Abstract
Background
There are several known autosomal genes responsible for Ras/MAPK pathway syndromes, including Noonan syndrome (NS) and related disorders (such as LEOPARD, neurofibromatosis type 1), although mutations of these genes do not explain all cases. Due to the important role played by the mitochondrion in the energetic metabolism of cardiac muscle, it was recently proposed that variation in the mitochondrial DNA (mtDNA) genome could be a risk factor in the Noonan phenotype and in hypertrophic cardiomyopathy (HCM), which is a common clinical feature in Ras/MAPK pathway syndromes. In order to test these hypotheses, we sequenced entire mtDNA genomes in the largest series of patients suffering from Ras/MAPK pathway syndromes analyzed to date (n = 45), most of them classified as NS patients (n = 42).
Methods/Principal Findings
The results indicate that the observed mtDNA lineages were mostly of European ancestry, reproducing in a nutshell the expected haplogroup (hg) patterns of a typical Iberian dataset (including hgs H, T, J, and U). Three new branches of the mtDNA phylogeny (H1j1, U5b1e, and L2a5) are described for the first time, but none of these are likely to be related to NS or Ras/MAPK pathway syndromes when observed under an evolutionary perspective. Patterns of variation in tRNA and protein genes, as well as redundant, private and heteroplasmic variants, in the mtDNA genomes of patients were as expected when compared with the patterns inferred from a worldwide mtDNA phylogeny based on more than 8700 entire genomes. Moreover, most of the mtDNA variants found in patients had already been reported in healthy individuals and constitute common polymorphisms in human population groups.
Conclusions/Significance
As a whole, the observed mtDNA genome variation in the NS patients was difficult to reconcile with previous findings that indicated a pathogenic role of mtDNA variants in NS.
Introduction
Noonan syndrome (NS) was first described by Noonan and Ehmke [1]. It refers to a pleiomorphic autosomal dominant disorder with short stature, facial dysmorphia, a webbed neck and heart defects, and its prevalence is about one in 1000–2500 live births [2]. Cardiovascular diseases including valvular pulmonary stenosis, atrial septal defect and hypertrophic cardiomyopathy (HCM) are generally observed in 50–80% of the patients, with HCM being one of the most common cardiac abnormalities in these patients [3], [4], [5], [6].
Several germ line gain-of-function mutations in several RAS pathway members, including PTPN11 (which encode tyrosine phosphatase SHP-2), KRAS, SOS1, BRAF, and RAF1, SHOC2, MEK1 (alias MAPP2K1) have been identified as being responsible for NS [7], [8], [9], [10], [11], [12]. It has been suggested that nuclear DNA (nDNA) mutations in PTPN11 account for about ∼50% of cases [13]. Mutations in KRAS, SOS1, and RAF1 make up ∼1–2%, ∼20%, and 3–5% of NS cases without PTPN11 mutations, respectively [14]. When combined, all the above mentioned nuclear genes would account for 70–85% of NS cases [15]. Thus far, seven genes have been causally related to NS but also to other closely related conditions, including LEOPARD syndrome and Noonan-like syndrome. Germline mutations in a subgroup of those genes and other genes encoding signal transducers participating in the same pathway (HRAS, KRAS, NF1, SPRED1, BRAF, MEK1 and MEK2, alias MAP2K2) have been identified to be implicated in other clinically related disorders such as Costello syndrome ore neurofibromatosis type 1 [16]. Some authors proposed to group these developmental diseases in a single family of disorders, which has been termed the neurocardiofacialcutaneous syndrome (NCFCS) family [16], the Ras/MAPK pathway syndromes or RASopathies [17].
The search for new causal genes responsible for Ras/MAPK pathway syndromes has motivated many authors to explore the potential role of mtDNA mutations in NS and HCM based on the assumption that the mitochondrion plays an essential role in the energetic metabolism of cardiac muscle. Thus, recently, Dhandapany et al. [18] reported nine mtDNA mutations in a Noonan Indian patient suffering hypertrophic obstructive cardiomyopathy. According to the authors, “Our case forms the first report, which emphasizes the importance of mtDNA mutations in Noonan syndrome and extends the scope for mitochondrial related syndromes” (p. 287) [18]. The study of Dhandapany et al. was based on the analysis of only one patient's complete mtDNA genome, and the full set of results was not reported by the authors: only a list of nine mutations observed in the patient's mtDNA genome was reported. Eight of these mutations were reported as novel, a finding that was interpreted by the authors as follows: “the identification of these mutations indicates that mutations in mtDNA may account for a significant portion of genetic etiology in Noonan syndrome” (p. 287) [18]. A year before the appearance of the study by Dhandapany's et al. [18], Prasad et al. [19] claimed that they had observed six novel mutations in an Indian HCM patient. Both studies attributed the pathogenic condition to their presumable “novel” variants without any further scientific support. The misconception of “novelty” being synonymous with “causality” for mtDNA variants is unfortunately all too common in medical literature. And this is particularly problematic in mtDNA studies due to the fact that the mtDNA molecule is highly variable in human populations; in fact, a large proportions of both rare and common variations in populations still remain to be discovered and are consequently unrecorded in databases. As discussed in Bandelt et al. [20], “…An observed mtDNA mutation or polymorphism is novel if it has not been observed before; that is, it has not been reported in the literature before or cannot be found in other publicly available source. This, however, is not the manner in which the novelty of mtDNA mutations is perceived and treated in practice by the working human geneticist.” (p. 1073) [20]. Novelty is almost always operationally defined by searching for mtDNA in the main reference mtDNA database in the field, namely, MITOMAP (http://www.mitomap.org/cgi-bin/mitomap/search.pl) [20]. However, MITOMAP, although useful for many medical applications, is deficient in few aspects [20], and has therefore been interpreted as a risk factor in medical studies [21], [22].
Other recent articles have contributed to the debate on the presumable association of mtDNA variants or haplogroups (hgs) with NS or HCM. For example, Castro et al. [23] claimed to have found an association between hg T and NS patients of European ancestry, while Rani et al. [24] reported a presumable association between hg R and NS cases in a very small cohort of seven Indian patients.
The present study was motivated by this controversy. We conducted a sequencing study of the whole mtDNA genome in a total of 45 patients suffering Ras/MAPK pathway syndromes; most of them were NS patients (93%). About 11% of the patients were also affected by HCM. This is the largest cohort of Ras/MAPK pathway syndromes and Noonan patients who were analyzed for mtDNA variations by far. An evolutionary approach was carried out in order to assist the interpretation of variations found in NS patients. This approach was shown to be very useful in a previous study dealing with the analysis of mtDNA variation in asthenozoospermic males [25]. The alternative method of using a mtDNA case-control population study would require a much larger sample size, which is unfeasible for rare traits such as NS [26], [27]. We aimed to address several issues in the present study: (i) to evaluate whether Ras/MAPK pathway syndromes (with especial focus in NS patients) cluster in particular hgs by comparing the data with data available from human populations of the same ancestry, (ii) to identify mutations that could explain the clinical phenotypes of our patients, (iii) to evaluate whether replacement substitutions accumulate to a greater extent in patients with respect to expectations derived from control individuals (represented by the worldwide phylogeny based on more than 7800 entire genomes); (iv) to see whether tRNA mutations are more prevalent in patients than in control individuals, and (v) to examine recurrent, private and heteroplasmic mutations for patterns that could explain the clinical phenotypes. Moreover, previous findings claiming an association between mtDNA mutations or hgs and NS are discussed here for the first time in view of the present evolutionary evidence.
Methods
Ethics statement
Written informed consent was required from all patients. Analysis of entire mtDNA genomes in patient samples was approved by the Ethical committee of the University of Santiago de Compostela. The study conforms to the Spanish Law for Biomedical Research (Law 14/2007- 3 of July).
