Abstract
Background
Poliovirus, the causative agent of poliomyelitis, is a human enterovirus and a member of the family of Picornaviridae and among the most rapidly evolving viruses known. Analysis of codon usage can reveal much about the molecular evolution of the viruses. However, little information about synonymous codon usage pattern of polioviruses genome has been acquired to date.
Methods
The relative synonymous codon usage (RSCU) values, effective number of codon (ENC) values, nucleotide contents and dinucleotides were investigated and a comparative analysis of codon usage pattern for open reading frames (ORFs) among 48 polioviruses isolates including 31 of genotype 1, 13 of genotype 2 and 4 of genotype 3.
Results
The result shows that the overall extent of codon usage bias in poliovirus samples is low (mean ENC = 53.754 > 40). The general correlation between base composition and codon usage bias suggests that mutational pressure rather than natural selection is the main factor that determines the codon usage bias in those polioviruses. Depending on the RSCU data, it was found that there was a significant variation in bias of codon usage among three genotypes. Geographic factor also has some effect on the codon usage pattern (exists in the genotype-1 of polioviruses). No significant effect in gene length or vaccine derived polioviruses (DVPVs), wild viruses and live attenuated virus was observed on the variations of synonymous codon usage in the virus genes. The relative abundance of dinucleotide (CpG) in the ORFs of polioviruses are far below expected values especially in DVPVs and attenuated virus of polioviruses genotype 1.
Conclusion
The information from this study may not only have theoretical value in understanding poliovirus evolution, especially for DVPVs genotype 1, but also have potential value for the development of poliovirus vaccines.
Background
When molecular sequence data started to be accumulated nearly 20 years ago, it was noted that synonymous codons are not used equally in different genomes, even in different genes of the same genome[1-3]. As an important evolutionary phenomenon, it is well known that synonymous codon usage bias exists in a wide range of biological systems from prokaryotes to eukaryotes [4,5]. Codon usage analysis has been applied to prokaryote and eukaryote, such as Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Caenorhabditis elegans and human beings [6-8]. These observed patterns in synonymous codon usage varied among genes within a genome, and among genomes. The codon usage is attributable to the equilibrium between natural selection and mutation pressure [9,10]. Recent studies of viral codon usage has shown that mutation bias may be a more important factor than natural selection in determining codon usage bias of some viruses, such as Picornaviridae, Pestivirus, plant viruses, and vertebrate DNA viruses [9,11-13]. Meanwhile, recent report also showed that the G+C compositional constraint is the main factor that determines the codon usage bias in iridovirus genomes[11,14]. Analysis of codon usage can reveal much about the molecular evolution or individual genes of the viruses.
Polioviruses belong to the family Picornaviridae and are classified as human enterovirus C (HEV-C) species in the genus Enterovirus according to the current taxonomy [15,16]. Polioviruses can be divided into three different genotypes: 1, 2 and 3. The genome of each genotypes contains a single positive-stranded RNA with a size of approximately 6 kb consisting of a single large open reading frame (ORF) flanked by 5' and 3' untranslated region [17].
As we known, the Sabin oral poliovaccine (OPV) was among the best known viral vaccines [18]. It has saved the lives and health of innumerable people, in particular children. However, poliovirus is highly genetically variable. OPV viruses may undergo transformation into circulating highly diverged VDPV, exhibiting properties hardly distinguishable from those of wild polioviruses [19]. So far, little information about synonymous codon usage pattern of polioviruses genome has been acquired to date. To our knowledge, this is the first report of the codon usage analysis on polioviruses (including wild strains, attenuated live vaccine strains and VDPV strains). In this study, we analyzed the codon usage data and base composition of 48 available representative complete ORFs of poliovirus to obtain some clues to the features of genetic evolution of the virus.
Methods
Sequence data
A total of 48 poliovirus genomes were used in this study (Table 1). The serial number (SN), genotype, length value, isolated region, GenBank accession numbers, and other detail information about these strains were listed in Table 1. All of the sequences were downloaded from NCBI http://www.ncbi.nlm.nih.gov/Genbank/, and 48 poliovirus genomes were selected in the study. The other sequences with >98% sequence identities were excluded.
Table 1.
