Abstract
Introduction
MicroRNAs are regulators of central cellular processes and are implicated in the pathogenesis and prognosis of human cancers. MicroRNAs also modulate responses to anti-cancer therapy. In the context of radiation oncology microRNAs were found to modulate cell death and proliferation after irradiation. However, changes in microRNA expression profiles in response to irradiation have not been comprehensively analyzed so far. The present study's intend is to present a broad screen of changes in microRNA expression following irradiation of different malignant cell lines.
Materials and methods
1100 microRNAs (Sanger miRBase release version 14.0) were analyzed in six malignant cell lines following irradiation with clinically relevant doses of 2.0 Gy. MicroRNA levels 6 hours after irradiation were compared to microRNA levels in non-irradiated cells using the "Geniom Biochip MPEA homo sapiens".
Results
Hierarchical clustering analysis revealed a pattern, which significantly (p = 0.014) discerned irradiated from non-irradiated cells. The expression levels of a number of microRNAs known to be involved in the regulation of cellular processes like apoptosis, proliferation, invasion, local immune response and radioresistance (e. g. miR-1285, miR-24-1, miR-151-5p, let-7i) displayed 2 - 3-fold changes after irradiation. Moreover, several microRNAs previously not known to be radiation-responsive were discovered.
Conclusion
Ionizing radiation induced significant changes in microRNA expression profiles in 3 glioma and 3 squamous cell carcinoma cell lines. The functional relevance of these changes is not addressed but should by analyzed by future work especially focusing on clinically relevant endpoints like radiation induced cell death, proliferation, migration and metastasis.
Introduction
MicroRNAs are small non-coding RNAs of typically 20 - 22 base pairs length. They are involved in gene regulation at the post-transcriptional level by silencing mRNA translation. To date more than 1000 microRNAs have been discovered. MicroRNAs are involved in the regulation of diverse cellular processes, including programmed cell death, proliferation, differentiation, metabolism, migration and stress responses (for review see [1]). Notably, a single microRNA potentially regulates a wide range of target genes resulting in a global impact on gene expression [2].
As central regulators of gene expression, microRNAs have been implicated in the pathogenesis of human cancers, acting either as tumor suppressors [3,4] or as oncogenes [5]. In fact, certain microRNA profiles of human cancers have been found to correlate with the malignant phenotype of cancer cells when compared to normal cells (for review see [6]). Clinically important, the expression of distinct microRNAs seems to be associated with the prognosis [7] and may also predict the efficacy of therapeutic interventions, including radiotherapy [8,9]. In fact, microRNAs have been shown to modulate the radiosensitivity of lung cancer cells in vitro [10] and breast cancer cells in vivo [11]. Moreover, normal cells show altered levels of microRNAs in response to ionizing radiation [12,13].
The response of cancer cells to ionizing radiation has been extensively studied resulting in the discovery of central regulators of radiosensitivity. However, irradiation-induced alterations in microRNA profiles have hitherto not been analyzed. This stimulated us to perform the present study: a microarray based analysis of irradiation-induced changes in all microRNAs published in the Sanger miRBase release version 14.0 (see http://microrna.sanger.ac.uk/sequences/index.shtml). We describe alterations in the abundance of 1100 microRNAs in six malignant cell lines following irradiation. To our knowledge this represents the broadest analysis of irradiation induced changes in microRNA patterns to date.