Sample collection and DNA extraction
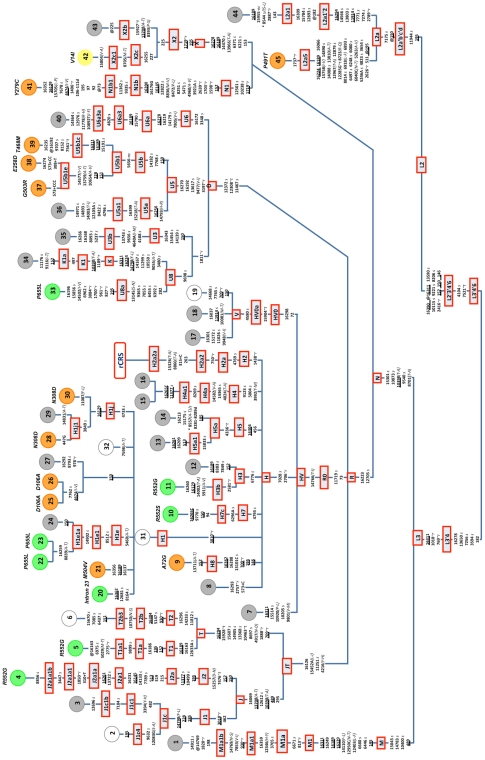
Blood samples were collected from all patients anonymously. A total of 45 samples were recruited for the present study. Our samples include 42 NS cases, two LEOPARD syndrome patients, and one neurofibromatosis type 1 patient. Note however, that the NS is clinically variable and a genetically heterogeneous developmental disorder; therefore our collection of patients was grouped more generally as patients suffering Ras/MAPK pathway syndromes. Among the 45 patients recruited, we included three pairs of brothers (namely, patients #15 and #16, #22 and #23, #25 and #26; see Figure 1). The DNA was extracted following standard phenol-chloroform protocols. Table 1 summarizes the clinical-pathological characteristics of our patients.
Figure 1. Maximum parsimony tree of 45 entire mtDNA genomes of patients suffering Ras/MAPK pathway syndromes.
The mutations are displayed along branches; the variant nomenclature is refered to was taken from the rCRS [30]. All mutations are transitions unless a suffix specifies a transversion (A, C, G, T), a deletion (d), an insertion (+), a synonymous substitution (s), a mutational change in tRNA (-t), a mutational change in rRNA (-r), stop codon (-stp), non-coding variant located in the mtDNA coding region (-nc) or amino acid replacement (indicated in round brackets). Recurrent mutational events are underlined. A prefix indicates a back mutation (@) or a position that is located in an overlapping region shared by two genes (*). Several mutational hotspot variants were not considered for phylogenetic reconstruction and therefore were eliminated from the tree; these included variants at the homopolymeric tracks around position 310, the microsatellite at m.523–524 d (aka m.522–523 d), the transversion m.16182A>C, m.16183A>C, m.16193+1C(C), m.16519T>C, and length or point heteroplasmies. Codes of the samples are indicated in colored circles at the terminal branches of the phylogeny: green indicates a mutation on gene SOS1, orange indicates a mutation on PTPN11, yellow indicates a mutation on KRAS, grey indicates lack of mutations on genes SOS1, PTPN11, KRAS, and RAF, and white indicates that data is not available for that sample.
Table 1. Clinico-pathological characteristics of the patients; numbers indicate percentages of the total sample.
| Phenotype | Sub-phenotype/Sub-classification | % |
| Facies | ||
| typical | 40.0 | |
| suggestive | 28.9 | |
| Cardiac features | ||
| typical ECG | 15.6 | |
| hypertrophic cardiomyopathy | 11.1 | |
| pulmonary valvular stenosis | 26.7 | |
| septal isolated defects | 2.2 | |
| bivalve aorta | 2.2 | |
| pulmonary artery dysplasia idiopathic dilatation | 4.4 | |
| septal atrial defects | 2.2 | |
| Height | ||
| percentile<3 | 35.6 | |
| percentile<10 | 17.8 | |
| Thoracic abnormalities | ||
| pectus escavatum/carinatum | 26.7 | |
| broad thorax | 33.3 | |
| Family history | ||
| first degree suggestive | 8.9 | |
| first degree definitive | 13.3 | |
| Others | ||
| mental retardation | 8.9 | |
| cryptorchidism | 20.0 | |
| lymphatic dysplasia | 6.7 | |
| Mutation in nuclear genes | ||
| PTPN11 | 24.4 | |
| KRAS | 2.2 | |
| SOS1 | 17.8 | |
| RAF | 0.0 | |
| Total | 44.4 |
Complete genome sequencing
The DNA from all patients was sequenced for the entire mtDNA molecule. We followed the sequencing protocols used by Álvarez-Iglesias et al. [28], which are briefly described here. The primers used for Polymerase Chain Reaction (PCR) amplification and sequencing were reported previously [29]. Polymerase Chain Reaction was performed in 10 µL of the reaction mixture, containing 4 µL of PCR Master Mix (Qiagen; Hilden, Germany), 0.5 µL 1 µM of each primer, 1 µL sample template and 4 µL of water. This PCR was carried out in a 9700 Thermocycler (AB) with one cycle of 95°C for 15 min and then 35 cycles of 94°C for 30 s, 58°C for 90 s and 72°C for 90 s with a full extension cycle of 72°C for 10 min. The sequencing reaction was performed in 11.5 µL of the reaction mixture, containing 2.5 µL of sequencing buffer (5X), 0.5 µL of BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), 1 µL of the corresponding primer (final concentration was 1 µM), 3 µL of the purified PCR product and water up to 11.5 µL. The automatic mtDNA sequencing was carried using capillary electrophoresis ABI3730 (Applied Biosystems).
Nomenclature and quality control
The revised Cambridge Reference Sequence or rCRS [30] was referred to for mtDNA variations. Haplogroup nomenclature was based on previous studies [28], [31], [32], [33], [34], [35]; the reference phylogeny is being updated by the project Phylotree [36]; see mtDNA tree Build 11 (7 Feb 2011) (http://www.phylotree.org/). In order to reduce the impact of sequencing artefacts [37], [38], [39], [40] we followed the phylogenetic procedures described in [40], [41], [42] which basically consisted of using the mtDNA worldwide tree as a reference to avoid artefactual profiles and documentation errors in mtDNA sequences and SNP genotypes as much as possible. This approach aimed to detect artificial patterns of mtDNA variations that significantly differed from the expected natural ones.
Statistical analysis
Counts of different types of mutational changes were carried out as in Elson et al. [43]. Pearson's chi-square test was applied to 2×2 contingency tables. A maximum parsimony tree was built using the genetic information from the entire mtDNA molecule (excluding the fastest mutational variants). Relative positional mutation rates were taken from Soares et al. [44].
All of the statistical analyses were carried out individually for carriers and non-carriers of nDNA mutations. However, the amount of mutations accumulated in both groups for the different mutational categories (nonsynonymous, synonymous, tRNA, and recurrent, among others) was statistically non-significant in all cases (Pearson's chi-square test, p-value>0.05). Therefore, given that mutational patterns were almost similar for carriers and non-carriers of nDNA mutations, the figures and tables presented in the main text refer to the total sample size of patients. However, for the sake of clarity, the analyses carried out separately for carriers and non-carriers are presented in Supplementary Figures S1, S2, S4, and S5. Analyses were also carried out for NS patients alone, and as expected, the results were virtually the same than those obtained for the whole sample (data not shown) given that NS cases represented 93% of the sample.
The dataset of mtDNA profiles reported by Álvarez-Iglesias et al. [28] representing a typical northern Iberian population was used as a control group for haplogroup frequency comparisons with patients.