The information of 48 polioviruses genomes used in this study
| SN | Strain | Gene type | Lengtha | Isolation | Note | Accession No. |
|---|---|---|---|---|---|---|
| 1 | CHN-Henan/91-3 | 1 | 6630 | China | W Virusb | AF111983 |
| 2 | CHN-Jiangxi/89-1 | 1 | 6630 | China | W Virusb | AF111984 |
| 3 | P1W/Bar65 (19276) | 1 | 6630 | Belarus | DVPV | AY278553 |
| 4 | HAI01008C2 | 1 | 6630 | Haiti | DVPV | AF405662 |
| 5 | HAI01007 | 1 | 6630 | Haiti | DVPV | AF405666 |
| 6 | HAI01002 | 1 | 6630 | Haiti | DVPV | AF405667 |
| 7 | HAI01001 | 1 | 6630 | Haiti | DVPV | AF405668 |
| 8 | HAI00003 | 1 | 6630 | Haiti | DVPV | AF405669 |
| 9 | DOR01012 | 1 | 6630 | Dominica | DVPV | AF405670 |
| 10 | DOR00041C3 | 1 | 6630 | Dominica | DVPV | AF405679 |
| 11 | DOR00028 | 1 | 6630 | Dominica | DVPV | AF405684 |
| 12 | 99/056-252-14 | 1 | 6630 | Russia | DVPV | AF462418 |
| 13 | RUS-1161-96-001 | 1 | 6630 | Russia | DVPV | AF462419 |
| 14 | HAI01-13 | 1 | 6630 | Haiti | DVPV | AF416342 |
| 15 | TCDCE01-135 | 1 | 6630 | C Taiwanc | DVPV | AF538840 |
| 16 | TCDC01-113 | 1 | 6630 | C Taiwanc | DVPV | AF538841 |
| 17 | TCDC01-330 | 1 | 6630 | C Taiwanc | DVPV | AF538842 |
| 18 | TCDC01-861 | 1 | 6630 | C Taiwanc | DVPV | AF538843 |
| 19 | Sabin 1 | 1 | 6630 | USA | Vaccined | AY184219 |
| 20 | Brunhilde | 1 | 6630 | China | W Virusb | AY560657 |
| 21 | USA10784 | 1 | 6630 | USA | DVPV | EF682356 |
| 22 | USA10785 | 1 | 6630 | USA | DVPV | EF682357 |
| 23 | USA10783 | 1 | 6630 | USA | DVPV | EF682358 |
| 24 | USA10786 | 1 | 6630 | USA | DVPV | EF682359 |
| 25 | CHN8229-3/GZ/CHN/2004 | 1 | 6630 | China | DVPV | FJ769381 |
| 26 | 10050 | 1 | 6630 | China | DVPV | FJ859058 |
| 27 | 10091c | 1 | 6630 | China | DVPV | FJ859060 |
| 28 | 10092c | 1 | 6630 | China | DVPV | FJ859061 |
| 29 | 10094c | 1 | 6630 | China | DVPV | FJ859062 |
| 30 | 10095c | 1 | 6630 | China | DVPV | FJ859063 |
| 31 | 10097c | 1 | 6630 | China | DVPV | FJ859064 |
| 32 | EGY88-074 | 2 | 6624 | Egypt | DVPV | AF448782 |
| 33 | EGY93-034 | 2 | 6624 | Egypt | DVPV | AF448783 |
| 34 | P2S/Mog65-3 (20120) | 2 | 6624 | Belarus | DVPV | AY278549 |
| 35 | P2S/Mog66-4 (21043) | 2 | 6624 | Belarus | DVPV | AY278551 |
| 36 | P2S/Mog65-2 (20077) | 2 | 7439 | Belarus | DVPV | AY278552 |
| 37 | NIE0210766 | 2 | 6624 | Nigeria | DVPV | DQ890385 |
| 38 | NIE0110767 | 2 | 6624 | Nigeria | DVPV | DQ890386 |
| 39 | USA9810768 | 2 | 6624 | USA | DVPV | DQ890387 |
| 40 | PER8310769 | 2 | 6624 | Peru | DVPV | DQ890388 |
| 41 | 32191 | 2 | 6624 | Belarus | DVPV | FJ460223 |
| 42 | 32189+AP1 | 2 | 6624 | Belarus | DVPV | FJ460224 |
| 43 | 31996 | 2 | 6624 | Belarus | DVPV | FJ460225 |
| 44 | PV2/Rus | 2 | 6624 | Russia | DVPV | FJ517649 |
| 45 | Sabin 3 | 3 | 6621 | USA | Vaccined | AY184221 |
| 46 | 33239 | 3 | 6621 | Belarus | DVPV | FJ460226 |
| 47 | 31974 | 3 | 6621 | Belarus | DVPV | FJ460227 |
| 48 | FIN84-60212 | 3 | 6621 | Finland | DVPV | FJ842158 |
Note: a the length values excluding non-coding sequence.
b means wild strain
c stands for China Taiwan
d stands for attenuated live vaccine strain
The actual and predicted values of the effective number of codon (ENC)
The ENC is used to measure the degree of departure from the equal use of synonymous codons of coding regions of polioviruses. The values of the effective number of codon (ENC) range from 20 to 61. In an extremely biased gene where only one codon is used for each amino acid, this value would be 20; if all codons are used equally, it would be 61; and if the value of ENC is greater than 40, the codon usage bias was regarded as low. The values of ENC were obtained by EMBOSS CHIPS program [20].
Genes, whose codon choice is constrained only by a mutation bias, will lie on or just below the curve of the predicted values. The predicted values of ENC were calculated as
where s represents the given (G+C)3% value [21].
The calculation of the relative synonymous codon usage (RSCU)
To investigate the pattern of relative synonymous codon usage (RSCU) without the influence of amino acid composition among all polioviruses samples, the RSCU values of codons in each ORF of polioviruses were calculated according to the formula of previous reports [22,23].
where gij is the observed number of the ith codon for jth amino acid which has ni type of synonymous codons. The codon with RSCU value more than 1.0 has positive codon usage bias, while the value <1.0 has relative negative codon usage bias. When RSCU value is equal to 1.0, it means that this codon is chosen equally and randomly.
