Abstract
B lymphopoiesis begins in fetal liver, switching to bone marrow after birth where it persists for life. The unique developmental outcomes of each phase are well documented, yet their molecular requirements are not. Here we describe two allelic X-linked mutations in mice that caused a cell-intrinsic arrest of adult B lymphopoiesis. Mutant fetal liver progenitors generated B cells in situ, but not in irradiated adult bone marrow, highlighting a necessity for the affected pathway only in the context of adult bone marrow. The causative mutation was ascribed to Atp11c, which encodes a P4-type ATPase with no previously described function. Our data establish an essential, cell-autonomous and context-sensitive function for ATP11C, a putative aminophospholipid flippase, in B cell development.
Keywords: B cell development, fetal liver, IL-7, flippase
Introduction
B cell progenitors first arise in fetal liver, then in bone marrow shortly after birth, and give rise to three major mature populations1. Marginal zone (MZ) B cells localize to the splenic marginal zone and respond to blood-borne antigens independently of T cell help2. Follicular B cells, by contrast, respond to protein antigens in a T cell-dependent manner, and progressively undergo immunoglobulin (Ig) isotype switching and affinity maturation. B-1 B cells comprise a much smaller population, which predominates in the pleural and peritoneal cavities and contributes most of the serum IgM during the early phases of infection3.
Whereas MZ and B-1 B cells are predominantly self-renewing, follicular B cells require constant replenishment from bone marrow, as demonstrated in mice with a conditional deletion of Rag2 (ref. 4). Briefly, deletion of Rag2 in newborn mice prevents almost all follicular B cell development, but leaves MZ and B-1 populations largely intact. Similarly in adult mice, Rag2 deletion causes a progressive decline in follicular B cell numbers, yet MZ and B-1 populations persist. A similar phenomenon has been reported for mice with a mutation of Il7 (ref. 5). While redundant with FLT3 in the fetal liver6–8, interleukin 7 (IL-7) becomes essential for B cell development in the bone marrow shortly after birth7. As a result, MZ and B-1 B cells predominate in the periphery of Il7 mutant mice, while follicular B cell numbers are severely reduced.
In the absence of IL-7 or its common gamma chain (γc)-associated receptor component IL-7R α, B cell development is arrested in the adult bone marrow shortly after cells acquiresurface B220, yet before they express CD19 or CD24 (refs. 8,9). Expression of the transcription factor early B cell factor (EBF) via STAT5 signaling appears to be important at this stage, since overexpression of EBF or constitutively active STAT5 can overcome the developmental arrest in Il7r mutant progenitors9,10. Other data suggests that rather than inducing transcription of Ebf1, STAT5 signaling instead promotes B cell progenitor survival through induction of Mcl1 (ref. 11). Whichever the mechanism, IL-7 and EBF are both key determinants of B cell differentiation12,13, and little else is known of the molecules and microenvironments that induce and maintain their expression in the adult.
We (and our colleagues14) describe here an essential role for the previously uncharacterized P4-type ATPase ATP11C in early B cell differentiation. ATP11C was redundant during B cell development in the fetal liver, yet essential in the context of adult bone marrow, where it was required for optimal responses to IL-7 and sustained expression of Ebf1.
Results
A heritable B cell deficiency
During a forward genetic screen for non-redundant regulators of lymphopoiesis (adapted from 15), we identified several mice from common N-ethyl-N-nitrosourea- (ENU) treated founders with low percentages of CD19+ cells in blood (Fig. 1a). The phenotype, named emptyhive, was transmitted as an X-linked recessive trait. Affected male mice were outwardly normal in behavior, fertility, lifespan and appearance, but were notably hyperbilirubinemic. This aspect of the phenotype, which stems from effects of the mutation in the liver, has been considered separately (O.M.S., B. Schnabl, B. Webb & B.B., submitted).