Results
Nuclear mutations and clinical features
Seven genes (PTPN11, SOS1, KRAS, RAF1, BRAF, SHOC2 and MEK1, alias MAP2K1) have been causally related to NS and closely related conditions (including LEOPARD syndrome) and clinically related disorders (e.g. neurofibromatosis type 1) [2]. All of our patients were screened for mutations in nuclear DNA [45; and author's unpublished data]. About 44% of them carried nDNA mutations; in particular, most of them (∼24%) harboured mutations on the PTPN11 gene, and some of them on SOS1 (∼18%), and KRAS (∼2%) (Figure 1 and Table 1). No mutations on the RAF1 gene were identified in negative cases of PTPN11, SOS1, and KRAS. Patients #39 and #41 were posteriorly diagnosed with suffering from LEOPARD syndrome, and in fact, they carried the characteristic mutations on genes PTPN11 (Figure 1, Supplementary Table S1). Patient #35 suffered from neurofibromatosis type 1 syndrome (NF1) and also carried a 6 Mb deletion at 17q11-12. Nearly half of the patients suffered from cardiopathies, especially HCM (∼11%) and pulmonary valvular stenosis (∼27%). The other clinico-pathological characteristics of the patients are summarized in Table 1.
Phylogeography and phylogeny of patient mtDNA genomes
Entire, complete genomes were obtained for our cohort of 45 patients (Supplementary Figure S1 and Table S1). Patient mtDNA lineages were allocated to their corresponding hgs: most of them were of European ancestry, and therefore included representatives of the main clades, H, V, U, K, T, J, X and N1b (Figure 1). Two patients (patients #37 and #38; Figure 1) belonged to a still unknown branch of haplogroup U5, here referred to as U5b1e, whereas two other patients (patients #28 and #29; Figure 1) belonged to a new branch within H1j, here referred to as H1j1. Two additional profiles belonged to the typical sub-Saharan hg L2 [46], [47], [48]. One of them fell within the sub-branch L2a1. The other one (patient #45) described a novel branch of the L2 phylogeny referred to here as L2a5; it shared a transition at position 7175 and a reversion at site 150 with hg L2a (Figure 1), and most of the variants were also shared with another entire genome uploaded in GenBank under accession number HM596745. Another mtDNA belonged to the predominantly North African clade M1, in particular to the branch M1a1b, a lineage very closely related to the Sardinian-specific M1a1b1 sub-clade [49]. The proportion of non-European lineages in our patients mirrored the proportion expected in a typical sample of healthy individuals from northern Iberia [28], [50], [51] (see Supplementary Figure S1). Furthermore, the distribution of hgs was almost identical in carriers and non-carriers of nDNA mutations (Supplementary Figure S1). The newly discovered branches of the mtDNA phylogeny (H1j1, U5b1e, L2a5) did not carry features indicating an association with the NS phenotype or more generally, with Ras/MAPK pathway syndromes (see more analyses below).
We did not observe a correlation between the mtDNA hg lineages of patients and whether they were positive or negative for nuclear gene mutations (Figure 1); in other words, mutations in nuclear genes do not seem to be correlated with the mtDNA background of an individual. For instance, within hg H, only half of the patients carried mutations on nuclear genes and almost all of the carriers of nuclear mutations belonged to different sub-branches of hg H.
In addition, patients negative for nDNA mutations showed different mtDNA backgrounds. Therefore, there is no evidence to indicate that basal mutations from the mtDNA tree are involved in Ras/MAPK pathway syndromes.
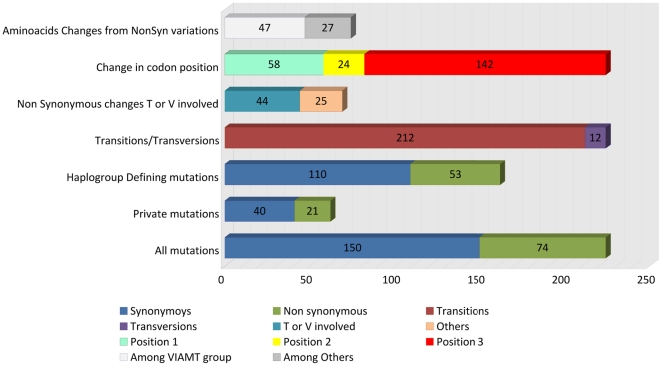
The phylogenetic tree in Figure 1 shows a total of 224 substitution events occurring at the coding region (sequence range 577–16023) of the mtDNA genomes of the patients analyzed. Ten of these were recurrent mutations (Table 2); of these, four were nonsynonymous changes, and two of them involved the threonine codon. The latter fits well with the estimation of Kivisild et al. [52], indicating that most of the nonsynonymous substitutions involved this codon. Nonsynonymous changes are more common in the amino acid groups V, I, A, M, and T (VIAMT group; see Figure 2), and most of the changes were between neutral apolar amino acids (Supplementary Figure S2), as previously noted by Pereira et al. [53] in natural populations, suggesting that these changes in the VIAMT group are more easily tolerated than other amino acid changes.
Table 2. Homoplasmic position in the coding region mtDNAs of Ras/MAPK pathway syndromes patients.
| Recurrent position | Sample ID1 | Nucleotide change | Gene Location | Syn/Nonsyn (aa substitution) | Hg | non-hg | Soares et al. 2 |
| 709 | #5*, #6, #9*, #19 | G-A | 12S rRNA | * | 3 (T, H8) | 1 | 59 |
| 930 | #1, #6 | G-A | 12S rRNA | * | 2 (M1a1b, T2b) | 0 | 5 |
| 1719 | #41*,#42*, #43 | G-A | 16S rRNA | * | 3 (N1, X2) | 0 | 31 |
| 3010 | #2, #3, #20*, #21*, #22*, #23*, #24; #25*,#26*, #27*, #28*, #29; #30*, #31, #32 | G-A | 16S rRNA | * | 13 (H1, J1) | 0 | 19 |
| 4674 | #41*; #25*, #26* | A-G | ND2 | Nonsyn (I-V) | 0 | 2 | 2 |
| 10398 | #1, #2, #3, #4*, #34, #44, #45* | A-G | ND3 | Nonsyn (T-A) | 7 (J, K1, N) | 0 | 18 |
| 11377 | #4*, #15, #16 | G-A | ND4 | Syn | 1 (J2a) | 1 | 9 |
| 11914 | #18, #44 | G-A | ND4 | Syn | 1 (L2a1'2) | 1 | 37 |
| 13708 | #2, #3, #4*, #43, | G-A | ND5 | Nonsyn(A-T) | 4 (X2b, J) | 0 | 24 |
| 14798 | #2, #3, #34 | T-C | CYT B | Nonsyn(F-L) | 3 (J1c, K) | 0 | 7 |
NOTE.
Starst identified samples carrying nDNA mutations;
Number of mutation hits in a worldwide phylogeny as recorded in Soares et al. [44].
Figure 2. Summary of the main features regarding different types of mtDNA changes in patients.
The percentages of changes at the first, second and third positions of the codons were ∼26%, ∼11%, and ∼63%, respectively (Figure 2). This pattern resembles the one obtained in the set of complete genomes analyzed by Pereira et al. [53], namely: 24%, 13%, and 63%. This finding confirmed that while the third position is under weaker evolutionary pressure than the first and the second positions, there is a significant bias against mutations at the second codon position.
The amino acid T and V codons were more frequently hit by nonsynonymous changes than other amino acid codons (Figure 2), in a proportion 1.76∶1; a similar figure to the one obtained by Kivisild et al. [52], namely: 1.7∶1.
The ratio of transitions:transversion was 17.6∶1 (Figure 2); this ratio also fits well with the one obtained by Pereira et al. [53], which was 22.2∶1, when considering polymorphism over 0.1%, which suggests the action of negative selection against transversion. The spectrum of transversions followed a bias towards a higher frequency of A and very low frequencies of G, C, and T (7∶1∶3∶1). A significant departure from this ratio could indicate documentation or genotyping errors in datasets [54].
The patterns observed for mutational changes in the total sample of patients (Figure 2) were also reproducible when the data were analyzed separately for carriers and non-carriers of nDNA mutations (Supplementary Figure S3).