Materials and methods
Cell culture
For the analysis of characteristic microRNA patterns, six different cell lines were used: Squamous cell carcinoma of the head and neck (SCC-4, SCC-25, CAL-27) and cell lines from brain tumors (LN229, T98G, U-87 MG). SCC-4, SCC-25 and CAL-27 were purchased from the "Deutsche Sammlung von Mikroorganismen und Zellkulturen" (DSMZ, Braunschweig Germany), T98G and U-87 MG were purchased from the European Collection of Cell Cultures (ECACC, UK). LN229 was a gift from the Department of Neurosurgery, University of Munich. SCC-4 and SCC-25 were grown in Dulbecco's MEM/Ham's F-12 medium supplemented with 20% fetal bovine serum (FBS) and Hydrocortisone (40 ng/ml) and Sodium Pyruvate (1 mM), respectively. CAL-27 were grown in Dulbecco's MEM, supplemented with 10% FBS. LN229, T98G, U-87 MG were grown in Earles buffered Salt Solution (EBSS), supplemented with 10% FBS, Sodium Glutamine (2 mM), Non Essential Amino Acids (1x) and Sodium Pyruvate (1 mM). All media and supplements were purchased from Biochrom, Germany.
Irradiation
Cells were seeded in culture flasks and grown for 5 - 10 passages. For the experiments, cells were grown to a confluency of ~70%. Cells were then irradiated with 6 MeV Photons at a dose rate of 3 Gy per minute using a linear accelerator (Siemens Mevatron) to a total dose of 2 Gy. After irradiation cells were incubated at 37°C for 6 hours before extraction of the total RNA. Non-irradiated cells were used as controls.
Isolation of total RNA
Total RNA was extracted using the mirVana™ microRNA Isolation Kit (Ambion) according to the manufacturer's instructions. Quantity and quality of the extracted RNA was analyzed with the Agilent 2100 Bioanalyzer (Agilent Technologies), using the company's RNA 6000 Nano Kit according to the manufacturer's instructions. RNA extracts were stored at - 20°C.
Analysis of the microRNAs
The RNAs patterns were analyzed by Febit (Heidelberg) using the company's "Geniom Biochip MPEA homo sapiens", generating five data points for each microRNA measured. To adjust for a systematic spatial variability on each microarray, the intensities of black probes were used for background correction. To test the hybridization process as well as positioning features additional hybridization controls were added to the array template (data not shown).
Statistical analysis
Hierarchical cluster analysis (bottom-up complete linkage clustering using Euclidean distance as a measure) was performed using the normalized data of the 65 most deregulated microRNAs to identify differentially expressed microRNAs following irradiation. The correlation between two dichotomous variables was assessed using the chi-square test. A two-tailed p-value < 0.05 was considered significant. MicroRNA changes in single cell lines were not analyzed because the statistical power was not sufficient.
Results
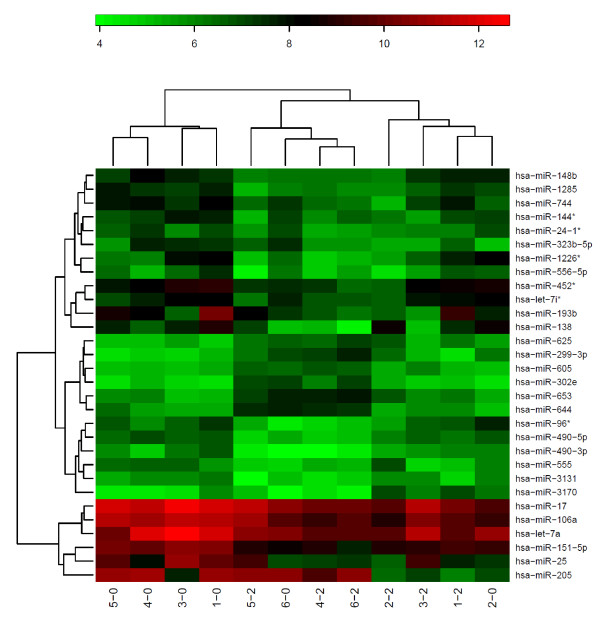
Hierarchical cluster analysis revealed two clusters of microRNAs which significantly (p = 0,014) differentiated between irradiated and non-irradiated cells. Figure 1 shows the heatmap comparing irradiated and non-irradiated cells.
Figure 1.