Relative dinucleotide abundance in polioviruses
Because dinucleotide biases can affect codon bias, the relative abundance of dinucleotides in the coding regions of polioviruse genomes was assessed using the method described by Karlin and Burge [24]. A comparison of actual and expected dinucleotide frequencies of the 16 dinucleotides in coding region of the 48 polioviruses genomes was also undertaken. The odds ratio ρxy=ƒxy/ƒyƒx, where ƒx denotes the frequency of the nucleotide X, ƒy denotes the frequency of the nucleotide Y, ƒyƒx the expected frequency of the dinucleotide XY and ƒxy the frequency of the dinucleotide XY, etc., for each dinucleotide were calculated. As a conservative criterion, for ρ xy > 1.23 (or < 0.78), the XY pair is considered to be of over-represented (or under-represented) relative abundance compared with a random association of mononucleotides.
Statistical analysis
Principal component analysis (PCA) was carried out to analyze the major trend in codon usage pattern in different genomes of polioviruses (excluding non-coding regions). It is a statistical method that performs linear mapping to extract optimal features from an input distribution in the mean squared error sense and can be used by self-organizing neural networks to form unsupervised neural preprocessing modules for classification problems [6]. In order to minimize the effect of amino acid composition on codon usage, each ORF is represented as a 59-dimensional vector. Each dimension corresponds to the RSCU value of one sense codon excluding Met, Trp and three stop codons.
Linear regression analysis was used to find the correlation between codon usage bias and gene length. Correlation analysis is used to identify the relationship between codon usage bias and synonymous codon usage pattern. This analysis is implemented based on the Spearman's rank correlation analysis way.
All statistical analyses were carried out using the statistical analysis software SPSS Version 17.0.
Results
The characteristics of synonymous codon usage in polioviruses
In order to investigate the extent of codon usage bias in polioviruses, all RSCU values of different codon in 48 polioviruses strains were calculated. There is only two preferred codons UUG (Leu) and GUG (Val), choosing G at the third position, and most of preferred codons are ended with A (Table 2). Moreover, polioviruses genome is A redundant with A content ranging from 29.739 to 30.826.11, with the mean value of 30.367 and S.D. of 0.234; in contrast, low content of G ranging from 21.723 to 22.401 (mean = 22.118, S.D. of 0.147), suggesting that nucleotide contents influence the patterns of synonymous codon usage (Table 3). The values of ENC among these polioviruses ORFs are similar, which vary from 52.609 to 55.105 with a mean value of 53.754 and S.D. of 0.545. The data showed that the extent of codon preference in polioviruses genes was kept basically stable.
Table 2.
Synonymous codon usage in the coding region of polioviruses
| AA a | Codon | RSCU b | AA | Codon | RSCU b |
|---|---|---|---|---|---|
| Phe | UUU | 1.020 | Gln | CAA | 1.075 |
| UUC | 0.980 | CAG | 0.925 | ||
| Leu | UUA | 0.914 | His | CAU | 0.787 |
| UUG | 1.349 | CAC | 1.213 | ||
| CUU | 0.566 | Asn | AAU | 0.900 | |
| CUC | 0.909 | AAC | 1.100 | ||
| CUA | 1.023 | Lys | AAA | 1.050 | |
| CUG | 1.072 | AAG | 0.95 | ||
| Val | GUU | 0.441 | Asp | GAU | 0.961 |
| GUC | 0.762 | GAC | 1.039 | ||
| GUA | 0.735 | Glu | GAA | 1.057 | |
| GUG | 1.657 | GAG | 0.943 | ||
| Ser c | UCU | 0.785 | Arg | AGA | 2.868 |
| UCC | 1.345 | AGG | 1.471 | ||
| UCA | 1.749 | CGU | 0.434 | ||
| UCG | 0.424 | CGC | 0.577 | ||
| AGU | 0.920 | CGA | 0.268 | ||
| AGC | 0.777 | CGG | 0.381 | ||
| Pro | CCU | 0.800 | Cys | UGU | 1.105 |
| CCC | 0.799 | UGC | 0.895 | ||
| CCA | 1.884 | Tyr | UAU | 0.847 | |
| CCG | 0.517 | UAC | 1.153 | ||
| Thr | ACU | 1.170 | Ala | GCU | 1.161 |
| ACC | 1.330 | GCC | 0.969 | ||
| ACA | 1.124 | GCA | 1.438 | ||
| ACG | 0.376 | GCG | 0.432 | ||
| Gly | GGU | 1.160 | Ile | AUU | 1.247 |
| GGC | 0.757 | AUC | 1.049 | ||
| GGA | 1.175 | AUA | 0.705 | ||
| GGG | 0.909 |
Note: The boldface means the preferred codon compare with other synonymous codon.
Table 3.