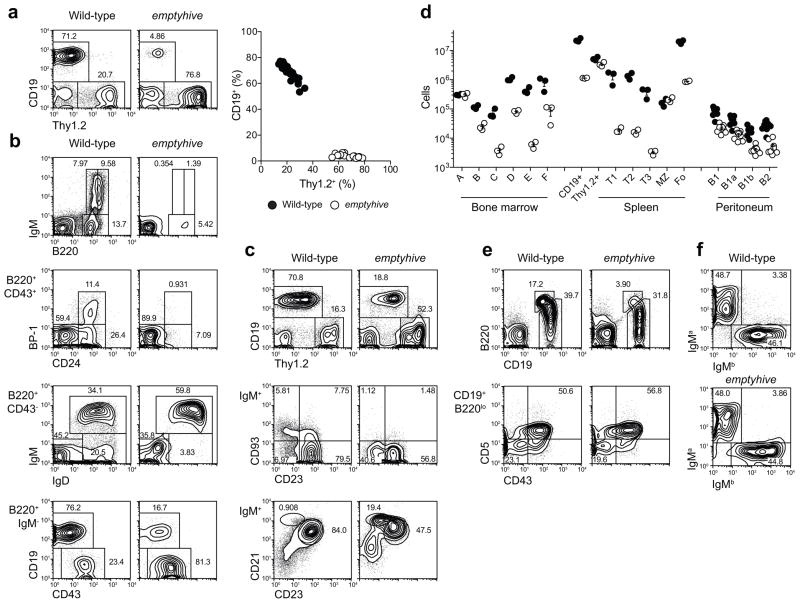
Figure 1.
A heritable B cell deficiency. (a) Frequencies of CD19+ and Thy1.2+ blood lymphocytes in the emptyhive pedigree. Percentages (b, c, e) and numbers (d) of B cell subsets in bone marrow (b), spleen (c) and peritoneal cavity (e). Hardy fractions in bone marrow (A-F) were gated as follows: A (B220+CD43+BP-1−CD24−); B (B220+CD43+BP-1−CD24+); C (B220+CD43+BP-1+CD24+); D (B220+CD43−IgM−IgD−); E (B220+CD43−IgM−IgD+); F (B220+CD43−IgM+IgD+). The C' fraction (B220+CD43+BP−1+CD24hi) was not resolved. Fractions A and B-D may also be gated as CD19− and CD19+ populations among B220+IgM−cells, respectively (b). IgM+ splenocytes were divided into the following subsets: T1 (CD93+CD23−); T2 (CD93+CD23+IgMint); T3 (CD93+CD23+IgMhi); MZ (CD93−CD23−IgMhiCD21hi); Fo (CD93−CD23+). Peritoneal lymphocytes were divided into B-2 (CD19+B220hi), B-1 (CD19+B220lo−int), B-1a (CD5+CD43+) and B-1b (CD5−CD43−) subsets. (f) IgM allotype expression on CD19+ blood lymphocytes from wild-type and emptyhive mice on a (C57BL/6 × BALB/c)F2 (IgMa/b) background. Data are representative of three (a to e), or one (f) independent experiments using three mice per genotype. Each symbol represents an individual mouse.
Among B cell progenitors in the bone marrow, emptyhive mutants had reduced numbers of cells beginning at the pre-pro-B to pro-B transition (Hardy fraction A [B220+CD43+BP-1−CD24−] to B [B220+CD43+BP-1−CD24+]16) (Fig. 1b), with a severe deficiency of immature B cells (Hardy fraction E [B220+CD43−IgM−IgD+]). In the spleen, emptyhive mice had one-tenth the normal number of CD19+ cells, largely due to a lack of follicular and transitional subsets (Fig. 1c,d), although numbers of MZ B cells and Thy1.2+ cells and were normal. Numbers of B-1 cells, the predominant population of B cells in the peritoneal cavity, were reduced by a factor of three in mutant mice (Fig. 1d,e), while numbers of peritoneal B-2 cells were reduced by a factor of ten. B cells in the blood of mutant mice had undergone normal allelic exclusion at the Igh locus (Fig. 1f).
