Abstract
Background
Several studies have shown that common variants in the MTNR1B gene were associated with fasting glucose level and type 2 diabetes. The purpose of this study was to examine whether tagging single nucleotide polymorphisms (SNPs) in the MTNR1B region were associated with type 2 diabetes and related traits in a Han Chinese population.
Methods
We investigated the association of polymorphisms in the MTNR1B gene with type 2 diabetes by employing a case-control study design (1118 cases and 1161 controls). Three tagging SNPs (rs10830963, rs3781637, and rs1562444) with R2>0.8 and minor allele frequency>0.05 across the region of the MTNR1B gene were studied. Genotyping was performed by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy using a MassARRAY platform.
Results
The polymorphism rs3781637 was associated with type 2 diabetes adjusted for age, sex and body mass index (BMI) in the additive model and recessive model (OR = 1.22, 95% CI 1.01-1.46, p = 0.038 and OR = 2.81, 95% CI 1.28-6.17, p = 0.01, respectively). In the non-diabetic controls, rs3781637 was nominally associated with plasma triglyceride, total cholesterol and low density lipoprotein cholesterol (LDL-C) levels in the recessive model (p = 0.018, 0.008 and 0.038, respectively). After adjustment for multiple comparisons, the associations of rs3781637 with total cholesterol and LDL-C remained significant in the recessive model (the empirical p = 0.024 and 0.045, respectively), but the association between rs3781637 and triglyceride became non-significant (the empirical p = 0.095). The associations of rs10830963 and rs1562444 with type 2 diabetes and related traits were not significant in the additive, dominant and recessive models.
Conclusions
The rs3781637 A/G polymorphism of the MTNR1B gene is associated with type 2 diabetes, plasma, total cholesterol and LDL-C levels in the Han Chinese population.
Background
Circadian rhythms are closely related to metabolism, and dysregulation of circadian rhythms may increase the risk of diabetes [1]. The MTNR1B gene encodes a high affinity receptor for melatonin, a hormone primarily secreted by the pineal gland to regulate circadian rhythm and sleep cycles [2]. Plasma melatonin follows an opposite circadian rhythm to plasma insulin and glucose, rising by night and falling by day [3]. There are favorable evidences that circadian rhythm of melatonin influences insulin secretion and glucose homeostasis via its islet-specific receptor [4]. Consistently, melatonin secretion and circadian rhythm are impaired in type 2 diabetes patients [2]. More importantly, MTNR1B inhibits insulin secretion through its effect on CGMP formation when activated by melatonin [5]. Therefore, the MTNR1B gene might be involved in glucose homeostasis and type 2 diabetes.
Several large-scale genome-wide association analyses demonstrated that common variants in or near the MTNR1B gene to be robustly associated with fasting glucose level in European populations [6-8], with polymorphism rs10830963 showing the most significant association signal [7]. Several replication studies in European [9-12], Indian [13], Sri Lankan and Japanese populations [14] confirmed that MTNR1B rs10830963 contributed to raised fasting glucose level and increased risk of type 2 diabetes.
Several studies investigated the associations of MTNR1B rs10830963 with type 2 diabetes and fasting glucose among Han Chinese populations [15-19]. Ling et al. first replicated the association of MTNR1B rs10830963 with type 2 diabetes and fasting glucose in a case-control study including 1165 type 2 diabetes cases and 1105 normoglycaemic controls [17]. After that, another four studies reported that MTNR1B rs10830963 was associated with type 2 diabetes and/or fasting glucose in Han Chinese populations [15,16,18,19]. Whether other common variants except rs10830963 in the MTNR1B gene are associated with type 2 diabetes and fasting glucose in Han Chinese population is unknown.
The aim of this study was to examine whether common polymorphisms in MTNR1B were associated with type 2 diabetes and related traits in a Han Chinese population.
Methods
Subjects
All participants were of Southern Han Chinese ancestry and resided in the Shanghai metropolitan area. Type 2 diabetic inpatients (n = 1118) were recruited from the Endocrinology and Metabolism Department of Zhongshan Hospital in Shanghai, which is affiliated to Fudan University. All type 2 diabetic patients met the 1999 WHO criteria for diabetes [20], had been diagnosed after the age of 29 years, and were treated with oral hypoglycaemic agents and/or insulin. 1161 unrelated non-diabetic control participants were people undergoing health examinations in Zhongshan Hospital, were older than 40 years, and had a fasting plasma glucose < 5.6 mmol/l.
Written informed consent was obtained from all participants and the study was approved by the ethnic committee of Zhongshan hospital, Fudan University, Shanghai, China.
Clinical measurements
Both the diabetic patients and the controls were extensively phenotyped for anthropometric and biochemical traits related to glucose metabolism. The phenotypes assessed in our study include height, weight, fasting glucose, total cholesterol, triglyceride, high density lipoprotein cholesterol (HDL-C) and LDL-C. BMI was calculated as weight (kg)/height2 (m2).