Heatmap and clustering of samples and probes for non-irradiated versus irradiated cells. The first number indicates the cell line (1 = T98G, 2 = U-87 MG, 3 = LN229, 4 = SCC-25, 5 = SCC-4, 6 = CAL-27) the second number indicates irradiation of the cell line (0 = non-irradiated; 2 = irradiated). On the left side, most of the non-irradiated cell lines cluster together while on the right side most of the irradiated cell lines form a cluster.
Although statistical interpretation of the data is difficult since commonly used p-values of 0.05 might lead to false positive results when analyzing more than 1000 microRNAs, the microRNAs displaying the most striking up- or downregulation after irradiation are presented in Table 1.
Table 1.
Most deregulated microRNAs in six malignant cell lines following irradiation
| microRNA | Fold change | Log2-value | p-value (unadjusted) |
|---|---|---|---|
| miR-24-1* | 3.07 | 1.12 | 0.01 |
| miR-144* | 2.93 | 1.07 | 0.01 |
| miR-1285 | 3.04 | 1.11 | 0.04 |
| miR-490-5p | 2.98 | 1.09 | 0.04 |
| miR-151-5p | 3.33 | 1.20 | 0.04 |
| let-7i* | 2.97 | 1.09 | 0.04 |
| miR-3131 | 3.11 | 1.14 | 0.06 |
| miR-323b-5p | 4.75 | 1.56 | 0.06 |
| miR-625 | 0.34 | -1.08 | 0.06 |
| miR-744 | 3.10 | 1.13 | 0.07 |
| miR-605 | 0.31 | -1.17 | 0.07 |
| miR-106a | 4.22 | 1.44 | 0.08 |
| miR-148b | 3.23 | 1.17 | 0.09 |
| miR-1226* | 5.81 | 1.76 | 0.10 |
| miR-452* | 2.89 | 1.06 | 0.10 |
| let-7a | 4.60 | 1.53 | 0.10 |
| miR-556-5p | 3.08 | 1.12 | 0.11 |
| miR-17 | 4.38 | 1.48 | 0.11 |
| miR-96* | 3.24 | 1.18 | 0.13 |
| miR-653 | 0.28 | -1.28 | 0.16 |
| miR-490-3p | 2.88 | 1.06 | 0.20 |
| miR-299-3p | 0.27 | -1.31 | 0.21 |
| miR-25 | 3.67 | 1.30 | 0.23 |
| miR-3170 | 0.32 | -1.14 | 0.23 |
| miR-205 | 9.66 | 2.27 | 0.26 |
| miR-555 | 3.13 | 1.14 | 0.28 |
| miR-138 | 4.15 | 1.42 | 0.29 |
| miR-193b | 3.71 | 1.31 | 0.36 |
| miR-302e | 0.34 | -1.08 | 0.44 |
Unadjusted p-values are presented.
When comparing irradiated cells with their non-irradiated counterparts, the levels of several microRNAs that target central regulators of cancer cells were found to be susceptible to ionizing radiation. Of note, miR-1285, which negatively regulates the expression of the crucial tumor suppressor p53 [14], was upregulated approximately 3-fold (p = 0.02, unadjusted). MiR-151-5p known to enhance migration and metastasis in human hepatocellular carcinoma [15] was another microRNA, whose level was found to be upregulated approximately 3-fold, thus potentially hampering the therapeutic effort.
On the contrary, miR-24-1, a member of the miR23b cluster [16] that interferes with Transforming Growth Factor β (TGFβ) expression, was also up-regulated approximately 3-fold (p = 0.0048, unadjusted). Elevated TGFβ expression in human malignancies has been reported to be associated with enhanced angiogenesis, local immune suppression and increased invasiveness (for review see [17]). Let-7i, a member of the let7-family negatively modulating tumor growth in human cancers [18] was also upregulated approximately 3-fold (p = 0.03, unadjusted). Notably, let-7i and its family member let-7a were the only microRNAs found in our screen that had already been described to be up-regulated following irradiation.
