hydrophobic. If there were no conformational change, a change in heat capacity on the order of -0.15 kcal/(K × mol) would have been expected; instead the measured heat capacity change is -1.0 kcal/(K × mol). It is then apparent that the successful development of an effective CD4/gp120 inhibitor requires the elimination of the unwanted viral infection enhancement, which originates from the conformational structuring of gp120 and is reflected in the thermodynamic signature.
Figure 2.
The thermodynamic signatures of sCD4 and NBD-556 at 25°C. The large conformational structuring of gp120 triggered by CD4 binding is reflected in a thermodynamic signature characterized by an unusually large favorable change in enthalpy and a very large unfavorable entropy change. Except for a lower affinity, the binding of NBD-556 to gp120 is also associated with enthalpy and entropy changes similar to those observed for CD4. ΔG is represented by blue bars, ΔH by green bars and -TΔS by red bars.
Viral Infection Enhancement and Thermodynamic Signature
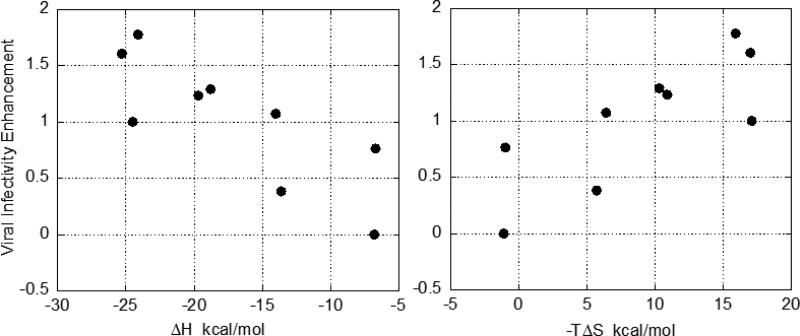
Viral infectivity assays performed on CD4 negative cells (19) for selected analogs of NBD-556 do in fact indicate that some analogs do not induce a viral infectivity enhancement in CD4-negative cells and that the unwanted effect is related to the magnitude of the enthalpy and entropy changes. Figure 3 shows the dependence of the unwanted infectivity enhancement with the enthalpy and entropy changes of a series of analogs. It is clear that compounds with small enthalpy and more favorable entropy changes exhibit low infectivity enhancement. It must be noted that this correlation cannot be perfect and has a characteristic spread because different analogs of the lead compound establish specific interactions with the target that contribute differently to the enthalpy and entropy changes.
Figure 3.
The correlation between viral infectivity enhancement in CD4 negative cell and the enthalpic and entropic contributions to the binding affinity. NBD-556 enhances viral infectivity in CD4 negative cells by three orders of magnitude (4). The effect of the compounds on infection with HIV-1 was studied by using recombinant, luciferase-expressing HIV-1 carrying the wild-type HIV-1YU2 envelope glycoproteins. HIV-1 was incubated with increasing concentrations of the compound and added to Cf2Th-CCR5 cells lacking CD4 receptor. Cells were lysed 48h later and luciferase activity measured (4). The area under the dose-response curve for each compound was calculated and normalized to the value obtained in presence of the reference compound, NBD-556. The enthalpy and entropy changes were determined at 25°C by isothermal titration calorimetry using a high-precision VP-ITC titration calorimetric system from MicroCal Inc. The calorimetric cell (~1.4 mL), containing gp120 from the YU2 strain dissolved in PBS (Roche Diagnostics GmbH), pH 7.4 with 2% DMSO, was titrated with the different inhibitors dissolved in the same buffer. The concentration of gp120 was ~2 μM and inhibitor at a concentration of 80 – 150 μM was added in aliquots of 10 μL until saturation was reached (usually in 20 – 30 injections). The compounds used in the plots were selected for measurements of viral infectivity enhancement based on the wide range of enthalpy and entropy values.
These experiments demonstrate that certain binding-related properties of drug candidates can be better correlated with the enthalpy/entropy balance rather than with the overall binding affinity. The overall binding energies (ΔG) of these compounds are all bracketed within 1.4 kcal/mol whereas the spread of ΔH or -TΔS is 18 kcal/mol. These results validate the use of thermodynamic signatures as optimization guidelines for drug properties other than binding affinity. They also demonstrate the feasibility of generating analogs that bind to the same target site but do not induce or induce to a lesser extent the unwanted conformational structuring. The wide spread of enthalpy/entropy values indicates the existence of an ensemble of partially structured conformations between the unstructured and the structured states. These observations can have general implications for the development of inhibitors of protein/protein interactions.