Genotyping
We selected tagging single nucleotide polymorphisms (SNPs) with the criteria of R2>0.8 and minor allele frequency (MAF)>0.05 across the region of the MTNR1B gene (include 3kb upstream and 3kb downstream of the gene) from the HapMap Phase II, using the pairwise tagging model. Pairwise tagging algorithm was developed by Carlson et al., and the detailed description was introduced previously [21]. Three tagging SNPs (rs10830963, rs3781637, and rs1562444) were selected. The genotyping was performed by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy using a MassARRAY platform (MassARRAY Compact Analyzer, Sequenom, San Diego, CA, USA).
Statistical analysis
Continuous variables are expressed as means ± standard error of mean (SEM) and median (interquartile range). Comparisons between groups were performed with T testing and χ2 testing for normally distributed continuous and categorical variables, respectively. Deviations from the Hardy-Weinberg equilibrium were assessed by means of χ2 testing. SNPs that were not in the Hardy-Weinberg equilibrium were excluded from further analysis. Additive, dominant and recessive models were assumed in the association analysis. We tested the association of the polymorphisms with type 2 diabetes by using logistic regression. Multivariate linear regression was used to test the association of the polymorphisms with quantitative traits. All models were adjusted for age, sex, and BMI. Permutations (10,000 times) were performed for each trait to assess empirical p values using PLINK[22] in order to adjust the multiple comparisons. Analysis was performed with SPSS software version 17.0.
Results
Baseline characteristics
The baseline characteristics of participants in this study are presented in Table 1. In 2279 participants, 1118 were type 2 diabetes patients and 1161 were non-diabetic controls. Diabetic cases were older and had higher BMI, fasting glucose and triglyceride levels, but lower cholesterol concentrations than the non-diabetic controls. There was no significant difference of the distribution of sex between diabetic cases and non-diabetic controls (p = 0.34).
Table 1.
Baseline characteristics of diabetic cases and non-diabetic controls
| Characteristic | Diabetic cases (n = 1118) |
Non-diabetic controls (n = 1161) |
P value* |
|---|---|---|---|
| Age (years) | 60.2 ± 0.37 | 56.5 ± 0.32 | <0.001 |
| Men (%) | 44.6 | 42.7 | 0.34 |
| BMI (kg/m2) | 24.4 ± 0.11 | 23.5 ± 0.09 | <0.001 |
| Fasting glucose (mmol/l) | 8.17 ± 0.04 | 4.86 ± 0.01 | <0.001 |
| Total cholesterol (mmol/l) | 4.37(3.78-5.07) | 5.10(4.50-5.80) | <0.001 |
| Triglyceride (mmol/l) | 1.47(1.00-2.24) | 1.40(1.00-1.90) | <0.001 |
| HDL-C (mmol/l) | 1.12(1.00-1.38) | 1.30(1.12-1.54) | <0.001 |
| LDL-C (mmol/l) | 2.38(1.92-3.01) | 3.07(2.50-3.60) | <0.001 |
Data are expressed as means ± SEM and median (interquartile range)
*p value for comparison between cases and controls
Associations of MTNR1B polymorphisms with type 2 diabetes
Overall, three SNPs (rs10830963, rs3781637, and rs1562444) were selected and genotyped in the present study. The call rates of rs10830963, rs3781637, and rs1562444 were 98.0%, 98.5% and 98.0%, respectively. The concordant rates of all SNPs based on 120 duplicates were 100%.
rs10830963, rs3781637, and rs1562444 were in Hardy-Weinberg equilibrium in the study population (Table 2). The polymorphism rs3781637 was nominally associated with type 2 diabetes adjusted for age, sex and BMI in the additive and recessive models (p = 0.038 and 0.01, respectively) (Table 3). After adjustment for multiple comparisons, the association of rs3781637 with type 2 diabetes remained significant in the additive and recessive models (the empirical p = 0.050 and 0.018, respectively). The carriers of GG genotype was associated with higher odds of type 2 diabetes compared with the carriers of AA and AG genotypes adjusted for age, sex and BMI (OR = 2.81, 95% CI 1.28-6.17, p = 0.01) (Table 3). The associations of rs10830963 and rs1562444 with type 2 diabetes were not significant in the additive, dominant and recessive models (Table 3).
Table 2.
Characteristics of SNPs genotyped in MTNR1B
| SNP identification | Chromosome Position |
Relation to the gene | Major allele | Minor allele | MAF | HWE P value |
|
|---|---|---|---|---|---|---|---|
| 1 | rs10830963 | 92348358 | Intron | C | G | 0.40 | 0.13 |
| 2 | rs3781637 | 92353418 | Intron | A | G | 0.14 | 0.13 |
| 3 | rs1562444 | 92355497 | 3'UTR | A | G | 0.31 | 0.14 |
Table 3.