Mutational changes in protein mtDNA genes of patients
About 33% of the variants were nonsynonymous, and they were almost homogeneously distributed between the different protein genes (Supplementary Figure S4). There is a quite common misconception in medical genetic studies that tends to interpret nonsynonymous variants as causal mutations by default. It is possible to compare the proportion of nonsynonymous variants in protein genes found in the mtDNA of patients with the proportion observed in healthy individuals. For instance, if we explore the dataset of Coble et al. [55] which consists of 241 complete genomes of mainly European ancestry (mimicking the hg background of our patients), a total of ∼33% of the variation occurred at nonsynonymous positions, as also occurred in the patients.
In our dataset, the ratio of nonsynonymous-synonymous positions in the coding region was about 1∶2.02 (Figure 2); this ratio fits very well with the proportion of 1∶1.97 that was obtained previously [53] in a survey of >5100 entire genomes (see caveats in [56]). According to Kivisild et al. [52]a “surplus of nonsynonymous mutations is a general feature of the young branches of the phylogenetic tree” (p.373) [52]. In the entire mtDNAs of patients, although there was an excess of nonsynonymous variants in young branches (n = 21; considering young branches to be the terminal ones, only) with respect to the older ones (n = 53)(see in Figure 2 “haplogroup defining mutations”), the difference was not statistically significant (Pearson's chi-square test, p-value = 0.869); these results are similar to the ones obtained by Pereira et al. [25]. The difference was not statistically significant when considering synonymous-nonsynonmous changes in the nDNA carriers (Pearson's chi-square test, p-value = 0.201) and nDNA non-carriers (Pearson's chi-square test, p-value = 0.847).
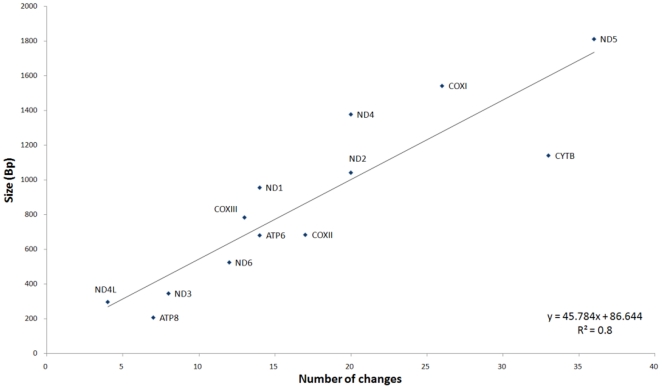
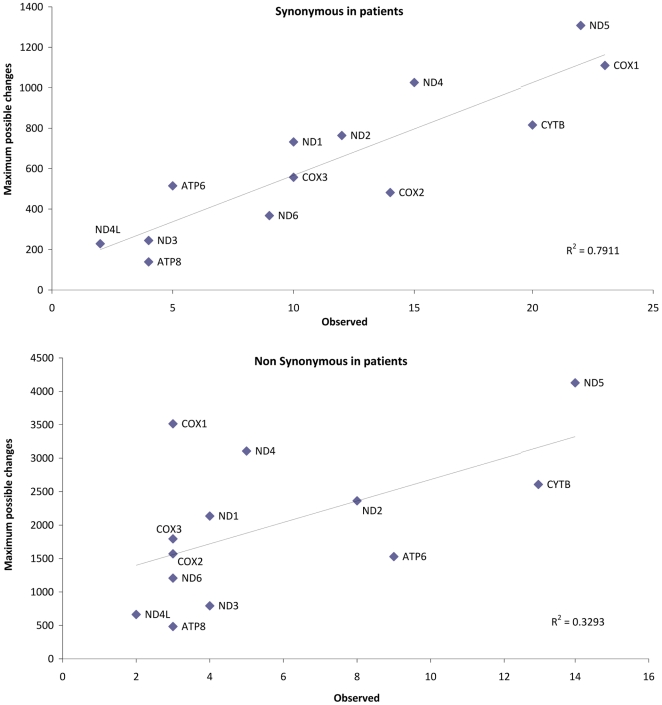
There was a high correlation (R2 = 0.8) between the number of changes that accumulated in the mtDNA coding region genes and the length of the gene (Figure 3). This correlation was also evident when only the synonymous changes were considered (R2 = 0.79), but not when looking only at the nonsynonymous substitutions (R2 = 0.33) (Figure 4). However, the pattern observed for the nonsynonymous changes fits again with the one described for the worldwide phylogeny [53].
Figure 3. Accumulation of mtDNA changes in the protein genes of patients versus size of the different genes.
Figure 4. Accumulation of synonymous and nonsynonymous mtDNA changes in the protein genes of patients versus the maximum number of possible changes per gene.
The accumulation of replacements per gene followed the same trend regarding the possible maximum number of changes per gene in carriers and non-carriers of nDNA mutations when considering all of the changes together (regarding the length of the genes) and when considering synonymous and nonsynonymous changes separately (Supplementary Figure S5).
Mutational changes in tRNA mtDNA genes of patients
Mutations in tRNA genes are commonly involved in mtDNA disorders, presumably due to their important role in protein translation. We found ten mutations located in tRNA, most of which were diagnostic of different hgs (Table 3). Some of these mutations are recorded in MITOMAP (http://www.mitomap.org/MITOMAP) as being related to some diseases, although none of them are labelled as “confirmed” pathogenic mutations. Note also that pathogenic indications in MITOMAP have to be considered with caution given the large amount of false positives in the literature and recorded in MITOMAP (see [20], [21]). The most conserved variants in our dataset were the m.12308A>G and m.7561G>A transitions, according to Helm et al. [57]; however, the former is a perfect diagnostic site for the frequent Eurasian haplogroup U and it is unlikely to be responsible for any rare disorder, whereas the latter transition has been previously reported in healthy individuals [58], [59]. The gene that showed the most variants in the mtDNA genomes analyzed was the tRNAThr; this finding also fits well with the prediction of Kivisild et al. [52], indicating that this gene bears significantly more substitutions than any other when observing the worldwide phylogeny. Although some of the tRNA mutations seem to be evolutionarily well conserved, none of them have a pattern of segregation with the disease, and none have been confirmed as pathogenic in the literature. Taking all these results together, tRNA mutations do not seem to play a pathogenic role in NS or related disorders [60].
Table 3. Variants observed at the mtDNA tRNA genes of Ras/MAPK pathway syndromes patients.
| Mutation position | Sample ID1 | Nucleotide change | tRNA | Location in secondary structure | Hg | MITOMAP | Conservation2 |
| 4336 | #13, #14 | T-C | tRNA-Gln | Acceptor stem | H5a, U6d | ADPD/hearing loss & migraine (unclear) | 50%<x<90% |
| 7476 | #4* | C-T | tRNA-SerUCN | Anticodon stem | J2 | Not reported | 50%<x<90% |
| 7521 | #44, #45* | G-A | tRNA-Asp | Acceptor stem | L3′4′6, G4, M76 | Not reported | Different in human and mammalian consensus |
| 7561 | #39* | T-C | tRNA-Asp | Variable loop | - | Not reported | 90%<x<100% |
| 10463 | #5*, #6 | T-C | tRNA-Arg | Acceptor stem | T, J1c1b1a, P4a | Not reported | 50%<x<90% |
| 122853 | #45* | T-G | tRNA-LeuCUN | DHU loop | L2a5 | Not reported | 50%<x<90% |
| 12308 | #33*, #34; #35, #36, #37*, #38*, #39*, #40 | A-G | tRNA-LeuCUN | Variable loop | U | CPEO/stroke/CM/renal & prostate cancer risk/altered brain pH | 100% |
| 15904 | #17, #18, #19 | C-T | tRNA-Thr | DHU loop | HV0a | Not reported | Natural variable site |
| 15927 | #43 | G-A | tRNA-Thr | Anticodon stem | X2b, B5b, U6a5, L0f2b, G3b, HV1a1 | Multiple sclerosis/DEAF1555 increased penetrance (P.M/possible helper mutation) | Different in human and mammalian consensus |
| 15928 | #5*, #6 | G-A | tRNA-Thr | Anticodon stem | T, L3x2b, C7b, Z3a, M25, M35b | Multiple sclerosis (P.M) | 50%<x<90% |
NOTE.