Nucleotide contents in ORFs of 48 poliovirus genomes
| No. | A | G | U | C | A3 | C3 | G3 | U3 | A+U | G+C | C3/G3 | ENC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.305 | 0.220 | 0.236 | 0.239 | 0.275 | 0.270 | 0.192 | 0.263 | 0.541 | 0.459 | 0.462 | 53.082 |
| 2 | 0.305 | 0.218 | 0.236 | 0.242 | 0.269 | 0.275 | 0.191 | 0.264 | 0.540 | 0.460 | 0.467 | 53.749 |
| 3 | 0.300 | 0.219 | 0.238 | 0.242 | 0.263 | 0.283 | 0.190 | 0.264 | 0.538 | 0.462 | 0.473 | 53.709 |
| 4 | 0.301 | 0.222 | 0.236 | 0.241 | 0.269 | 0.279 | 0.189 | 0.263 | 0.537 | 0.463 | 0.468 | 54.085 |
| 5 | 0.308 | 0.221 | 0.233 | 0.238 | 0.285 | 0.268 | 0.189 | 0.258 | 0.541 | 0.459 | 0.457 | 53.936 |
| 6 | 0.300 | 0.223 | 0.233 | 0.244 | 0.265 | 0.289 | 0.192 | 0.253 | 0.533 | 0.467 | 0.481 | 53.506 |
| 7 | 0.300 | 0.223 | 0.236 | 0.241 | 0.267 | 0.279 | 0.191 | 0.263 | 0.536 | 0.464 | 0.470 | 53.929 |
| 8 | 0.305 | 0.220 | 0.233 | 0.242 | 0.281 | 0.283 | 0.185 | 0.250 | 0.538 | 0.462 | 0.469 | 53.506 |
| 9 | 0.301 | 0.222 | 0.232 | 0.244 | 0.270 | 0.287 | 0.189 | 0.253 | 0.533 | 0.467 | 0.477 | 53.592 |
| 10 | 0.303 | 0.220 | 0.232 | 0.244 | 0.275 | 0.288 | 0.186 | 0.252 | 0.535 | 0.465 | 0.474 | 53.308 |
| 11 | 0.303 | 0.221 | 0.234 | 0.242 | 0.273 | 0.283 | 0.187 | 0.257 | 0.537 | 0.463 | 0.470 | 53.389 |
| 12 | 0.302 | 0.219 | 0.240 | 0.238 | 0.270 | 0.264 | 0.188 | 0.278 | 0.542 | 0.458 | 0.452 | 54.486 |
| 13 | 0.304 | 0.220 | 0.237 | 0.240 | 0.274 | 0.268 | 0.188 | 0.270 | 0.541 | 0.459 | 0.456 | 54.637 |
| 14 | 0.301 | 0.222 | 0.234 | 0.243 | 0.268 | 0.284 | 0.191 | 0.257 | 0.535 | 0.465 | 0.474 | 53.640 |
| 15 | 0.305 | 0.222 | 0.235 | 0.238 | 0.279 | 0.267 | 0.193 | 0.261 | 0.540 | 0.460 | 0.460 | 52.948 |
| 16 | 0.307 | 0.221 | 0.236 | 0.237 | 0.281 | 0.264 | 0.191 | 0.263 | 0.543 | 0.457 | 0.456 | 53.822 |
| 17 | 0.305 | 0.222 | 0.236 | 0.237 | 0.279 | 0.264 | 0.195 | 0.263 | 0.541 | 0.459 | 0.458 | 53.194 |
| 18 | 0.305 | 0.222 | 0.240 | 0.233 | 0.279 | 0.254 | 0.194 | 0.273 | 0.545 | 0.455 | 0.448 | 53.054 |
| 19 | 0.308 | 0.219 | 0.231 | 0.241 | 0.282 | 0.274 | 0.189 | 0.254 | 0.540 | 0.460 | 0.464 | 53.359 |
| 20 | 0.305 | 0.219 | 0.236 | 0.240 | 0.274 | 0.277 | 0.190 | 0.259 | 0.540 | 0.460 | 0.467 | 54.470 |
| 21 | 0.305 | 0.222 | 0.234 | 0.240 | 0.276 | 0.273 | 0.194 | 0.257 | 0.538 | 0.462 | 0.467 | 53.840 |
| 22 | 0.305 | 0.221 | 0.234 | 0.239 | 0.276 | 0.272 | 0.193 | 0.258 | 0.539 | 0.461 | 0.466 | 53.705 |
| 23 | 0.305 | 0.222 | 0.234 | 0.239 | 0.276 | 0.273 | 0.194 | 0.257 | 0.539 | 0.461 | 0.466 | 53.745 |
| 24 | 0.306 | 0.221 | 0.233 | 0.240 | 0.280 | 0.273 | 0.191 | 0.256 | 0.539 | 0.461 | 0.464 | 53.546 |
| 25 | 0.305 | 0.222 | 0.232 | 0.241 | 0.276 | 0.273 | 0.193 | 0.257 | 0.537 | 0.463 | 0.466 | 53.349 |
| 26 | 0.303 | 0.223 | 0.233 | 0.241 | 0.273 | 0.275 | 0.196 | 0.256 | 0.535 | 0.465 | 0.471 | 53.914 |
| 27 | 0.303 | 0.223 | 0.232 | 0.241 | 0.273 | 0.275 | 0.196 | 0.256 | 0.535 | 0.465 | 0.471 | 53.800 |
| 28 | 0.302 | 0.224 | 0.231 | 0.243 | 0.273 | 0.279 | 0.196 | 0.252 | 0.533 | 0.467 | 0.475 | 54.002 |
| 29 | 0.304 | 0.222 | 0.233 | 0.241 | 0.275 | 0.274 | 0.194 | 0.257 | 0.536 | 0.464 | 0.468 | 53.752 |
| 30 | 0.303 | 0.224 | 0.231 | 0.242 | 0.273 | 0.278 | 0.196 | 0.253 | 0.534 | 0.466 | 0.474 | 53.895 |