Moreover, our screen identified different other microRNAs with so far unknown function to be up- or downregulated in response to irradiation. For many of them this is first time that they are reported to be susceptible to irradiation, like for instance, miR-144*, which was upregulated approximately 3-fold (p = 0.0026, unadjusted). A detailed comparison of the levels of all 1100 microRNAs before and after irradiation can be found in the additional file 1.
Discussion
The data presented here show that ionizing radiation with clinically relevant doses of 2 Gy stimulate significant changes in microRNA expression patterns in different cancer cell lines. These changes might well influence the clinical outcome, since the expression of microRNAs, which are known to regulate apoptosis, migration and proliferation was altered in response to irradiation.
Although the functional role of single microRNAs can not be established from the data presented here, several hypotheses can by generated which should be addressed by future experiments.
In this context, the up-regulation of the Epidermal Growth Factor Receptor (EGF-R) following ionizing radiation might serve as a paradigm. Irradiation-induced EGF-R expression leads to increased radioresistance of malignant cells in subsequent treatment sessions during the course of fractionated radiotherapy [19]. Similarly, an increased expression of mirR-1285, a negative regulator of the cardinal tumor suppressor p53, as it was observed in the present study might possibly lead to increased radioresistance in subsequent radiotherapy sessions. Furthermore, irradiation-induced changes in microRNA expression levels might also affect migration and metastasis of surviving cells. In this context, ionizing radiation-induced over-expression of miR-151-5p as described here might enhance dissemination and migration of malignant cells during a course of radiation therapy, since miR151-5p was found to increase migration and intra-hepatic metastasis in hepatocellular carcinoma [15]. Indeed, enhanced migration of malignant glioma cells was observed in response to radiotherapy [20,21].
Another candidate for regulating responsiveness to anticancer therapy is the let-7 family, although certain members of the let-7 family had different effects on radiation sensitivity in A549 lung cancer cells [10]. Let-7 family members are down-regulated in lung cancer cells [9] which possibly increases cell growth by increasing KRAS levels [22]. Over-expression of let-7a, which in the present study was up-regulated 4,6-fold (n. s.), was shown to increase radiation sensitivity in lung cancer cells [23]. Moreover, low levels of let-7a correlated with poor survival in patients with lung cancer [8,9], while over-expression of let-7a inhibited growth of lung cancer cells in vitro [9]. On the other hand, let-7i, which in the present study was observed to be up-regulated following irradiation, might increase growth, since it was shown that decreased levels of let-7i decreased growth of malignant cells and increased drug potency [18].
Another mechanism, through which ionizing radiation exerts its effects, involves changes in the tumor microenvironment. Interestingly, miR-24-1 levels were increased following irradiation and miR-24-1 might influence angiogenesis, invasion and local immune response through down-regulation of TGFβ [16].
In summary, the present study revealed altered expression levels of microRNAs known to influence apoptosis, migration and proliferation, angiogenesis and local immune response in response to irradiation. Moreover, a number of microRNAs with unknown functions were found to be radiation-responsive.
The power of the present study is based upon the huge number of investigated microRNAs (>1000) and the combined analysis of different malignant cells lines. Although adjusted p-values of changes in the expression levels of single microRNAs were not significant because of the huge number of microRNAs analyzed, the changes observed allow the generation of hypotheses and the design of further experiments validating the initial findings presented here and investigating the functional relevance of microRNA level alterations in the context of radiation oncology.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
OMN: RNA-Isolation and Purification, writing Manuscript, MN: Statistics and critical revision of the manuscript, SC: Cell culture and critical revision of the manuscript, FZ: Organization and Negotiation with Febit and critical revision of the manuscript, ML: Irradiation and critical revision of the manuscript, KL: Support concerning all technical questions, planning of experiments and critical revision of the manuscript.
CB: Development of the concept and critical revision of the manuscript.
All authors read and approved the final manuscript.
Supplementary Material
Changes in 1100 microRNAs of six malignant cell lines following irradiation. Raw data of the levels of all 1100 microRNAs before and after irradiation.