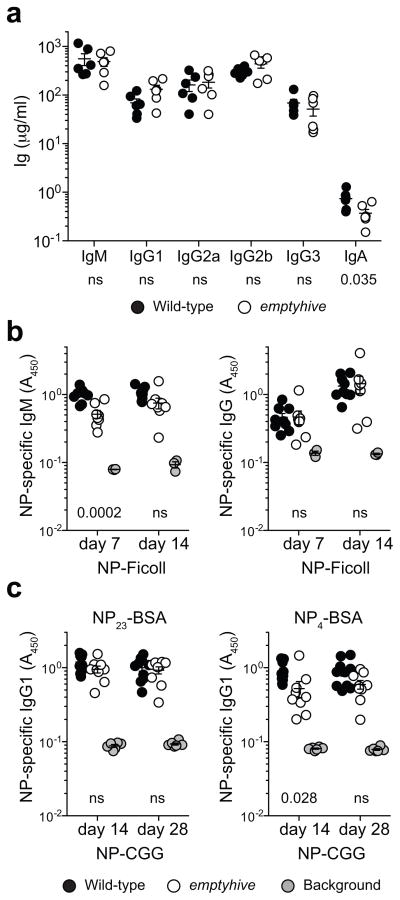
Despite a reduction in follicular B cell numbers, the B cells that remained appeared largely functional, and retained the capacity to produce all major immunoglobulin isotypes (Fig. 2a), as well as the ability to generate specific antibodies to T-independent and T-dependent immunogens (here, 4-hydroxy-3-nitrophenylacetyl (NP)-conjugated Ficoll and NP-chicken gamma globulin, NP-CGG, Fig. 2b,c, respectively). However, 50% less NP-specific IgM and high-affinity IgG1 was produced in response to NP-Ficoll and alum-precipitated NP-CGG immunization, respectively.
Figure 2.
Immunoglobulin secretion in emptyhive mice. (a) Total immunoglobulins as measured in the serum of 12–24 week-old naive mice. NP-specific antibodies were measured 7, 14 or 28 days after immunization of 12 week-old mice with NP-Ficoll (b) or alum-precipitated NP-CGG (c). Different capture antigens were used to discriminate between total (NP23-BSA) and high-affinity (NP4-BSA) IgG1 in response to NP-CGG. Each symbol represents an individual mouse from one experiment. P values indicated under plots were calculated by unpaired t-tests comparing wild-type and emptyhive groups. ns, not significant (P > 0.05).
Cell-intrinsic failure of adult B lymphopoiesis
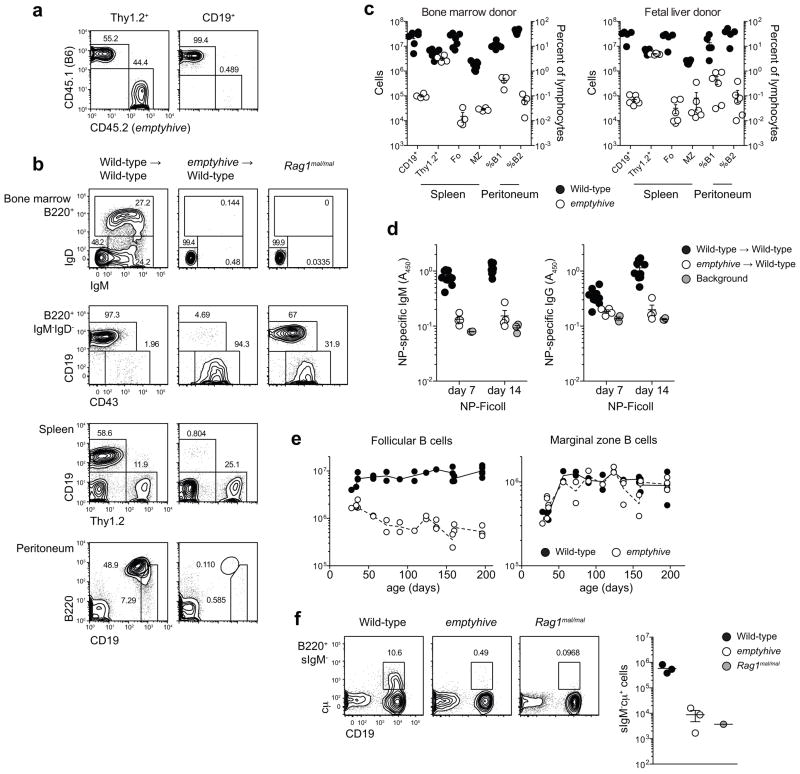
To determine the cellular origin of the emptyhive phenotype, irradiated Rag1 mutant mice were reconstituted with an equal mix of mutant and wild-type bone marrow. Although the Thy1.2+ compartment was reconstituted equally, less than 1% of CD19+ cells were of mutant origin (Fig. 3a), indicating that the defect was intrinsic to emptyhive B cell progenitors but did not appreciably affect T cell development. Even in the absence of competition, bone marrow cells from emptyhive donors were unable to repopulate any peripheral B cell compartment in irradiated recipients (Fig. 3b,c), and unlike their non-irradiated counterparts (Fig. 2b), recipients of mutant bone marrow also failed to produce specific antibody after immunization with NP-Ficoll (Fig. 3d). These data suggested a failure of adult B cell development in emptyhive mice, consistent with the severe reduction of immature B cells in mutant bone marrow (Fig. 1b) and the age-dependent decline of follicular, but not MZ, B cells in mutant spleens (Fig. 3e). This effect was not due to an intrinsic difference between adult and fetal progenitors, since neither bone marrow nor fetal liver cells could reconstitute the B cell compartment in irradiated recipients (Fig. 3c). Rather, the results suggest that emptyhive B cell progenitors, irrespective of their source, cannot develop in the microenvironment of adult bone marrow.