Genotypic and allelic distribution of MTNR1B polymorphisms and association with type 2 diabetes
| SNP | Alleles Major/minor |
MAF | Genotype Frequency T2DM cases/controls |
ORadd (95%CI) a |
P value* | ORdom (95%CI)b |
P value* | ORrec (95%CI) c |
P value* |
|---|---|---|---|---|---|---|---|---|---|
| rs10830963 | C/G | 0.40 | 0.361/0.481/0.159 | 0.92 | 0.22 | 0.85 | 0.08 | 1.01 | 0.96 |
| 0.348/0.508/0.144 | (0.81-1.05) | (0.70-1.02) | (0.78-1.30) | ||||||
| rs3781637 | A/G | 0.14 | 0.717/0.260/0.022 | 1.22 | 0.038 | 1.17 | 0.13 | 2.81 | 0.01 |
| 0.736/0.252/0.012 | (1.01-1.46) | (0.96-1.43) | (1.28-6.17) | ||||||
| rs1562444 | A/G | 0.31 | 0.472/0.432/0.096 | 1.06 | 0.42 | 1.02 | 0.83 | 1.27 | 0.14 |
| 0.457/0.456/0.088 | (0.92-1.22) | (0.85-1.22) | (0.92-1.74) |
a Calculated using logistic regression, assuming an additive model adjusted for age, sex and BMI
b Calculated using logistic regression, assuming a dominant model adjusted for age, sex and BMI
c Calculated using logistic regression, assuming a recessive model adjusted for age, sex and BMI
* P values shown are not corrected for multiple comparisons
MAF: minor allele frequency in control samples; T2DM: type 2 diabetes.
Associations of MTNR1B polymorphisms with quantitative traits in non-diabetic controls
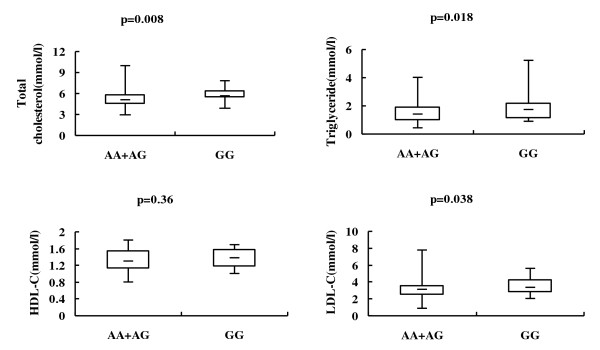
In the non-diabetic controls, the polymorphism rs3781637 was nominally associated with plasma triglyceride, total cholesterol and LDL-C levels adjusted for age, sex and BMI in the recessive model (p = 0.018, 0.008 and 0.038, respectively) (Table 4, Figure 1). The p values of the associations of rs3781637 with plasma triglyceride, total cholesterol and LDL-C levels were 0.011, 0.006 and 0.036 after additional adjustment for fasting glucose. After adjustment for multiple comparisons, the associations of rs3781637 with total cholesterol and LDL-C remained significant in the recessive model (the empirical p = 0.024 and 0.045, respectively), but the association between rs3781637 and triglyceride became non-significant (the empirical p = 0.095). The carriers of GG genotype had a significantly higher plasma total cholesterol and LDL-C levels compared with the carriers of AA and AG genotypes adjusted for age, sex and BMI, and the effect sizes for the GG genotype were 0.841 and 0.585 mmol/l, respectively (Table 4). The polymorphism rs3781637 was not associated with fasting plasma glucose and HDL-C levels in the additive, dominant and recessive models (Table 4). The associations of rs10830963 and rs1562444 with fasting plasma glucose, plasma triglyceride, total cholesterol, LDL-C and HDL-C levels were not significant in all three models (Table 4).
Table 4.
Association between MTNR1B polymorphisms and quantitative traits in non-diabetic controls
| rs10830963 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | CC | CG | GG | βadd ± SEM a | Padda* | βdom ± SEM b | Pdomb* | βrec ± SEM c | Precc* |
| n | 396 | 578 | 164 | ||||||
| Fasting glucose (mmol/l) | 4.86 ± 0.02 | 4.87 ± 0.02 | 4.84 ± 0.03 | -0.009 ± 0.017 | 0.59 | 0.0004 ± 0.024 | 0.99 | -0.033 ± 0.032 | 0.30 |
| Triglyceride (mmol/l) | 1.40(1.10-1.90) | 1.40(1.00-1.90) | 1.40(1.10-1.80) | -0.062 ± 0.049 | 0.21 | -0.079 ± 0.070 | 0.25 | -0.080 ± 0.094 | 0.40 |
| Total cholesterol (mmol/l) | 5.20(4.50-5.90) | 5.10(4.50-5.73) | 5.10(4.50-5.80) | -0.015 ± 0.045 | 0.74 | -0.042 ± 0.063 | 0.50 | 0.022 ± 0.084 | 0.79 |
| LDL-C (mmol/l) | 3.10(2.54-3.64) | 3.09(2.50-3.57) | 3.09(2.57-3.67) | -0.002 ± 0.039 | 0.97 | -0.020 ± 0.055 | 0.72 | 0.030 ± 0.075 | 0.69 |
| HDL-C (mmol/l) | 1.30(1.10-1.60) | 1.40(1.13-1.53) | 1.40(1.14-1.50) | -0.003 ± 0.014 | 0.84 | -0.011 ± 0.019 | 0.57 | 0.010 ± 0.026 | 0.70 |
| rs3781637 | |||||||||
| Genotype | AA | AG | GG | βadd ± SEM a | Padda* | βdom ± SEM b | Pdomb* | βrec ± SEM c | Precc* |
| n | 841 | 288 | 14 | ||||||
| Fasting glucose (mmol/l) | 4.87 ± 0.01 | 4.84 ± 0.02 | 4.78 ± 0.10 | -0.037 ± 0.025 | 0.14 | -0.035 ± 0.026 | 0.18 | -0.129 ± 0.124 | 0.30 |