Starst identified samples carrying nDNA mutations;
According to Helm et al. [57];
Transversion 12285T>G is not actually a private variant if we consider that a new branch, L2a5, has been defined in the present article based on this entire genome and another one previously described in the literature under the GenBank entry HM596745.
Recurrent mutations in mtDNAs of patients
As indicated in Table 2, the homoplasmic mutations did not concentrate in particular hgs or genes; some of them were found at the tips of the phylogeny (see next section: private variants). Most of these mutations are well-known hotspots in the phylogeny [44], with the only exception of m.4674A>G that appears as a private substitution in two patients and which received two hits in Soares et al. [44] (Table 2). The apparent overrepresentation of tRNA and nonsynonymous mutations among recurrent mutations was previously observed [52] for the worldwide mtDNA phylogeny (Table 2).
Private mutations in mtDNAs of patients
Some other variants observed in our cases were private (Table 4) if we consider the private changes regarding their status in the phylogeny of Figure 1 (mutations located at the terminal branches). Nonsynonymous changes were more common among private variants (nonsynonymous:synonymous ratio: 1∶1.91) than within haplogroup defining mutations (1∶2.08); this is because natural selection had more time to filter out deleterious changes from the older branches than the younger branches (see above).
Table 4. Private coding region mutations observed in the entire mtDNA genomes of the patients (see Figure 1) that are “novel” or are recorded in MITOMAP as (confirmed or unconfirmed) disease-associated variants.
| Positions | Sample ID1 | Location | Nucleotide change | Synonymous/nonsynonymous(aa change) | Haplogroup2 | mtDNA Mutations with reports of disease-associations in MITOMAP3 |
| 827 | #33* | 12s rRNA | A-G | – | G1a1a1, D4h1a2, R0a1, B4b’d’e | Maternally inherited deafness or aminoglycoside-induced deafness (conflicting reports-B4b'd marker) |
| 961 | #33* | 12s rRNA | T-C | – | M7a2b, M44, D4h2, N9a2, A5b, R6a1a, B2i, U5a1c2, U4a1a, L0a1b1a1, L6, M2a1a2a1a | Maternally inherited deafness or aminoglycoside-induced deafness/possibly left ventricular non-compaction-associated (unclear) |
| 1820 | #1 | 16s rRNA | A-G | – | ‘Novel’ | Novel |
| 4796*4 | #36 | ND2 | C-T | Synonymous | – | Novel |
| 5029 | #5* | ND2 | T-C | (M/T) Neutral apolar-neutral polar | ‘Novel’ | Novel |
| 5911 | #11* | COX I | C-T | (A-V) Neutral apolar-neutral apolar | R8a1, L0a1b | Prostate cancer (reported) |
| 8544 | #44 | ATP6 | C-T | Synonymous | ‘Novel’ | Novel |
| 8544 | #44 | ATP8 | C-T | (S-L) Neutral polar-neutral apolar | ‘Novel’ | Novel |
| 100814 | #18 | ND3 | T-C | (M/T) Neutral apolar-neutral polar | – | Novel |
| 10205 | #7 | ND3 | C-T | Synonymous | ‘Novel’ | Novel |
| 11026 | #17 | ND4 | A-G | Synonymous | ‘Novel’ | Novel |
| 117785 | #40 | ND4 | G-A | (R-H) Basic polar-basic polar | – | LHON (confirmed); progressive dystonia (confirmed) |
| 12103A4 | #36 | ND4 | C-A | Synonymous | – | Novel |
| 14502 | #33* | ND6 | T-C | (I-V) Neutral apolar-neutral apolar | M10, X2a, R8b2, P7, N11a | LHON (reported-possible synergistic) |
| 14668 | #19 | ND6 | C-T | Synonymous | Z2, D4, L5a1b | Major depressive disorder-associated (reported) |
| 14831 | #29 | CYTB | G-A | (A-T) Neutral apolar-neutral polar | L1c3b2 | LHON (reported) |
| 15175 | #14 | CYTB | C-T | Synonymous | M9a1a1d | Novel |
NOTE.
Starst identified samples carrying nDNA mutations;
Mutations defining haplogroup(s) according to Phylotree and the data obtained here; “novel” means a variant that was not found in Phylotree [36], mtDB [62], HmtDB [63], and Google searches as executed in [20], [22];
The ‘novel’ condition is as indicated in MITOMAP;
Note that m.4796C>T and m.12103C>A were reported by Gasparre et al. [64] as novel changes in oncocytoma and CCRCC, m.4796C>T pop-up in HmtDB as reported by Porcelli et al. [65], although this variant does not appear in the original publication, and m.10081T>C appears in Zheng et al. [66] but as generated by human pol γ in vitro;
m.11778G>A is a well-confirmed mutation responsible for LHON and progressive dystonia, and, it fact, this pathogenic mutation appeared in a NS patient who also suffered from LHON (see Figure 1, #40); aa: amino acid.
Most of the private variants had already been reported in the literature in healthy individuals, with some of them appearing sporadically in different hg backgrounds. Some private variants were even reported as possibly pathogenic in MITOMAP, but this was never confirmed, with the exception of m.11778G>A, a well-known mutation responsible for Leber hereditary optic neuropathy LHON and progressive dystonia (patient #40; Table 4). In addition, all of these variants appear simultaneously as polymorphisms in MITOMAP (obviously with the exception of m.11778G>A). Only some of the variants listed in Table 4 are actually private and were referred to here as “novel”, in the understanding that “novel” means a variant that could not been found in the main mtDNA databases and does not show up on Google searches (Table 4). This “novel” condition alone cannot be used to attribute a causal role to these variants; in fact, any dataset of either healthy or unhealthy individuals will contain an large proportion of private variants, even taking into account the fact that there are more than 8700 complete or semi-complete genomes available in the literature to date (http://www.phylotree.org/). For instance, in this large dataset of entire complete genomes, more than 60% of the transversions and 28% of the transitions were only observed once (private substitutions). Taking all of this evidence together, it seems unlikely that any of the private variants observed in the patients are involved in the NS or Noonan-like phenotypes.
Heteroplasmic variants in mtDNAs of patients
Quite often, common mtDNA diseases are related to mutations with a heteroplasmic status. Six different heteroplasmies were found in the 45 patients analyzed (Table 5). Two of them fell in the control region and were highly recurrent in the phylogeny, especially position 16093 [44]. Only one of the positions, 15924, fell in the tRNAThr, but this is also a well-known mtDNA hotspot. Patient #11 carried two heteroplasmic variants (positions 4992 and 5144), but both were synonymous changes on gene ND2. Another position (10784) fell in the ND4 gene and was a nonsynonymous variant that changed the amino acid isoleucine to valine, but it appeared in a healthy individual belonging to haplogroup U6a1b (GenBank accession number: EF064320).
Table 5. Features of heteroplasmic variants found in patients.
| Position | Heteroplasmy | rCRS | Loci | Sample ID | GenBank and/or otherdatabase searches (hg) | Soares et al.Score |
| 4992 | G>A | A | ND2 | #11 | AP010974 (D4b2b1) | 0 |
| 5144 | C>T | C | ND2 | #11 | – | 0 |
| 10784 | G-A | A | ND4 | #9 | EF064320 (U6a1b) | 1 |
| 15924 | A>G | A | tRNAThr | #15, #16 | Common polymorphism | 30 |
| 16286 | T>C | C | D-loop | #37 | Common polymorphism | 5 |
| 16093 | T-C | T | D-loop | #32 | Common polymorphism | 79 |
Discussion
The patients analyzed in the present study (together with other related disorders) represented the largest cohort of patients analyzed to date by far for variations in the mtDNA molecule. The analysis of entire, complete genomes has, for the first time, enabled the implementation of an evolutionary approach aimed at discovering the potential pathogenicity of mtDNA changes in patients. The known human mtDNA phylogeny is based on 8731 complete mtDNA genomes (Phylotree), and, therefore, it provides a solid background to compare the variation observed in the mtDNA genomes of patients against.