| 31 | 0.303 | 0.223 | 0.233 | 0.241 | 0.274 | 0.274 | 0.195 | 0.258 | 0.536 | 0.464 | 0.469 | 53.803 |
| 32 | 0.303 | 0.220 | 0.234 | 0.244 | 0.273 | 0.281 | 0.180 | 0.265 | 0.537 | 0.463 | 0.462 | 53.837 |
| 33 | 0.304 | 0.220 | 0.237 | 0.239 | 0.280 | 0.273 | 0.180 | 0.267 | 0.541 | 0.459 | 0.453 | 53.339 |
| 34 | 0.303 | 0.221 | 0.237 | 0.238 | 0.276 | 0.269 | 0.183 | 0.272 | 0.541 | 0.459 | 0.452 | 54.287 |
| 35 | 0.298 | 0.222 | 0.235 | 0.245 | 0.260 | 0.284 | 0.184 | 0.271 | 0.534 | 0.466 | 0.469 | 53.712 |
| 36 | 0.297 | 0.222 | 0.238 | 0.242 | 0.274 | 0.272 | 0.181 | 0.273 | 0.535 | 0.465 | 0.453 | 55.105 |
| 37 | 0.302 | 0.221 | 0.236 | 0.240 | 0.271 | 0.276 | 0.184 | 0.269 | 0.539 | 0.461 | 0.460 | 54.092 |
| 38 | 0.302 | 0.222 | 0.235 | 0.241 | 0.270 | 0.276 | 0.187 | 0.266 | 0.537 | 0.463 | 0.464 | 54.774 |
| 39 | 0.305 | 0.220 | 0.236 | 0.240 | 0.280 | 0.272 | 0.178 | 0.269 | 0.541 | 0.459 | 0.450 | 54.418 |
| 40 | 0.304 | 0.222 | 0.237 | 0.237 | 0.274 | 0.266 | 0.191 | 0.270 | 0.541 | 0.459 | 0.457 | 53.926 |
| 41 | 0.304 | 0.221 | 0.237 | 0.239 | 0.277 | 0.267 | 0.185 | 0.272 | 0.540 | 0.460 | 0.452 | 54.478 |
| 42 | 0.303 | 0.221 | 0.237 | 0.239 | 0.276 | 0.268 | 0.184 | 0.273 | 0.541 | 0.459 | 0.452 | 54.450 |
| 43 | 0.304 | 0.220 | 0.237 | 0.239 | 0.276 | 0.268 | 0.184 | 0.272 | 0.541 | 0.459 | 0.453 | 54.463 |
| 44 | 0.306 | 0.219 | 0.236 | 0.239 | 0.280 | 0.271 | 0.181 | 0.268 | 0.542 | 0.458 | 0.452 | 52.838 |
| 45 | 0.300 | 0.224 | 0.235 | 0.241 | 0.270 | 0.274 | 0.194 | 0.263 | 0.535 | 0.465 | 0.467 | 52.609 |
| 46 | 0.303 | 0.221 | 0.238 | 0.238 | 0.272 | 0.267 | 0.190 | 0.271 | 0.541 | 0.459 | 0.457 | 52.735 |
| 47 | 0.301 | 0.222 | 0.237 | 0.240 | 0.270 | 0.271 | 0.191 | 0.268 | 0.538 | 0.462 | 0.462 | 54.245 |
| 48 | 0.303 | 0.223 | 0.232 | 0.242 | 0.278 | 0.281 | 0.192 | 0.248 | 0.535 | 0.465 | 0.474 | 53.968 |
Compositional properties of ORFs of 48 polioviruses genomes
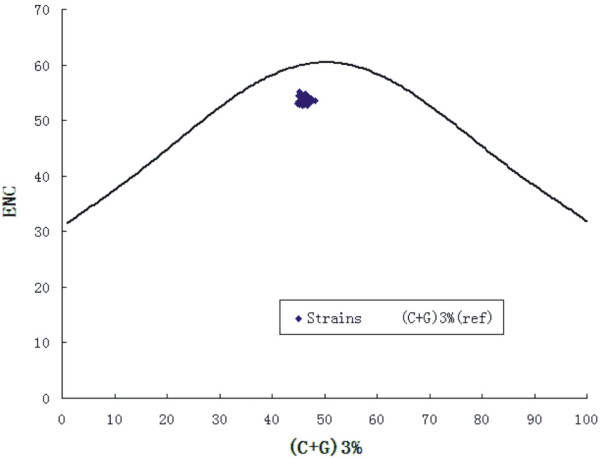
The values of A, U, C, G and C+G were compared with the values of A3, C3, G3, U3, (G+C) 3, respectively. An interesting and complex correlation was observed. In detail, the (C+G)3 have highly significant correlations with A, U, C, G and C+G, respectively, indicating C+G may reflect interaction between mutation pressure and natural selection. However, the A have no correlation with A3, G3 and C3, and U have no correlation with A3 (Table 4). Both cases suggested that the nucleotide constraint possibly influence synonymous codon usage of polioviruses. In addition, the correlation between the Axis 1 (calculated by PCA) and the values of A, C, G, U, A3, C3, G3, U3, (G+C), (G+C)3 of each strain was also analyzed. The significant correlation was found between nucleotide compositions and synonymous codon usage to some extent excluding Axis 1 and the value of A (Table 4). The analysis revealed that most of the codon usage bias among ORFs of polioviruses strains was directly related to the base composition. Finally, the ENC-plot [ENC plotted against (G+C)3%] was used as a part of general strategy to investigate patterns of synonymous codon usage and all of the spots lie below the expected curve (Figure 1). These imply that the codon bias can be explained mainly by an uneven base composition, in other words, by mutation pressure rather than natural selection.
Table 4.