| Triglyceride (mmol/l) | 1.40(1.00-1.90) | 1.40(1.00-1.90) | 1.70(1.13-2.18) | 0.032 ± 0.071 | 0.65 | 0.002 ± 0.075 | 0.98 | 0.742 ± 0.314 | 0.018 |
| Total cholesterol (mmol/l) | 5.10(4.50-5.80) | 5.10(4.40-5.80) | 5.60(5.40-6.38) | -0.022 ± 0.065 | 0.74 | -0.062 ± 0.069 | 0.367 | 0.841 ± 0.316 | 0.008 |
| LDL-C (mmol/l) | 3.07(2.55-3.62) | 3.02(2.50-3.56) | 3.36(2.81-4.26) | -0.021 ± 0.057 | 0.71 | -0.050 ± 0.060 | 0.40 | 0.585 ± 0.281 | 0.038 |
| HDL-C (mmol/l) | 1.30(1.13-1.53) | 1.30(1.11-1.60) | 1.38(1.17-1.59) | -0.001 ± 0.020 | 0.95 | 0.003 ± 0.021 | 0.89 | -0.086 ± 0.095 | 0.36 |
| rs1562444 | |||||||||
| Genotype | AA | AG | GG | βadd ± SEM a | Padda* | βdom ± SEM b | Pdomb* | βrec ± SEM c | Precc* |
| n | 520 | 518 | 100 | ||||||
| Fasting glucose (mmol/l) | 4.86 ± 0.02 | 4.86 ± 0.02 | 4.87 ± 0.04 | 0.005 ± 0.018 | 0.79 | 0.005 ± 0.023 | 0.83 | 0.009 ± 0.042 | 0.83 |
| Triglyceride (mmol/l) | 1.40(1.00-1.90) | 1.40(1.00-1.90) | 1.40(0.90-1.90) | 0.009 ± 0.047 | 0.85 | 0.046 ± 0.060 | 0.44 | -0.108 ± 0.109 | 0.32 |
| Total cholesterol (mmol/l) | 5.10(4.50-5.80) | 5.10(4.50-5.80) | 5.10(4.40-5.90) | -0.004 ± 0.047 | 0.94 | -0.019 ± 0.060 | 0.75 | 0.046 ± 0.110 | 0.67 |
| LDL-C (mmol/l) | 3.11(2.57-3.64) | 3.03(2.50-3.60) | 3.00(2.59-3.58) | -0.022 ± 0.042 | 0.60 | -0.041 ± 0.053 | 0.44 | 0.020 ± 0.097 | 0.84 |
| HDL-C (mmol/l) | 1.30(1.13-1.51) | 1.30(1.11-1.54) | 1.36(1.16-1.65) | 0.010 ± 0.014 | 0.51 | 0.001 ± 0.018 | 0.97 | 0.049 ± 0.033 | 0.14 |
Data are expressed as means ± SEM and median (interquartile range)
a Calculated using multivariate linear regression, assuming an additive model adjusted for age, sex and BMI
b Calculated using multivariate linear regression, assuming a dominant model adjusted for age, sex and BMI
c Calculated using multivariate linear regression, assuming a recessive model adjusted for age, sex and BMI
* P values shown are not corrected for multiple comparison
Figure 1.
Associations of rs3781637 with lipid levels. The upper, middle and lower lines represent the maximum, the median and the minimum, respectively. The bottom and the top of the box represent the 25th and 75th percentiles, respectively. All p values shown are calculated using multivariate linear regression, assuming a recessive model adjusted for age, sex and BMI, and are not corrected for multiple comparisons.
Discussion
Here we reported that a common genetic variant rs3781637 in MTNR1B was associated with type 2 diabetes in a Han Chinese population. The association of rs3781637 with type 2 diabetes was found to be significant in the additive and recessive models. In the recessive model, the carriers of GG genotype were associated to higher odds of type 2 diabetes compared with the carriers of AA and AG genotypes (OR = 2.81). In addition, rs3781637 showed nominal association with plasma triglyceride, total cholesterol and LDL-C levels in the recessive model in non-diabetic controls, and the associations of rs3781637 with total cholesterol and LDL-C levels remained significant after adjustment for multiple comparisons. The carriers of GG genotype were associated to a higher total cholesterol and LDL-C levels compared with the carriers of AA and AG genotypes. In addition, the associations of rs3781637 with lipid levels were independent of glucose metabolism. The association between MTNR1B variants and lipid profile was not reported before, and the mechanism underlying this association remains to be determined. However, many evidences showed that melatonin played an important role in the lipid metabolism. Animal studies showed that melatonin treatment significantly improved dyslipidemia in diabetic rats, with reductions in triglyceride and LDL-C levels [23,24]. The administration of melatonin and zinc was found to improve the lipid profile in type 2 diabetic patients [25]. A novel melatonin agonist, NEU-P11, decreased total cholesterol and triglyceride levels, while increased HDL-C level in obese rats [26]. Therefore, the MTNR1B gene which encoded a high affinity receptor for melatonin may be involved in the lipid metabolism.