Analysis of the data showed that the pattern of mutations in tRNA genes and the pattern of nonsynonymous changes in protein genes fit well with the variation observed in natural human populations. In other words, replacements, substitutions and tRNA mutations are not more prevalent in the mtDNA of patients than expected. In addition, most of the nonsynonymous mutations that were observed in the genomes of patients are common polymorphisms widely distributed throughout the global mtDNA tree. The recurrent and private mutations inferred from the phylogeny of entire mtDNAs from patients are also as expected, according to the patterns observed in human populations. Heteroplasmic mutations were also rare among the patients, and the few found did not seem to play a pathogenic role given their presence in healthy individuals too. Therefore, the evolutionary view of entire mtDNA genomes of patients does not support a role of mtDNA variants in the NS phenotype or in Ras/MAPK pathway syndromes. Moreover, the pattern observed for carriers of nuclear DNA mutations was very similar to that of non-carriers (Supplementary Figures S1, S2, S4, and S5).
In addition, there was no prevalent mutation in our patients nor a hg background apparently associated with the clinical phenotypes. Therefore, the theory of a highly penetrant mtDNA mutation being responsible for NS or Ras/MAPK pathway syndromes can be completely disregarded by the data obtained in the present study. In contrast to the Mendelian-like dominant pattern observed in most Ras/MAPK pathway syndromes cases (involving nDNA mutations), one could alternatively hypothesize a multi-factorial (genetic) complex scenario where some phenotypes could be explained by the sum (or interactions) of small effects contributed by different nuclear and/or mtDNA genes. Although the evolutionary approach employed here does not reveal the existence of a predominant variant in patients, a population-based approach (e.g. case-control study) could be used instead to reveal the existence of such low risk mtDNA variants. However, such an approach would need proper population sample sizes (in order to obtain reasonable power to detect any positive associations, e.g. 80%), monitorization of population stratification (which is particularly problematic in mtDNA studies) [27], [61], and adequate corrections for multiple tests, amongst others. Deficient study designs or wrong statistical treatments of the data could easily lead to false positives of an association. With this in mind, one could retrospectively look to the previous evidence suggesting the weak association between mtDNA variants and NS. The case-control study by Castro et al. [23] represents a paradigmatic example. These authors genotyped eight mtDNA variants in 130 Spanish HCM patients and 300 healthy controls; note that HCM is one of the characteristic phenotypes in NS patients (Table 1). According to the authors, “Because multiple comparisons were taken into account (9 haplogroups and 8 SNPs), we used the Bonferroni's correction and a p<0.01 was considered as the level of statistical significance.” It is not clear from the text whether the p-value mentioned refers to the initial nominal value or the one adjusted using Bonferroni. Either way, if one assume an standard nominal significant value of α = 0.05, an adjusted p-nominal value using Bonferroni for 17 independent tests (mtSNPs) would lead to a threshold for significance of 0.0029 (but not 0.01). However, Castro et al. claimed to have found a positive association between hg T (variant G13368A) and HCM, supported by a p-value = 0.007, which is above the correct adjusted nominal value.
Recently, Rani et al. [24] analyzed the complete genome of seven NS patients lacking PTPN11 mutations. They found that all of them belonged to different sub-lineages of hg R (including R7b1b, R30a1, R30c, T2b7, and U9a1), but the common factor in all of them was the lack of transitions at positions 12705 and 16223 (that lead from hg N to hg R). Since the authors only screened their patients for mutations at PTPN11 (which, worldwide, explains about 50% of the NS cases), their patients could have carried mutations at any of the other genes commonly held responsible for NS [2]. The study by Rani et al. [24] does not explain why the transitions at 12705 and 16223 should be responsible for the NS condition. Note that hg R represents the most common macro-hg in Europe (e.g. ∼92% in northern Iberia [28] and ∼87% in our cohort of northern Iberian patients; Pearson's chi-square test, p-value = 0.939); and that it includes the macro-hg R0 (where hg H is nested), hg U, hg J and T, amongst others. Therefore, the observations by Rani et al. contradict the scenario observed for our European patients: (i) our cases showed a lower frequency of hg R than a typical control group (although the difference was not statistically significant), and (ii) the supposed pathogenic variants (the ones defining hg R) are the predominant ones in Europe, an observation that is difficult to reconcile with the prevalence of NS worldwide. As also mentioned by Rani et al., India is very complex genetically; this means that any study aiming to evaluate the association between mtDNA variants and any disease should take the confounding effect of population sub-structure into account. Finally, Rani et al. deliberately considered hg R to be R minus U : “…followed by hgs R and U with frequencies of 14.27%, 15.23%, respectively…”) (p. 169) [24] and minus T (see Table 4 and Figure 3 in Rani et al. [24]) in the controls, but they included U and T within R in cases (as one of their patients belonged to hg U9a1 and another one to T2b7), and thereby they artificially created more differences in the apparent frequency between the samples than was actually the case. Moreover, apart from U and T, their R category should also have included the controls who belonged to hgs H2 and J1b, because all of them shared the feature common to the rest of the sub-lineages of hg R, which is the lack of mutations at positions 12705 and 16223. Finally, independent of possible population stratification, any case-control study based on seven cases and 105 controls has a very low a priori power for detecting a positive statistical association when the risk effect being looked for is weak. Therefore, their main conclusion “The haplogroup R by itself may be susceptible to disease phenotype or different environmental background or some of the unidentified nuclear gene might render susceptibility to disease phenotype…” (p. 171) [24] have little support in view of the contradictions mentioned above.
On the other hand, the results and conclusions of the studies by Dhandapany et al. [18] and Prasad et al. [19] were also critically questioned by Bandelt et al. [21] based on two main arguments: (i) their “novel” mutations were, in reality, not novel at the time of publication, and (ii) there is little support in favour of the causal role of these mutations in NS because most of them (if not all) are common polymorphisms, for example, the transition of m.2755A>G characterize hg R8. Our results agree with the conclusions of Bandelt et al. [21]: that authors tend to overstate the novelty of particular mtDNA variants and impute them a pathogenic role based on this “novel” condition. In reality, most of these variants were polymorphisms already known and which are unlikely to constitute pathogenic mutations. In case-control association studies, spurious positive associations generally show up when using deficient statistical approaches, or when under the presence of the population stratification, which is particularly problematic in mtDNA association studies because its reduced effective population sizes in comparison to average nuclear genes.
Conclusions
The analyses of replacement substitutions and other variants observed in the patients suffering Ras/MAPK pathway syndromes (tRNA, private, recurrent and heteroplasmic mutations), as well as the pattern of hg frequencies indicated that this variation can be fully expected as in any typical European dataset. Changes in mtDNA genomes of patients are therefore unlikely to be related to NS phenotype or Ras/MAPK pathway syndromes. The combined evolutionary and phylogeographic approach employed here seems more appropriate for evaluating the potential pathogenicity of mtDNA variants than a case-control study when the risk effect and the sample size are too low to provide reasonable statistical power or when it is under the presence of a population sub-structure.
Supporting Information
Haplogroup frequencies in the patients and in a typical Iberian sample of healthy individuals [28]. For the sake of clarity, some macro-haplogroups were sub-divided into main sub-haplogroups and other aggregated paragroup categories (e.g. phylogenetically, hg R0 should be considered as the sum of H+V+other-R0; and U should be considered as the sum of U5+K+other-U); the phylogenic relationships are clarified in Figure 1 and, more generally, in the worldwide phylogeny of Phylotree.