Thermodynamic Optimization Plot
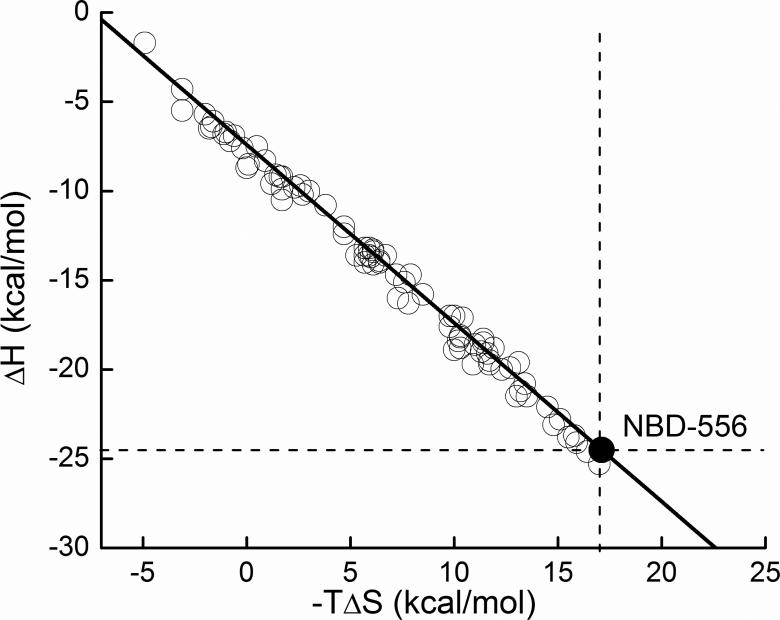
The thermodynamic optimization plot (21) was presented earlier as a convenient tool to identify the enthalpic and entropic consequences of introducing modifications at specific positions of a lead compound. As such, its first applications were directed at the optimization of the binding affinity. Here we show that the thermodynamic optimization plot can also be employed to modify the thermodynamic signature of a compound. The thermodynamic optimization plot (21) is constructed by plotting the measured ΔH and -TΔS values of the lead compound (in this case NBD-556) as shown in Figure 4. A straight line of slope -1 is drawn over the experimental point. As discussed before, all compounds that fall onto the optimization line have the same Gibbs energy (ΔG) and correspondingly the same binding affinity as the lead compound. Likewise, all compounds that fall above the optimization line have a lower binding affinity (more positive ΔG), and all compounds that fall below the optimization line have a higher binding affinity (more negative ΔG). In addition, all compounds above the dashed horizontal line have less favorable enthalpy than the parent compound and all compounds to the left of the dashed vertical line have a more favorable entropy contribution. Vice versa, compounds below the horizontal line have a more negative enthalpy and compounds to the right of the vertical line a less favorable entropy change. The thermodynamic optimization plot allows mapping of the enthalpic, entropic and affinity consequences of specific chemical modifications to precise locations within the lead compound.
Figure 4.
Thermodynamic Optimization Plot. ΔH, -TΔS pairs for each compound were determined by ITC as described in the figure legend to Figure 3.
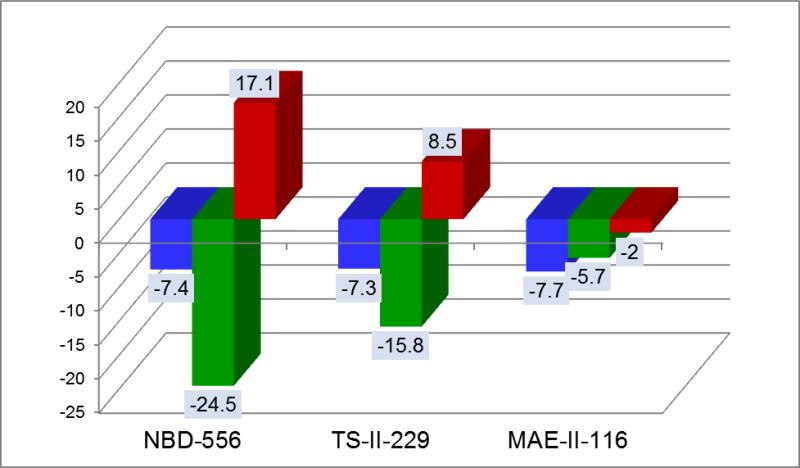
For the development of CD4/gp120 inhibitors, the design goals are: 1) Elimination of unwanted conformational structuring effects; and, 2) Improvement of binding affinity. Since the unwanted conformational structuring effects are proportional to a large negative enthalpy and a large unfavorable entropy, the lead optimization strategy is to modify the parent compound in such a way that the resulting new compounds will move above the dashed horizontal line and to the left of the dashed vertical line. Figure 4 shows the results achieved through this exercise. Several conclusions can be obtained. First, it is clear that the parent compound can be modified in such a way as to generate analog compounds characterized by small favorable enthalpy changes (~ -5 kcal/mol) and favorable rather than unfavorable entropy changes, as expected for small molecular weight compounds that bind without inducing a structuring effect. Second, under those circumstances, binding affinity optimization should be achieved by compound modifications that improve the entropy change. Since, in this particular case, a more favorable enthalpy is not only related to better compound/gp120 interactions but also to the unwanted structuring effect, the main affinity optimization criteria should be the entropy change. Figure 5 shows the evolution of the thermodynamic signature along the optimization path. The three compounds shown in the figure belong to the same chemical scaffold and have similar molecular weights (<340) and similar number of rotatable bonds indicating that the entropic changes are not due to the compounds themselves. As observed previously for other protein targets (8, 22-25) these studies confirm that different ligands can bind to the same site with vastly different thermodynamic signatures. Experience from other drug targets (22, 24, 26) indicate that entropic contributions usually account for more than -9 kcal/mol to the binding affinity if the enthalpy change is close to zero. The thermodynamic optimization plot provides a clear map of the effects of different modifications in different regions of the compound on the binding enthalpy and entropy changes. As such, the thermodynamic optimization plot provides a blueprint for optimization of the affinity within the required enthalpy/entropy constraints.