Correlation analysis between the A, U, C, G contents and A 3, U 3, C 3, G 3 contents in ORF of 48 polioviruses genomes
| A3 | U3 | G3 | C3 | (G+C) 3 | Axis 1 | |
|---|---|---|---|---|---|---|
| A | r = -0.093N | r = -0.303* | r = -0.169N | r = -0.185N | r = -0.287* | r = -0.126 |
| U | r = -0.078N | r = 0.905** | r = -0.422** | r = -0.573** | r = -0.706** | r = -0.782** |
| G | r = -0.285* | r = -0.341* | r = 0.641** | r = 0.195N | r = 0.777** | r = 0.556** |
| C | r = -0.529** | r = -0.509** | r = -0.014N | r = 0.913** | r = 0.461** | r = 0.466** |
| G+C | r = -0.544** | r = -0.599** | r = 0.307 * | r = 0.807** | r = -0.851** | r = 0.708** |
| Axis 1 | r = 0.541** | r = -0.700** | r = 0.360* | r = 0.401** | r = 0.502** |
Note: ** Means p < 0.01
* Means 0.01 < p < 0.05
N Means no correlation
Figure 1.
Graphs showing the relationship between the effective number of codons (ENC) and the GC content of the third codon position (GC3). The curve indicates the expected codon usage if GC compositional constraints alone account for codon usage bias.
Effect of other potential factors on codon usage
Principal component analysis was carried out to identify the codon usage bias among ORFs. From which we could detect one major trend in the Axis 1 which accounted for 20.815% of the total variation, and another major trend in the Axis 2 for 16.273% of the total variation. A plot of the Axis 1 and the Axis 2 of each gene was shown in Additional file 1, Figure S1. Obviously, those polioviruses belong to the same genotype tends to come together (except strain 48, isolated from Finland). Compared with the scattered groups of polioviruses genotype 1, genotype 2 and 3 strains aggregated more tightly to some degree. Although this graph is a little complex, it seems that there is a clear geographical demarcation in the polioviruses genotype 1 such as the VDPV strains isolated from USA, Dominica, China mainland and Taiwan. These may indicate that geographic is another factor on codon usage bias.
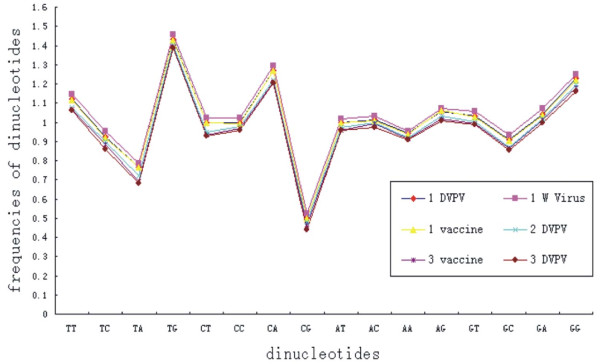
The frequencies of occurrence for dinucleotides were not randomly distributed and no dinucleotides were present at the expected frequencies. And the frequency of CpG and TpA was significantly low at all codon positions for coding region of 48 polioviruses genomes (mean ± S.D. = 0.490 ± 0.012; and mean ± S.D. = 0.748 ± 0.034. both < 0.78). The relative abundance of CpA and TpG also showed slight deviation from the ''normal range'' (mean ± S.D. = 1.253 ± 0.032 and 1.423 ± 0.023, respectively) (Table 5). In addition, the RSCU values of the eight codons containing CpG (CCG, GCG, UCG, ACG, CGC, CGG, CGU, and CGA) were analyzed, to reveal the possible effects of CpG under-represented on codon usage bias. All of these eight codons were not preferential codons and were markedly suppressed. The six codons containing TpA (UUA, CUA, GUA, UAU, UAC and AUA) were suppressed too. Conversely, the RSCU values of the eight codons containing CpA (UCA, CCA, ACA, GCA, CAA, CAG, CAU, CAC) and five codons containing UpG (UUG, GUG, UGU, UGC, CUG) are high, and most of them (8 out of 13) were preferential codons (Table 2 and Table 5). In addition, compared with DVPVs and live attenuated strain of polioviruses genotype 1, the wild viruses has higher frequencies of dinucleotides including CpG (Figure 2 and Table 5).
Table 5.
Relative abundance of the 16 dinucleotides in ORF of 48 polioviruses
| Dinucleotides | Rangea | Mean ± S.Db | DVPV 1 | Wild viruses | Vaccine |
|---|---|---|---|---|---|
| ApA | 0.909-0.965 | 0.935 ± 0.016 | 0.943 | 0.955 | 0.937 |
| ApG | 0.993-1.083 | 1.048 ± 0.019 | 1.058 | 1.073 | 1.060 |
| ApT | 0.955-1.041 | 0.989 ± 0.019 | 0.999 | 1.018 | 0.999 |
| ApC | 0.960-1.040 | 1.007 ± 0.016 | 1.013 | 1.030 | 1.010 |
| GpA | 0.987-1.094 | 1.035 ± 0.020 | 1.043 | 1.071 | 1.038 |
| GpG | 1.140-1.266 | 1.215 ± 0.024 | 1.228 | 1.249 | 1.221 |
| GpT | 0.986-1.079 | 1.023 ± 0.021 | 1.033 | 1.057 | 1.028 |
| GpC | 0.846-0.950 | 0.895 ± 0.028 | 1.228 | 1.249 | 1.221 |
| CpA | 1.204-1.308 | 1.253 ± 0.032 | 1.270 | 1.293 | 1.268 |
| CpG | 0.418-0.538 | 0.490 ± 0.012 | 0.499 | 0.522 | 0.496 |
| CpT | 0.920-1.039 | 0.979 ± 0.033 | 0.995 | 1.022 | 0.997 |
| CpC | 0.952-1.035 | 0.991 ± 0.012 | 0.999 | 1.021 | 0.995 |
| TpA | 0.669-0.801 | 0.748 ± 0.034 | 0.766 | 0.787 | 0.766 |
| TpG | 1.386-1.474 | 1.423 ± 0.023 | 1.434 | 1.457 | 1.427 |
| TpT | 1.059-1.164 | 1.106 ± 0.029 | 1.122 | 1.144 | 1.118 |
| TpC | 0.824-0.973 | 0.914 ± 0.026 | 0.925 | 0.953 | 0.916 |
Note: The boldface means that the dinucleotide was over-represented or under-represented.