(TIF)
Distribution of synonymous and nonsynonymous changes in the mtDNA protein genes of all patients, and also considering carriers and non-carriers of nuclear mutations separately.
(TIF)
Number of different types of amino acid changes regarding nonsynonymous substitutions.
(TIF)
Summary of the main features regarding different types of mtDNA changes in the patients divided into carriers and non-carriers of nDNA mutations.
(TIF)
For carriers and non-carriers of nDNA mutations: accumulation of mtDNA changes in protein genes versus the size of the different genes, and accumulation of synonymous and nonsynonymous mtDNA changes in the protein genes versus the maximum number of possible changes per gene.
(TIF)
Mitochondrial DNA variants observed in the 45 entire complete genomes of the patients. The notation of the variants is explained in the legend of Figure 1. Heteroplasmic positions were also included.
(XLS)
Acknowledgments
We thank Yong Gang Yao and Hans Jürgen Bandelt for their useful comments and suggestions on a preliminary version of this article. The 45 complete mtDNA genomes analyzed in the present study were submitted to GenBank; accession numbers HQ384171-HQ384215.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research received support from two grants from the Fundación de Investigación Médica Mutua Madrileña, and a grant from the Ministerio de Ciencia e Innovación (SAF2008-02971), given to AS. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Noonan J, Ehmke D. Associated non cardiac malformations in children with congenital heart disease. J Pediatr. 1963:468–470. [Google Scholar]
- 2.Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Kogaki S, Kurotobi S, Nasuno S, Ohta M, et al. A novel mutation in the PTPN11 gene in a patient with Noonan syndrome and rapidly progressive hypertrophic cardiomyopathy. Eur J Pediatr. 2005;164:497–500. doi: 10.1007/s00431-005-1679-y. [DOI] [PubMed] [Google Scholar]
- 4.Bertola DR, Kim CA, Sugayama SM, Albano LM, Wagenfuhr J, et al. Cardiac findings in 31 patients with Noonan's syndrome. Arq Bras Cardiol. 2000;75:409–412. doi: 10.1590/s0066-782x2000001100005. [DOI] [PubMed] [Google Scholar]
- 5.Burch M, Sharland M, Shinebourne E, Smith G, Patton M, et al. Cardiologic abnormalities in Noonan syndrome: phenotypic diagnosis and echocardiographic assessment of 118 patients. J Am Coll Cardiol. 1993;22:1189–1192. doi: 10.1016/0735-1097(93)90436-5. [DOI] [PubMed] [Google Scholar]
- 6.Marino B, Digilio MC, Toscano A, Giannotti A, Dallapiccola B. Congenital heart diseases in children with Noonan syndrome: An expanded cardiac spectrum with high prevalence of atrioventricular canal. J Pediatr. 1999;135:703–706. doi: 10.1016/s0022-3476(99)70088-0. [DOI] [PubMed] [Google Scholar]
- 7.Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 8.Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 9.Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 10.Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 11.Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 12.Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 13.Gelb BD, Tartaglia M. Noonan syndrome and related disorders: dysregulated RAS-mitogen activated protein kinase signal transduction. Hum Mol Genet 15 Spec No. 2006;2:R220–226. doi: 10.1093/hmg/ddl197. [DOI] [PubMed] [Google Scholar]
- 14.Araki T, Chan G, Newbigging S, Morikawa L, Bronson RT, et al. Noonan syndrome cardiac defects are caused by PTPN11 acting in endocardium to enhance endocardial-mesenchymal transformation. Proc Natl Acad Sci U S A. 2009;106:4736–4741. doi: 10.1073/pnas.0810053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shchelochkov OA, Patel A, Weissenberger GM, Chinault AC, Wiszniewska J, et al. Duplication of chromosome band 12q24.11q24.23 results in apparent Noonan syndrome. Am J Med Genet A. 2008;146A:1042–1048. doi: 10.1002/ajmg.a.32215. [DOI] [PubMed] [Google Scholar]
- 16.Tartaglia M, Zampino G, Gelb BD. Noonan syndrome: clinical aspects and molecular pathogenesis. Mol Syndromol. 2010;1:2–26. doi: 10.1159/000276766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhandapany PS, Sadayappan S, Vanniarajan A, Karthikeyan B, Nagaraj C, et al. Novel mitochondrial DNA mutations implicated in Noonan syndrome. Int J Cardiol. 2007;120:284–285. doi: 10.1016/j.ijcard.2006.07.229. [DOI] [PubMed] [Google Scholar]
- 19.Prasad GN, Vanniarajan A, Emmanuel C, Cherian KM, Singh L, et al. Novel mitochondrial DNA mutations in a rare variety of hypertrophic cardiomyopathy. Int J Cardiol. 2006;109:432–433. doi: 10.1016/j.ijcard.2005.07.065. [DOI] [PubMed] [Google Scholar]
- 20.Bandelt H-J, Salas A, Bravi CM. What is a ‘novel’ mtDNA mutation―and does ‘novelty’ really matter? J Hum Genet. 2006;51:1073–1082. doi: 10.1007/s10038-006-0066-5. [DOI] [PubMed] [Google Scholar]
- 21.Bandelt H-J, Yao Y-G, Salas A. The search of ‘novel’ mtDNA mutations in hypertrophic cardiomyopathy: MITOMAPping as a risk factor. Int J Cardiol. 2008;126:439–442. doi: 10.1016/j.ijcard.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 22.Bandelt H-J, Salas A, Taylor RW, Yao Y-G. The exaggerated status of "novel" and "pathogenic" mtDNA sequence variants due to inadequate database searches. Hum Mutat. 2009;30:191–196. doi: 10.1002/humu.20846. [DOI] [PubMed] [Google Scholar]
- 23.Castro MG, Huerta C, Reguero JR, Soto MI, Domenech E, et al. Mitochondrial DNA haplogroups in Spanish patients with hypertrophic cardiomyopathy. Int J Cardiol. 2006;112:202–206. doi: 10.1016/j.ijcard.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Rani DS, Dhandapany PS, Nallari P, Govindaraj P, Singh L, et al. Mitochondrial DNA haplogroup ‘R’ is associated with Noonan syndrome of south India. Mitochondrion. 2010;10:166–173. doi: 10.1016/j.mito.2009.12.146. [DOI] [PubMed] [Google Scholar]
- 25.Pereira L, Goncalves J, Franco-Duarte R, Silva J, Rocha T, et al. No evidence for an mtDNA role in sperm motility: data from complete sequencing of asthenozoospermic males. Mol Biol Evol. 2007;24:868–874. doi: 10.1093/molbev/msm004. [DOI] [PubMed] [Google Scholar]
- 26.Samuels DC, Carothers AD, Horton R, Chinnery PF. The power to detect disease associations with mitochondrial DNA haplogroups. Am J Hum Genet. 2006;78:713–720. doi: 10.1086/502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosquera-Miguel A, Álvarez-Iglesias V, Vega A, Milne R, Cabrera de León A, et al. Is mitochondrial DNA variation associated with sporadic breast cancer risk? Cancer Res. 2008;68:623–625. doi: 10.1158/0008-5472.CAN-07-2385. [DOI] [PubMed] [Google Scholar]
- 28.Álvarez-Iglesias V, Mosquera-Miguel A, Cerezo M, Quintáns B, Zarrabeitia MT, et al. New population and phylogenetic features of the internal variation within mitochondrial DNA macro-haplogroup R0. PLoS ONE. 2009;4:e5112. doi: 10.1371/journal.pone.0005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, et al. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet. 2001;69:1348–1356. doi: 10.1086/324511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 31.Palanichamy Mg, Sun C, Agrawal S, Bandelt H-J, Kong Q-P, et al. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet. 2004;75:966–978. doi: 10.1086/425871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Achilli A, Rengo C, Magri C, Battaglia V, Olivieri A, et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loogväli E-L, Roostalu U, Malyarchuk BA, Derenko MV, Kivisild T, et al. Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia. Mol Biol Evol. 2004;21:2012–2021. doi: 10.1093/molbev/msh209. [DOI] [PubMed] [Google Scholar]