Figure 3.
A cell-intrinsic failure of adult B cell development. (a) A 1:1 mix of emptyhive (CD45.2) and wild-type (CD45.1) bone marrow was used to reconstitute irradiated Rag1 mutant recipients, and lymphocyte repopulation was measured eight weeks later in blood. Unmixed bone marrow (b, c) or E16.5 fetal liver cells (c) from either mutant or wild-type donors (CD45.1) was used to reconstitute irradiated wild-type recipients (CD45.2), and donor-derived reconstitution of bone marrow, spleen and peritoneal B cell subsets was analyzed eight weeks later. (d) Bone marrow chimeras were immunized with NP-Ficoll 8 weeks after reconstitution, and NP-specific immunoglobulin titers measured 7 and 14 days later. (e) Absolute numbers of follicular and marginal zone B cells as a function of age. (f) Intracellular IgM (cμ) expression in B220+ bone marrow cells negative for surface IgM. Plots are representative of 6 (a), 4–7 (b), or 3 (f) mice per genotype, from one (a-e) or two (f) independent experiments. Each symbol represents an individual mouse.
Immature and mature B cells were also entirely absent from the bone marrow of emptyhive-reconstituted recipients (Fig. 3b), with very few progressing beyond the B220+CD19− pre-pro-B stage. This developmental arrest was even more severe than that observed in non-irradiated emptyhive or Rag1 mutant mice, implying that a failure of IgM rearrangement was not the cause of developmental arrest in emptyhive mutants (Fig. 3b). Few mutant progenitors were capable of inducing expression of cytoplasmic IgM (cμ, Fig. 3f), consistent with the interpretation that the emptyhive phenotype originated from a defect upstream of Rag-mediated recombination.
Compromised responsiveness to IL-7
Because IL-7 is known to be essential for adult, but not fetal B cell development5, we hypothesized that a specific failure of IL-7R signaling in B cell progenitors could explain the emptyhive phenotype. To examine the competence of IL-7R signaling in mutant progenitors, surface IgM− precursors were purified by flow cytometry and cultured ex vivo in the presence of IL-7. emptyhive progenitors formed lymphoblasts with similar kinetics to wild-type in response to IL-7, indicating that IL-7R-induced proliferation was largely intact (Fig. 4a). In vitro IL-7 treatment also leads to the differentiation of low frequencies of surface IgM+ cells, which unexpectedly also emerged in emptyhive mutant cultures (Fig. 4b). Absolute numbers of IL-7-induced IgM+ cells were nonetheless lower than wild-type: a deficiency exaggerated by as much as two-fold at high IL-7 concentrations (Fig. 4b).