a The range of coding region of 48 polioviruses's relative dinucleotide ratios
b Mean values of coding region of 48 polioviruses's relative dinucleotide ratios ± S.D
Figure 2.
Comparison the relative dinucleotide abundance in polioviruses DVPVs genotype 1, live attenuated virus genotype 1, wild viruses genotype 1, DVPVs genotype 2, DVPVs genotype 3 and live attenuated virus genotype 3.
Furthermore, we also performed a linear regression analysis on ENC value and gene length of ORFs of 48 polioviruses genomes. However, there was no significant correlation between codon usage and gene length in these virus genes (Spearman P > 0.05).
Discussion
Studies of synonymous codon usage in viruses can reveal much about viral genomes [25]. The overall codon usage among 48 ORFs of polioviruses was analyzed in this study. First, the ENC values of all the poliovirus samples were analyzed, and the results showed that the majority of polioviruses do not have a strong codon bias (mean ENC = 53.754 > 40). In addition, together with published data on codon usage bias among some RNA viruses, such as BVDV, H5N1 influenza virus and SARS-covs with mean values of 51.43, 50.91 and 48.99, respectively, one possible explanation for this is that the weak codon bias of RNA virus is advantageous to replicate efficiently in vertebrate host cells, with potentially distinct codon preferences [26-28].
Natural selection and mutation pressure are thought to be the main factors that account for codon usage variation in different organisms [29-31]. In this study, the general association between codon usage bias and base composition suggests that mutational pressure, rather than natural selection is the mainly factors on codon usage pattern of polioviruses.
Codon usage can also be strongly influenced by underlying biases in dinucleotide frequency, which differs greatly among organisms. Specifically, after accounting for dinucleotide biases, the proportion of codon usage bias explained by mutation pressure often increases, as seen in human RNA viruses [25]. Our study revealed that CpG and the eight CpG-containing codons are notably deficient in ORFs of 48 poliovirus genomes. The explanation for the CpG deficiency is immunologic escape. A high CpG content may be detrimental to small DNA (or RNA) viruses, as unmethylated CpGs are recognized by the host's innate immune system (Toll-like receptor 9) as a pathogen signature [32]. As with vertebrate genomes, methylated viral genomes would face a high chance of mutation at CpGs, that would result in a reduction of this dinucleotide [9,33]. We found that DVPVs and live attenuated virus of genotype 1 have lower frequencies of CpG dinucleotide compare with wild viruses of polioviruses genotype 1. The most popular explanation for lower frequencies of CpG in ORFs of DVPV genomes is that when OPV viruses turning into VDPV genotype 1, a lower frequencies of CpG dinucleotide maybe help VDPV out of the host immunity.
Although it seems speculative and complex, some researchers have found that reduction of the rate of poliovirus protein synthesis through large-scale utilization of codons that are not optimal has caused attenuation of viral virulence by lowering specific infectivity [34]. Therefore, the information from this study may not only have theoretical value in understanding poliovirus evolution (especially for DVPVs genotype 1), but also have practical value for the development the poliovirus vaccine. However, a more comprehensive analysis is needed to reveal more information about codon usage bias variation within poliovirus and other responsible factors.
Conclusions
The information from this study may not only help to understand the evolution of the poliovirus, especially for DVPVs genotype 1, but also have potential value for the development of poliovirus vaccines.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JZ, MW and YL designed the study and drafted the manuscript. WL, JZ, HC and LM collected the data and participated in the sequence alignment. YD and YG performed the statistical analysis. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. A plot of the values of the Axis1a (20.82%) and the Axis2a (16.27%) of each ORF in principle component analysis.
Contributor Information
Jie Zhang, Email: liuyongshengvip@sina.com.cn.
Meng Wang, Email: 64878893@163.com.
Wen-qian Liu, Email: mao49@163.com.
Jian-hua Zhou, Email: zhoujianhuazjh@163.com.
Hao-tai Chen, Email: haotaichen@163.com.
Li-na Ma, Email: frmln@126.com.
Yao-zhong Ding, Email: dyz1953@126.com.
Yuan-xing Gu, Email: guzi.123@163.com.
Yong-sheng Liu, Email: liuyongshengvip@sina.com.cn.
Acknowledgements
This work was supported in parts by grants from National Key Technologies R&D Program (2009ZX08007-006B), International Science & Technology Cooperation Program of China (No.2010DFA32640) and Science and Technology Key Project of Gansu Province (No.0801NKDA034). This study was also supported by National Natural Science foundation of China (No.30700597 and No. 31072143).