- 34.Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, et al. The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet. 1999;64:232–249. doi: 10.1086/302204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards M, Macaulay V, Hickey E, Vega E, Sykes B, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 36.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 37.Bandelt H-J, Olivieri A, Bravi C, Yao Y-G, Torroni A, et al. ‘Distorted’ mitochondrial DNA sequences in schizophrenic patients. Eur J Hum Genet. 2007;15:400–402; author reply 402-404. doi: 10.1038/sj.ejhg.5201781. [DOI] [PubMed] [Google Scholar]
- 38.Bandelt H-J, Achilli A, Kong Q-P, Salas A, Lutz-Bonengel S, et al. Low "penetrance" of phylogenetic knowledge in mitochondrial disease studies. Biochem Biophys Res Commun. 2005;333:122–130. doi: 10.1016/j.bbrc.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 39.Salas A, Yao Y-G, Macaulay V, Vega A, Carracedo A, et al. A critical reassessment of the role of mitochondria in tumorigenesis. PLoS Med. 2005;2:e296. doi: 10.1371/journal.pmed.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salas A, Carracedo Á, Macaulay V, Richards M, Bandelt H-J. A practical guide to mitochondrial DNA error prevention in clinical, forensic, and population genetics. Biochem Biophys Res Commun. 2005;335:891–899. doi: 10.1016/j.bbrc.2005.07.161. [DOI] [PubMed] [Google Scholar]
- 41.Bandelt H-J, Quintana-Murci L, Salas A, Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet. 2002;71:1150–1160. doi: 10.1086/344397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong Q-P, Bandelt H-J, Sun C, Yao Y-G, Salas A, et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- 43.Elson JL, Turnbull DM, Howell N. Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection. Am J Hum Genet. 2004;74:229–238. doi: 10.1086/381505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soares P, Ermini L, Thomson N, Mormina M, Rito T, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heredia CE, Balboa E, Castro-Feijóo L, Rica I, Barreiro J, et al. PTPN11, SOS1, KRAS and RAF: genotype-phenotype correlations in Noonan syndrome. Horm Res. 2009;72(Suppl.3):317. [Google Scholar]
- 46.Salas A, Richards M, De la Fé T, Lareu MV, Sobrino B, et al. The making of the African mtDNA landscape. Am J Hum Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Behar DM, Villems R, Soodyall H, Blue-Smith J, Pereira L, et al. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kivisild T, Reidla M, Metspalu E, Rosa A, Brehm A, et al. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am J Hum Genet. 2004;75:752–770. doi: 10.1086/425161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olivieri A, Achilli A, Pala M, Battaglia V, Fornarino S, et al. The mtDNA legacy of the Levantine early Upper Palaeolithic in Africa. Science. 2006;314:1767–1770. doi: 10.1126/science.1135566. [DOI] [PubMed] [Google Scholar]
- 50.Crespillo M, Luque JA, Paredes M, Fernández R, Ramirez E, et al. Mitochondrial DNA sequences for 118 individuals from northeastern Spain. Int J Legal Med. 2000;114:130–132. doi: 10.1007/s004140000158. [DOI] [PubMed] [Google Scholar]
- 51.Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo Á. mtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet. 1998;6:365–375. doi: 10.1038/sj.ejhg.5200202. [DOI] [PubMed] [Google Scholar]
- 52.Kivisild T, Shen P, Wall DP, Do B, Sung R, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereira L, Freitas F, Fernandes V, Pereira JB, Costa MD, et al. The diversity present in 5140 human mitochondrial genomes. Am J Hum Genet. 2009;84:628–640. doi: 10.1016/j.ajhg.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bandelt H-J, Kong Q-P, Richards M, Macaulay V. Estimation of mutation rates and coalescence times: some caveats. In: Bandelt H-J, Richards M, Macaulay V, editors. Human mitochondrial DNA and the evolution of Homo sapiens. Berlin: Springer-Verlag; 2006. pp. 47–90. [Google Scholar]
- 55.Coble MD, Just RS, O'Callaghan JE, Letmanyi IH, Peterson CT, et al. Single nucleotide polymorphisms over the entire mtDNA genome that increase the power of forensic testing in Caucasians. Int J Legal Med. 2004;118:137–146. doi: 10.1007/s00414-004-0427-6. [DOI] [PubMed] [Google Scholar]
- 56.Yao Y-G, Salas A, Logan I, Bandelt H-J. mtDNA data mining in GenBank needs surveying. Am J Hum Genet. 2009;85933:929–933; author reply 933. doi: 10.1016/j.ajhg.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helm M, Brule H, Friede D, Giege R, Putz D, et al. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 59.Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences from the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McFarland R, Elson JL, Taylor RW, Howell N, Turnbull DM. Assigning pathogenicity to mitochondrial tRNA mutations: when "definitely maybe" is not good enough. Trends Genet. 2004;20:591–596. doi: 10.1016/j.tig.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 61.Salas A, Carracedo Á. Studies of association in complex diseases: statistical problems related to the analysis of genetic polymorphisms. Rev Clin Esp. 2007;207:563–565. doi: 10.1157/13111575. [DOI] [PubMed] [Google Scholar]
- 62.Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Attimonelli M, Accetturo M, Santamaria M, Lascaro D, Scioscia G, et al. HmtDB, a human mitochondrial genomic resource based on variability studies supporting population genetics and biomedical research. BMC Bioinformatics. 2005;6(Suppl 4):S4. doi: 10.1186/1471-2105-6-S4-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gasparre G, Hervouet E, de Laplanche E, Demont J, Pennisi LF, et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet. 2008;17:986–995. doi: 10.1093/hmg/ddm371. [DOI] [PubMed] [Google Scholar]
- 65.Porcelli AM, Ghelli A, Ceccarelli C, Lang M, Cenacchi G, et al. The genetic and metabolic signature of oncocytic transformation implicates HIF1alpha destabilization. Hum Mol Genet. 2010;19:1019–1032. doi: 10.1093/hmg/ddp566. [DOI] [PubMed] [Google Scholar]
- 66.Zheng W, Khrapko K, Coller HA, Thilly WG, Copeland WC. Origins of human mitochondrial point mutations as DNA polymerase gamma-mediated errors. Mutat Res. 2006;599:11–20. doi: 10.1016/j.mrfmmm.2005.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Haplogroup frequencies in the patients and in a typical Iberian sample of healthy individuals [28]. For the sake of clarity, some macro-haplogroups were sub-divided into main sub-haplogroups and other aggregated paragroup categories (e.g. phylogenetically, hg R0 should be considered as the sum of H+V+other-R0; and U should be considered as the sum of U5+K+other-U); the phylogenic relationships are clarified in Figure 1 and, more generally, in the worldwide phylogeny of Phylotree.
(TIF)
Distribution of synonymous and nonsynonymous changes in the mtDNA protein genes of all patients, and also considering carriers and non-carriers of nuclear mutations separately.
(TIF)
Number of different types of amino acid changes regarding nonsynonymous substitutions.
(TIF)
Summary of the main features regarding different types of mtDNA changes in the patients divided into carriers and non-carriers of nDNA mutations.
(TIF)
For carriers and non-carriers of nDNA mutations: accumulation of mtDNA changes in protein genes versus the size of the different genes, and accumulation of synonymous and nonsynonymous mtDNA changes in the protein genes versus the maximum number of possible changes per gene.
(TIF)
Mitochondrial DNA variants observed in the 45 entire complete genomes of the patients. The notation of the variants is explained in the legend of Figure 1. Heteroplasmic positions were also included.
(XLS)