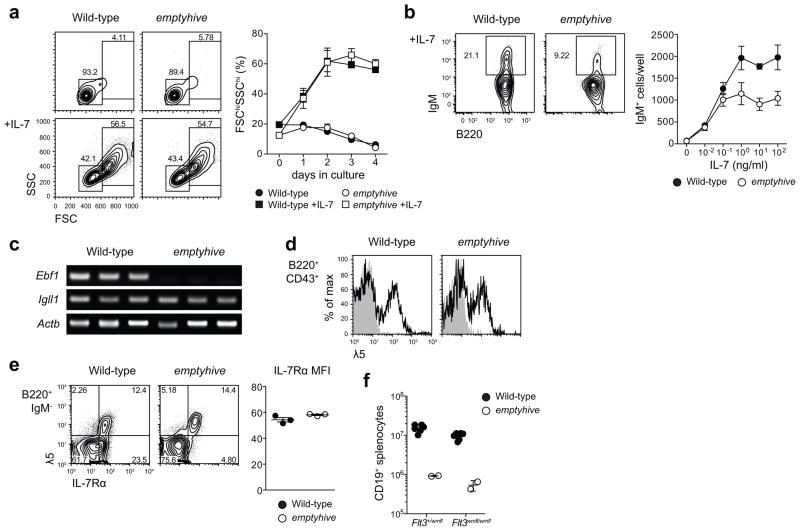
Figure 4.
Sensitivity to IL-7 and a failure to sustain expression of Ebf1. (a) B220+ surface IgM−bone marrow cells were sorted by flow cytometry and cultured ex vivo in the presence or absence of 100 ng/ml IL-7, and percentages of 7-AAD− lymphoblasts (FSChiSSChi) were measured at daily intervals. Symbols and error bars represent the mean and standard error of three mice per genotype. (b) B220+ surface IgM− bone marrow cells sorted from wild-type (CD45.1) or emptyhive (CD45.2) mice were cocultured in the presence of various concentrations of IL-7. Following four days of culture, frequencies and numbers of 7-AAD−surface IgM+ cells were measured. Symbols and error bars represent the mean and standard error of three mice per genotype. (c) RT-PCR PCR of cDNA from pre-pro-B cells (7-AAD−B220+IgM/IgD−CD19−NK1.1−Ly6C−). Each lane represents an individual mouse. (d) λ5 (CD179b) expression on the surface of B220+CD43+ bone marrow lymphocytes. Shaded histograms represent rat IgG2a isotype control, while solid histograms show λ5 staining. (e) IL-7Rα (CD127) expression on the surface of B220+IgM− bone marrow lymphocytes (dot plots, left), or B220+IgM−λ5+ (mean fluorescence intensity, right). (f) Splenic B cell numbers in emptyhive mice with combined mutation of Flt3. All data are representative of two independent experiments, each with three mice per genotype.
Since these data suggested a qualitative or quantitative difference in the IL-7 response of emptyhive progenitors, we examined expression of EBF: an early transcriptional regulator of B cell differentiation12 whose expression is lost in the absence of IL-7R signaling9. We measured Ebf1 transcription in sorted pre-pro-B cells by RT-PCR, and found it to be severely reduced in emptyhive mutant progenitors (Fig. 4c). We also measured transcription of Igll1: a target of EBF12 encoding the λ5 component of the surrogate light chain (SLC) portion of the pre-BCR17. Neither transcription of Igll1 (Fig. 4c), nor surface expression of λ5 (Fig. 4d) was compromised in emptyhive mutant progenitors, suggesting that the lack of Ebf1 transcript was either transient (and isolated to adult pre-pro-B cells or a subset thereof), or insufficient to affect Igll1 expression.
Consistent with their intact IL-7 responsiveness in vitro, IL-7Rα was expressed in equivalent amounts on the surface of emptyhive mutant progenitors (Fig. 4e). Furthermore, in the absence of IL-7, FLT3 signaling is thought to account for residual B lymphopoiesis in fetal liver18. While Flt3−/−Il7−/−and Flt3−/−Il7r−/−double mutants lack all peripheral B cells6,18, emptyhive Flt3−/−double mutant mice had similar numbers of CD19+ splenocytes as emptyhive single mutants (Fig. 4e). This result implied that IL-7R signaling remained intact in emptyhive fetal liver progenitors, at least to a degree sufficient for fetal B lymphopoiesis in the absence of FLT3.