References
- Grantham R, Gautier C, Gouy M, Mercier R, Pave A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK. Molecular evolution of synonymous codon usage in Populus. BMC Evol Biol. 2008;8:307. doi: 10.1186/1471-2148-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Vlahovicek K. Comparison of codon usage measures and their applicability in prediction of microbial gene expressivity. BMC Bioinformatics. 2005;6:182. doi: 10.1186/1471-2105-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archetti M. Codon usage bias and mutation constraints reduce the level of error minimization of the genetic code. J Mol Evol. 2004;59:258–266. doi: 10.1007/s00239-004-2620-0. [DOI] [PubMed] [Google Scholar]
- Liu X, Wu C, Chen AY. Codon usage bias and recombination events for neuraminidase and hemagglutinin genes in Chinese isolates of influenza A virus subtype H9N2. Arch Virol. 2010;155:685–693. doi: 10.1007/s00705-010-0631-2. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Kinouchi M, Abe T, Kudo Y, Yamada Y, Nishi T, Mori H, Ikemura T. Analysis of codon usage diversity of bacterial genes with a self-organizing map (SOM): characterization of horizontally transferred genes with emphasis on the E. coli O157 genome. Gene. 2001;276:89–99. doi: 10.1016/S0378-1119(01)00673-4. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrazek J. What drives codon choices in human genes? J Mol Biol. 1996;262:459–472. doi: 10.1006/jmbi.1996.0528. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16:8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton LA, Parrish CR, Holmes EC. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J Mol Evol. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Stenico M, Peden JF, Lloyd AT. Codon usage: mutational bias, translational selection, or both? Biochem Soc Trans. 1993;21:835–841. doi: 10.1042/bst0210835. [DOI] [PubMed] [Google Scholar]
- Zhong J, Li Y, Zhao S, Liu S, Zhang Z. Mutation pressure shapes codon usage in the GC-Rich genome of foot-and-mouth disease virus. Virus Genes. 2007;35:767–776. doi: 10.1007/s11262-007-0159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao P, Dai L, Luo M, Tang F, Tien P, Pan Z. Analysis of synonymous codon usage in classical swine fever virus. Virus Genes. 2009;38:104–112. doi: 10.1007/s11262-008-0296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M. Codon usage bias in herpesvirus. Arch Virol. 2010;155:391–396. doi: 10.1007/s00705-010-0597-0. [DOI] [PubMed] [Google Scholar]
- Tsai CT, Lin CH, Chang CY. Analysis of codon usage bias and base compositional constraints in iridovirus genomes. Virus Res. 2007;126:196–206. doi: 10.1016/j.virusres.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Savolainen-Kopra C, Blomqvist S. Mechanisms of genetic variation in polioviruses. Rev Med Virol. 2010;20:358–371. doi: 10.1002/rmv.663. [DOI] [PubMed] [Google Scholar]
- Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol. 2005;59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- Minor PD, Almond J. Poliovirus vaccines: molecular biology and immune response. Molecular biology of picornaviruses. 2002. pp. 381–390.
- Agol VI. Vaccine-derived polioviruses. Biologicals. 2006;34:103–108. doi: 10.1016/j.biologicals.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Lu H, Zhao WM, Zheng Y, Wang H, Qi M, Yu XP. Analysis of synonymous codon usage bias in Chlamydia. Acta Biochim Biophys Sin (Shanghai) 2005;37:1–10. doi: 10.1093/abbs/37.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. The 'effective number of codons' used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986;14:7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JH, Zhang J, Chen HT, Ma LN, Liu YS. Analysis of synonymous codon usage in foot-and-mouth disease virus. Vet Res Commun. 2010;34:393–404. doi: 10.1007/s11259-010-9359-4. [DOI] [PubMed] [Google Scholar]
- Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/S0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Holmes EC. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/S0168-1702(02)00309-X. [DOI] [PubMed] [Google Scholar]
- Wang M, Zhang J, Zhou JH, Chen HT, Ma LN, Ding YZ, Liu WQ, Liu YS. Analysis of codon usage in bovine viral diarrhea virus. Arch Virol. 2011;156:153–160. doi: 10.1007/s00705-010-0848-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhou T, Ma J, Sun X, Lu Z. Analysis of synonymous codon usage in SARS Coronavirus and other viruses in the Nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Gu W, Ma J, Sun X, Lu Z. Analysis of synonymous codon usage in H5N1 virus and other influenza A viruses. Biosystems. 2005;81:77–86. doi: 10.1016/j.biosystems.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Schultz ST, Willis JH. Individual variation in inbreeding depression: the roles of inbreeding history and mutation. Genetics. 1995;141:1209–1223. doi: 10.1093/genetics/141.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. Bioessays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Krieg AM. CpG DNA: trigger of sepsis, mediator of protection, or both? Scand J Infect Dis. 2003;35:653–659. doi: 10.1080/00365540310015999. [DOI] [PubMed] [Google Scholar]
- Ambinder RF, Robertson KD, Tao Q. DNA methylation and the Epstein-Barr virus. Semin Cancer Biol. 1999;9:369–375. doi: 10.1006/scbi.1999.0137. [DOI] [PubMed] [Google Scholar]
- Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A plot of the values of the Axis1a (20.82%) and the Axis2a (16.27%) of each ORF in principle component analysis.