We failed to replicate the association of rs10830963 with type 2 diabetes or fasting plasma glucose in our Han Chinese population. However, another three case-control studies in Shanghai Han Chinese populations have reported the significant association of rs10830963 with type 2 diabetes and/or fasting plasma glucose [16-18]. Inconsistent results are often seen between different genetic association studies. Here, population heterogeneity and different environmental factors are unlikely responsible for the inconsistency between studies. Because all study populations had Han ancestry, were from Shanghai metropolitan area, and had similar lifestyle. In addition, the allele frequency of rs10830963 in our study is similar to those in other studies, and the call rate and concordant rate of rs10830963 genotyping in our study are 98.0% and 100%, respectively. Genotyping error could not be the reason for the discrepancy between our study and other studies. Therefore, more replication studies of MTNR1B polymorphisms in Han Chinese and meta-analysis are needed in the future.
Recently, MTNR1B polymorphism rs10830963 was shown to be associated with fasting glucose and homeostasis model assessment of βcell function (HOMA-β) in Caucasian overweight and obese children and adolescents[27], which suggests that the effects of MTNR1B polymorphisms on glucose metabolism were not limited to adult populations.
As diabetes is one of the risk equivalents of coronary heart disease (CAD), and the MTNR1B polymorphism is associated with type 2 diabetes and lipid levels in the present study, do they also contribute to the development of CAD? Until now, there is no report about the association between MTNR1B polymorphisms and CAD. Some other diabetes-related gene polymorphisms such as KCNQ1 gene polymorphisms [28-30]and uncoupling protein 2 (UCP2) gene polymorphisms [31] have been proved to be associated with CAD or the risk factors of CAD. Recently, a polymorphism rs1333049 on chromosome 9p21.3 was found to be associated with CAD and coronary plaque progression in non-diabetic but not in type 2 diabetic patients[32], implying that it affects the development of CAD through a novel biological pathway other than interaction with diabetes or glucose metabolism. Whether polymorphisms in MTNR1B contribute to the susceptibility of CAD, and whether they contribute to diabetes and CAD through a common pathway should be investigated in the future study.
There is strong epidemiologic, clinical and phenotypic overlap between type 2 diabetes and some insulin resistance-related conditions, such as polycystic ovary syndrome (PCOS) [33]. One study investigated the relationship between MTNR1B polymorphisms and PCOS among Chinese women, and found that rs10830963 in MTNR1B is not only associated with susceptibility to PCOS, but also contribute to the PCOS phenotype[34]. However, the authors didn't provide any information about the difference of plasma glucose between the PCOS patients and control subjects. It is possible that the association of MTNR1B polymorphism with PCOS was confounded by the plasma glucose level which tends to be higher in PCOS patients. Further studies are needed to determine whether the association of MTNR1B polymorphisms with PCOS is mediated through its effect on glucose metabolism or is independent of glucose metabolism.
Recent studies also found that MTNR1B polymorphisms are associated with adolescent idiopathic scoliosis (AIS) [35,36], a complex deformity of the spine that most commonly occurs in girls at the peripubertal period between 10 and 16 years of age. This finding is not so surprising because of the fact that melatonin can induce osteoblast differentiation from human mesenchymal stem cells and plays an important role in osteogenesis[37,38]. Studies with animal models, such as pinealectomized chickens[39], pinealectomized bipedal rats[40], and bipedal C57BL/6J mice with genetically low circulating melatonin levels[41], suggested that melatonin deficiency could have a significant role in the development of AIS. Besides melatonin, hormones like leptin[42], adiponectin[43], glucagon-like peptide-1[44] and 1, 25-dihydroxy-vitamin D[45] all effect glucose metabolism and bone metabolism simultaneously.
The associations of MTNR1B rs3781637 with type 2 diabetes and lipids levels found in our study were not reported in the previous studies, and the function of rs3781637 is unknown. The limitation of our study is that we didn't do functional study of rs3781637. It is located in the intron of MTNR1B. It may be associated with alternative splicing of mRNA or the binding of transcription factor and affects the expression level of protein. Future functional study is needed to demonstrate this. There is also no evidence that rs3781637 is linked with any functional variant now, but future fine mapping and resequencing of the MTNR1B gene may detect such functional variants.