Contributor Information
Olivier M Niemoeller, Email: Olivier.Niemoeller@med.uni-muenchen.de.
Maximilian Niyazi, Email: Maximilian.Niyazi@med.uni-muenchen.de.
Stefanie Corradini, Email: Stefanie.Corradini@med.uni-muenchen.de.
Franz Zehentmayr, Email: Franz.Zehentmayr@med.uni-muenchen.de.
Minglun Li, Email: Minglun.Li@med.uni-muenchen.de.
Kirsten Lauber, Email: Kirsten.Lauber@med.uni-muenchen.de.
Claus Belka, Email: Claus.Belka@med.uni-muenchen.de.
References
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- He L, He XY, Lim LP, De Stanchina E, Xuan ZY, Liang Y, Xue W, Zender L, Magnus J, Ridzon D. et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–U1116. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M. et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y. et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Research. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Weidhaas JB, Babar L, Nallur SM, Trang P, Roush S, Boehm N, Gillespie E, Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Research. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Paranjape T, Ullrich R, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes OC, An J, Sarojini H, Wu HL, Wang E. Changes in MicroRNA Expression Patterns in Human Fibroblasts After Low-LET Radiation. Journal of Cellular Biochemistry. 2008;105:824–834. doi: 10.1002/jcb.21878. [DOI] [PubMed] [Google Scholar]
- Ilnytskyy Y, Koturbash I, Kovalchuk O. Radiation-Induced Bystander Effects In Vivo are Epigenetically Regulated in a Tissue-Specific Manner. Environmental and Molecular Mutagenesis. 2009;50:105–113. doi: 10.1002/em.20440. [DOI] [PubMed] [Google Scholar]
- Tian S, Huang S, Wu S, Guo W, Li J, He X. MicroRNA-1285 inhibits the expression of p53 by directly targeting its 3' untranslated region. Biochem Biophys Res Commun. pp. 435–439. [DOI] [PubMed]
- Ding J, Huang SL, Wu SQ, Zhao YJ, Liang LH, Yan MX, Ge C, Yao J, Chen TY, Wan DF. et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nature Cell Biology. 2010;12:390–U208. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, Rogler LE. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- Blobe GC, Schiemann WP, Lodish HF. Mechanisms of disease: Role of transforming growth factor beta in human disease. New England Journal of Medicine. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- Blower PE, Chung JH, Verducci JS, Lin SL, Park JK, Dai ZY, Liu CG, Schmittgen TD, Reinhold WC, Croce CM. et al. MicroRNAs modulate the chemosensitivity of tumor cells. Molecular Cancer Therapeutics. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- Schmidtullrich RK, Valerie KC, Chan W, McWilliams D. Altered Expression of Epidermal Growth-Factor Receptor and Estrogen-Receptor In Mcf-7 Cells After Single and Repeated Radiation Exposures. International Journal of Radiation Oncology Biology Physics. 1994;29:813–819. doi: 10.1016/0360-3016(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Goetze K, Scholz M, Taucher-Scholz G, Mueller-Klieser W. The impact of conventional and heavy ion irradiation on tumor cell migration in vitro. International Journal of Radiation Biology. 2007;83:889–896. doi: 10.1080/09553000701753826. [DOI] [PubMed] [Google Scholar]
- Wild-Bode C, Weller M, Rimner A, Dichgans J, Wick W. Sublethal irradiation promotes migration and invasiveness of glioma cells: Implications for radiotherapy of human glioblastoma. Cancer Research. 2001;61:2744–2750. [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 MicroRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Oh JS, Kim JJ, Byun JY, Kim IA. Lin28-Let7 Modulates Radiosensitivity of Human Cancer Cells With Activation of K-Ras. International Journal of Radiation Oncology Biology Physics. 2010;76:5–8. doi: 10.1016/j.ijrobp.2009.08.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Changes in 1100 microRNAs of six malignant cell lines following irradiation. Raw data of the levels of all 1100 microRNAs before and after irradiation.