We also found no major role for programmed cell death as the cause of the emptyhive phenotype. Homozygosity for the Faslpr mutation19 or a null allele of Bcl2l11 (ref. 20) could not correct the B cell deficiency in blood or bone marrow (Supplementary Fig. 1a-c), although partial rescue was observed in the presence of the B lineage-restricted EμBCL2-22 transgene21 (Supplementary Fig. 1a-c). Despite a relative increase of early B cell progenitors (B220+IgM−IgD−, Supplementary Fig. 1b,c), numbers of mature B cells (B220+IgM+IgD+) in the bone marrow of EμBCL2 transgenic mutants were nevertheless reduced as severely as they were in the absence of the transgene (Supplementary Fig. 1c). In the spleen, numbers of follicular and marginal zone B cells were increased (Supplementary Fig. 1d), likely due to an increased half-life4,21, which was consistent with the increased frequency of recirculating B cells observed in blood (Supplementary Fig. 1a).
Partial correction of B cell deficiency by a BCR transgene
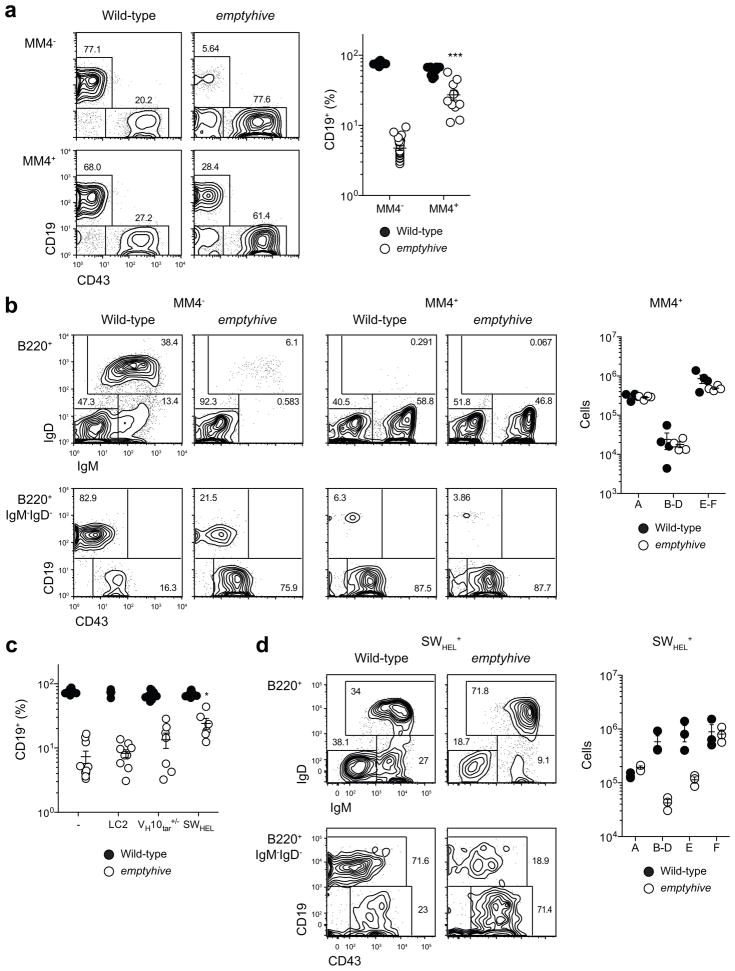
pre-BCR signaling is an essential step in the progression of B cell development, signaling productive rearrangement at the Igh locus, triggering proliferation and differentiation of preB cells, and initiating recombination of the kappa light chain allele22. The presence of recombined heavy and light chain transgenes renders pre-BCR signaling redundant, and effectively bypasses the developmental stages that surround it. The hen egg lysozyme (HEL)-specific MM4 transgene23, for example, is induced at the pre-pro-B stage, bypassing Rag-mediated recombination and expression of endogenous heavy and light chain genes and diverting progenitors directly to the surface IgM+ stage (Fig. 5a). Combining the emptyhive mutation with the MM4 transgene partially increased CD19+ cell frequencies in the blood (Fig. 5a) as well as numbers of IgM+ cells in the bone marrow (Fig. 5b). Since the MM4 transgene is a co-integration of heavy and light chain elements, this correction could reflect the effect of either recombined chain, or both in combination. We directly tested whether recombined heavy or light chains could rescue the emptyhive B cell defect in isolation using the ‘switch- HEL’ (SWHEL) system24, which consists of a HEL-specific VH10 heavy chain knockin and the LC2 light chain transgene. Significant correction of the emptyhive B cell deficiency was achieved only in the presence of both a heavy chain knockin allele and light chain transgene (Fig. 5c). This result is in contrast with SLC or Rag1-deficient mutants, whose respective B cell deficiencies can be fully or partially corrected by light or heavy chain transgenes alone25,26, and reveals that bypassing intermediate stages of B cell development (namely those within the B220+CD19+IgM−IgD− subset) could partially, but not completely, overcome the B cell deficiency in emptyhive mutant mice. IL-7R and pre-BCR signaling are both important during these stages22, and this may therefore reflect a failure of one or both pathways during the development of emptyhive mutant precursors.