Conclusions
Our study demonstrated that MTNR1B rs3781637 A/G polymorphism is associated with type 2 diabetes, total cholesterol and LDL-C levels in the Han Chinese population. These observations suggest that MTNR1B variants may be involved in both glucose and lipid metabolism, which warrants further studies using larger, independent cohorts and a population-based approach.
Abbreviations
SNP: single nucleotide polymorphism; MAF: minor allele frequency; LD: linkage disequilibrium; SEM: standard error of mean; BMI: body mass index; CI: confidence interval; OR: odds ratio; HDL-C: high density lipoprotein cholesterol; LDL-C: low density lipoprotein cholesterol; T2DM: type 2 diabetes; HWE: Hardy-Weinberg equilibrium; HOMA-β: homeostasis model assessment of βcell function; CAD: coronary heart disease; UCP2: uncoupling protein 2; PCOS: polycystic ovary syndrome; AIS: adolescent idiopathic scoliosis.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YL participated in the design of the study, carried out the SNP genotyping and the statistical analysis of the genotype data, and drafted the manuscript. XG contributed to the design and coordination of the study, to the statistical analysis, interpreted the findings and drafted the manuscript. XL, QG, HC and DL participated in the design of the study and the SNP genotyping. All authors read and approved the final manuscript.
Contributor Information
Yan Ling, Email: ling.yan@zs-hospital.sh.cn.
Xiaomu Li, Email: li.xiaomu@zs-hospital.sh.cn.
Qian Gu, Email: gu.qian@zs-hospital.sh.cn.
Hongyan Chen, Email: hychen@fudan.edu.ch.
Daru Lu, Email: drlu@fudan.edu.cn.
Xin Gao, Email: gao.xin@zs-hospital.sh.cn.
Acknowledgements
This work was supported by a grant from the Ministry of Science and Technology of China to XG (grant number: 2008BAI52B03) and a grant from the Major Project of Subject Construction of Shanghai Bureau of Health to XG (grant number: 08GWZX0203).
We are grateful to all subjects who participated in this study.
References
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke E. Melatonin, endocrine pancreas and diabetes. J Pineal Res. 2008;44(1):26–40. doi: 10.1111/j.1600-079X.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- Muhlbauer E, Gross E, Labucay K, Wolgast S, Peschke E. Loss of melatonin signalling and its impact on circadian rhythms in mouse organs regulating blood glucose. Eur J Pharmacol. 2009;606(1-3):61–71. doi: 10.1016/j.ejphar.2009.01.029. [DOI] [PubMed] [Google Scholar]
- Korkmaz A, Topal T, Tan DX, Reiter RJ. Role of melatonin in metabolic regulation. Rev Endocr Metab Disord. 2009;10(4):261–270. doi: 10.1007/s11154-009-9117-5. [DOI] [PubMed] [Google Scholar]
- Mulder H, Nagorny CL, Lyssenko V, Groop L. Melatonin receptors in pancreatic islets: good morning to a novel type 2 diabetes gene. Diabetologia. 2009;52(7):1240–1249. doi: 10.1007/s00125-009-1359-y. [DOI] [PubMed] [Google Scholar]
- Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, Marchand M, Delplanque J, Lobbens S, Rocheleau G, Durand E. et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41(1):89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y. et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41(1):77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, Brodsky J, Jones CG, Zaitlen NA, Varilo T, Kaakinen M. et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spegel P, Bugliani M, Saxena R, Fex M, Pulizzi N. et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41(1):82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenberg C, Pascoe L, Mari A, Tura A, Laakso M, Frayling TM, Barroso I, Loos RJ, Wareham NJ, Walker M. Common genetic variation in the melatonin receptor 1B gene (MTNR1B) is associated with decreased early-phase insulin response. Diabetologia. 2009;52(8):1537–1542. doi: 10.1007/s00125-009-1392-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling E, van 't Riet E, Groenewoud MJ, Welschen LM, van Hove EC, Nijpels G, Maassen JA, Dekker JM, t Hart LM. Combined effects of single-nucleotide polymorphisms in GCK, GCKR, G6PC2 and MTNR1B on fasting plasma glucose and type 2 diabetes risk. Diabetologia. 2009;52(9):1866–1870. doi: 10.1007/s00125-009-1413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparso T, Bonnefond A, Andersson E, Bouatia-Naji N, Holmkvist J, Wegner L, Grarup N, Gjesing AP, Banasik K, Cavalcanti-Proenca C. et al. G-allele of intronic rs10830963 in MTNR1B confers increased risk of impaired fasting glycemia and type 2 diabetes through an impaired glucose-stimulated insulin release: studies involving 19,605 Europeans. Diabetes. 