Figure 5.
Evolution of thermodynamic signatures throughout the optimization path. The suppression of the unwanted side effect requires elimination of the conformational change that structures the coreceptor site. This goal is accomplished by searching for an analog of the lead compound, which binds without eliciting large favorable enthalpy and large unfavorable entropy changes. In fact, the last compound in the series already exhibits the characteristic binding signature of a small ligand (26). The three compounds belong to the same scaffold as NBD-556 and have similar molecular weights and number of rotatable bonds. ΔG is represented by blue bars, ΔH by green bars and -TΔS by red bars.
CONCLUSIONS
The studies presented here indicate that important properties of drug candidates, other than binding affinity, may be related to the proportion in which the binding enthalpy and binding entropy contribute to the binding affinity, and that modification of their thermodynamic signatures can be used to optimize those properties. During the development of small molecule inhibitors of protein/protein interactions there is always the danger that the inhibitor may act as a surrogate protein ligand, triggering the signal that needs to be inhibited. The results presented here suggest that, at least in some cases, those unwanted effects can be avoided by modifying the location and type of interactions that determine inhibitor binding. In analogy to the so-called binding “hot spots” (27), protein/protein interfaces may also have allosteric “hot spots” that initiate signaling transmission. Depending on the design goals, those allosteric “hot spots” will need to be targeted or avoided.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM56550 and GM57144) and the National Science Foundation (MCB0641252).
REFERENCES
- 1.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 2.Berg T. Small-molecule inhibitors of protein-protein interactions. Curr Opin Drug Discov Devel. 2008;11:666–674. [PubMed] [Google Scholar]
- 3.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 4.Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, et al. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry. 2006;45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 6.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, et al. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 8.Ohtaka H, Freire E. Adaptive inhibitors of the HIV-1 protease. Prog Biophys Mol Biol. 2005;88:193–208. doi: 10.1016/j.pbiomolbio.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Leavitt SA, Schön A, Klein JC, Manjappara U, Chaiken IM, Freire E. Interactions of HIV-1 proteins gp120 and Nef with cellular partners define a novel allosteric paradigm. Curr Protein Pept Sci. 2004;5:1–8. doi: 10.2174/1389203043486955. [DOI] [PubMed] [Google Scholar]
- 10.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, et al. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci U S A. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 15.Luque I, Freire E. Structure-based prediction of binding affinities and molecular design of peptide ligands. Methods Enzymol. 1998;295:100–127. doi: 10.1016/s0076-6879(98)95037-6. [DOI] [PubMed] [Google Scholar]
- 16.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 17.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mobley DL, Dill KA. Binding of small-molecule ligands to proteins: “what you see” is not always “what you get”. Structure. 2009;17:489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madani N, Schon A, Princiotto AM, Lalonde JM, Courter JR, Soeta T, et al. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure. 2008;16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Q, Ma L, Jiang S, Lu H, Liu S, He Y, et al. Identification of N-phenyl-N'-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology. 2005;339:213–225. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Freire E. A thermodynamic approach to the affinity optimization of drug candidates. Chem Biol Drug Des. 2009;74:468–472. doi: 10.1111/j.1747-0285.2009.00880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbonell T, Freire E. Binding thermodynamics of statins to HMG-CoA reductase. Biochemistry. 2005;44:11741–11748. doi: 10.1021/bi050905v. [DOI] [PubMed] [Google Scholar]
- 23.Muzammil S, Armstrong AA, Kang LW, Jakalian A, Bonneau PR, Schmelmer V, et al. Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations. J Virol. 2007;81:5144–5154. doi: 10.1128/JVI.02706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarver RW, Peevers J, Cody WL, Ciske FL, Dyer J, Emerson SD, et al. Binding thermodynamics of substituted diaminopyrimidine renin inhibitors. Anal Biochem. 2007;360:30–40. doi: 10.1016/j.ab.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Sarver RW, Bills E, Bolton G, Bratton LD, Caspers NL, Dunbar JB, et al. Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Med Chem. 2008;51:3804–3813. doi: 10.1021/jm7015057. [DOI] [PubMed] [Google Scholar]
- 26.Freire E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discov Today. 2008;13:869–874. doi: 10.1016/j.drudis2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]