2009;58(6):1450–1456. doi: 10.2337/db08-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Zhang W, Zabaneh D, Sehmi J, Jain P, McCarthy MI, Froguel P, Ruokonen A, Balding D, Jarvelin MR. et al. Common genetic variation near melatonin receptor MTNR1B contributes to raised plasma glucose and increased risk of type 2 diabetes among Indian Asians and European Caucasians. Diabetes. 2009;58(11):2703–2708. doi: 10.2337/db08-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Katsuya T, Chakrewarthy S, Yamamoto K, Fujioka A, Serizawa M, Fujisawa T, Nakashima E, Ohnaka K, Ikegami H. et al. Common variants at the GCK, GCKR, G6PC2-ABCB11 and MTNR1B loci are associated with fasting glucose in two Asian populations. Diabetologia. 2010;53(2):299–308. doi: 10.1007/s00125-009-1595-1. [DOI] [PubMed] [Google Scholar]
- Liu C, Wu Y, Li H, Qi Q, Langenberg C, Loos RJ, Lin X. MTNR1B rs10830963 is associated with fasting plasma glucose, HbA1C and impaired beta-cell function in Chinese Hans from Shanghai. BMC Med Genet. 2010;11:59. doi: 10.1186/1471-2350-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Zhang R, Wang C, Yu W, Lu J, Ma X, Wang J, Jiang F, Tang S, Bao Y. et al. Effects of GCK, GCKR, G6PC2 and MTNR1B variants on glucose metabolism and insulin secretion. PLoS One. 2010;5(7):e11761. doi: 10.1371/journal.pone.0011761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronn T, Wen J, Yang Z, Lu B, Du Y, Groop L, Hu R, Ling C. A common variant in MTNR1B, encoding melatonin receptor 1B, is associated with type 2 diabetes and fasting plasma glucose in Han Chinese individuals. Diabetologia. 2009;52(5):830–833. doi: 10.1007/s00125-009-1297-8. [DOI] [PubMed] [Google Scholar]
- Kan MY, Zhou DZ, Zhang D, Zhang Z, Chen Z, Yang YF, Guo XZ, Xu H, He L, Liu Y. Two susceptible diabetogenic variants near/in MTNR1B are associated with fasting plasma glucose in a Han Chinese cohort. Diabet Med. 2010;27(5):598–602. doi: 10.1111/j.1464-5491.2010.02975.x. [DOI] [PubMed] [Google Scholar]
- Tam CH, Ho JS, Wang Y, Lee HM, Lam VK, Germer S, Martin M, So WY, Ma RC, Chan JC. et al. Common polymorphisms in MTNR1B, G6PC2 and GCK are associated with increased fasting plasma glucose and impaired beta-cell function in Chinese subjects. PLoS One. 2010;5(7):e11428. doi: 10.1371/journal.pone.0011428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agil A, Navarro-Alarcon M, Ruiz R, Abuhamadah S, El-Mir MY, Vazquez GF. Beneficial effects of melatonin on obesity and lipid profile in young Zucker diabetic fatty rats. J Pineal Res. 2010. in press . [DOI] [PubMed]
- Anwar MM, Meki AR. Oxidative stress in streptozotocin-induced diabetic rats: effects of garlic oil and melatonin. Comp Biochem Physiol A Mol Integr Physiol. 2003;135(4):539–547. doi: 10.1016/S1095-6433(03)00114-4. [DOI] [PubMed] [Google Scholar]
- Kadhim HM, Ismail SH, Hussein KI, Bakir IH, Sahib AS, Khalaf BH, Hussain SA. Effects of melatonin and zinc on lipid profile and renal function in type 2 diabetic patients poorly controlled with metformin. J Pineal Res. 2006;41(2):189–193. doi: 10.1111/j.1600-079X.2006.00353.x. [DOI] [PubMed] [Google Scholar]
- She M, Deng X, Guo Z, Laudon M, Hu Z, Liao D, Hu X, Luo Y, Shen Q, Su Z. et al. NEU-P11, a novel melatonin agonist, inhibits weight gain and improves insulin sensitivity in high-fat/high-sucrose-fed rats. Pharmacol Res. 2009;59(4):248–253. doi: 10.1016/j.phrs.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Holzapfel C, Siegrist M, Rank M, Langhof H, Grallert H, Baumert J, Irimie C, Klopp N, Wolfarth B, Illig T. et al. Association of a MTNR1B gene variant with fasting glucose and HOMA-B in children and adolescents with high BMI-SDS. Eur J Endocrinol. 2011;164(2):205–212. doi: 10.1530/EJE-10-0588. [DOI] [PubMed] [Google Scholar]
- Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K, Jorgensen T. et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40(9):1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhou DZ, Zhang D, Chen Z, Zhao T, Zhang Z, Ning M, Hu X, Yang YF, Zhang ZF. et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes in the population of mainland China. Diabetologia. 2009;52(7):1315–1321. doi: 10.1007/s00125-009-1375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yin Q, Ma G, Qian Q. KCNQ1 gene polymorphisms are associated with lipid parameters in a Chinese Han population. Cardiovasc Diabetol. 2010;9:35. doi: 10.1186/1475-2840-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapice E, Pinelli M, Pisu E, Monticelli A, Gambino R, Pagano G, Valsecchi S, Cocozza S, Riccardi G, Vaccaro O. Uncoupling protein 2 G(-866)A polymorphism: a new gene polymorphism associated with C-reactive protein in type 2 diabetic patients. Cardiovasc Diabetol. 2010;9:68. doi: 10.1186/1475-2840-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Peng W, Zhang X, Lu L, Zhang R, Zhang Q, Wang L, Chen Q, Shen W. Chromosome 9p21.3 polymorphism in a Chinese Han population is associated with angiographic coronary plaque progression in non-diabetic but not in type 2 diabetic patients. Cardiovasc Diabetol. 2010;9:33. doi: 10.1186/1475-2840-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelusi B, Gambineri A, Pasquali R. Type 2 diabetes and the polycystic ovary syndrome. Minerva Ginecol. 2004;56(1):41–51. [PubMed] [Google Scholar]
- Li C, Shi Y, You L, Wang L, Chen ZJ. Association of rs10830963 and rs10830962 SNPs in the melatonin receptor (MTNR1B) gene among Han Chinese women with polycystic ovary syndrome. Mol Hum Reprod. 2011;17(3):193–198. doi: 10.1093/molehr/gaq087. [DOI] [PubMed] [Google Scholar]
- Qiu XS, Tang NL, Yeung HY, Lee KM, Hung VW, Ng BK, Ma SL, Kwok RH, Qin L, Qiu Y. et al. Melatonin receptor 1B (MTNR1B) gene polymorphism is associated with the occurrence of adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2007;32(16):1748–1753. doi: 10.1097/BRS.0b013e3180b9f0ff. [DOI] [PubMed] [Google Scholar]
- Morocz M, Czibula A, Grozer ZB, Szecsenyi A, Almos PZ, Rasko I, Illes T. Association study of BMP4, IL6, Leptin, MMP3, and MTNR1B gene promoter polymorphisms and adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2011;36(2):E123–130. doi: 10.1097/BRS.0b013e318a511b0e. [DOI] [PubMed] [Google Scholar]
- Zhang L, Su P, Xu C, Chen C, Liang A, Du K, Peng Y, Huang D. Melatonin inhibits adipogenesis and enhances osteogenesis of human mesenchymal stem cells by suppressing PPARgamma expression and enhancing Runx2 expression. J Pineal Res. 2010;49(4):364–372. doi: 10.1111/j.1600-079X.2010.00803.x. [DOI] [PubMed] [Google Scholar]
- Sethi S, Radio NM, Kotlarczyk MP, Chen CT, Wei YH, Jockers R, Witt-Enderby PA. Determination of the minimal melatonin exposure required to induce osteoblast differentiation from human mesenchymal stem cells and these effects on downstream signaling pathways. J Pineal Res. 2010;49(3):222–238. doi: 10.1111/j.1600-079X.2010.00784.x. [DOI] [PubMed] [Google Scholar]
- Machida M, Dubousset J, Satoh T, Murai I, Wood KB, Yamada T, Ryu J. Pathologic mechanism of experimental scoliosis in pinealectomized chickens. Spine (Phila Pa 1976) 2001;26(17):E385–391. doi: 10.1097/00007632-200109010-00006. [DOI] [PubMed] [Google Scholar]
- Machida M, Murai I, Miyashita Y, Dubousset J, Yamada T, Kimura J. Pathogenesis of idiopathic scoliosis. Experimental study in rats. Spine (Phila Pa 1976) 1999;24(19):1985–1989. doi: 10.1097/00007632-199910010-00004. [DOI] [PubMed] [Google Scholar]
- Oyama J, Murai I, Kanazawa K, Machida M. Bipedal ambulation induces experimental scoliosis in C57BL/6J mice with reduced plasma and pineal melatonin levels. J Pineal Res. 2006;40(3):219–224. doi: 10.1111/j.1600-079X.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- Gruodyte R, Jurimae J, Cicchella A, Stefanelli C, Passariello C, Jurimae T. Adipocytokines and bone mineral density in adolescent female athletes. Acta Paediatr. 2010;99(12):1879–1884. doi: 10.1111/j.1651-2227.2010.01905.x. [DOI] [PubMed] [Google Scholar]
- Jiang X, Song D, Ye B, Wang X, Song G, Yang S, Hu J. Effect of intermittent administration of adiponectin on bone regeneration following mandibular osteodistraction in rabbits. J Orthop Res. 2011. in press . [DOI] [PubMed]
- Nuche-Berenguer B, Lozano D, Gutierrez-Rojas I, Moreno P, Marinoso ML, Esbrit P, Villanueva-Penacarrillo ML. GLP-1 and exendin-4 can reverse hyperlipidic-related osteopenia. J Endocrinol. 2011. in press . [DOI] [PubMed]
- Frost M, Abrahamsen B, Nielsen TL, Hagen C, Andersen M, Brixen K. Vitamin D status and PTH in young men: a cross-sectional study on associations with bone mineral density, body composition and glucose metabolism. Clin Endocrinol (Oxf) 2010;73(5):573–580. doi: 10.1111/j.1365-2265.2010.03847.x. [DOI] [PubMed] [Google Scholar]