Figure 5.
Partial correction of the emptyhive phenotype by a BCR transgene. The emptyhive mutation was combined with the MM4 transgene (a, b) or the LC2 light chain transgene, VH10tar knockin allele, or both (SWHEL) (c, d). Frequencies of CD19+ cells in blood (a, c), or B cell subsets in bone marrow (b, d) as determined by flow cytometry. An unpaired t-test was performed in (a) with comparison to the emptyhive non-transgenic group. For (c), a one-way ANOVA followed by Bonferroni post test was used, with reference to the emptyhive non-transgenic group. *P < 0.05, ***P < 0.001. Each symbol represents an individual mouse, and data are representative of two independent experiments.
Mutation of an uncharacterized P4-type ATPase
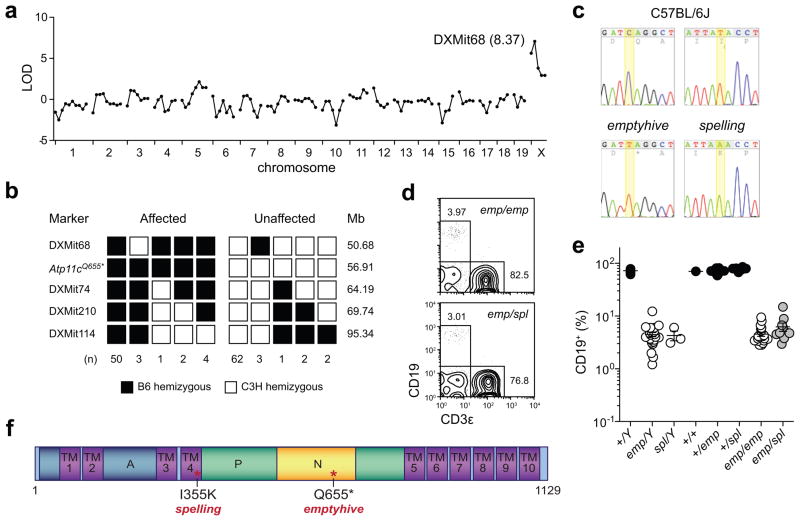
To identify the mutation responsible for the emptyhive phenotype, mutant males were outcrossed to C3H/HeN females and backcrossed to their F1 daughters. A total of 25 F2 males (11 mutant and 14 wild-type phenotype) were typed at 128 polymorphic microsatellite markers. Phenotypic linkage to the X chromosome was detected (maximum LOD score of 8.37 at DXMit68) (Fig. 6a), and fine mapping with a further 105 meioses confined the mutation to a 13.51Mb interval between DXMit68 and DXMit74 (Fig. 6b). Coding exons and flanking splice junctions of the 81 annotated protein-encoding genes in the mutant interval were sequenced by capillary sequencing, with 85.6% high-quality coverage (Phred score>30) of all nucleotides (129589/151336). A single hemizygous transition mutation was identified in Atp11c (C2113T, CAG→TAG, Q655*) (Fig. 6c). Since this mutation (in exon 19 of 30) occurs before the penultimate exon, the mutant transcript is most likely targeted by the nonsense-mediated decay pathway shortly after transcription27, although this has yet to be confirmed. In addition, we subsequently identified a mutant pedigree which phenocopied emptyhive (named spelling), and after sequencing Atp11c in these mice identified a second mutant allele (T1214A, ATA→AAA, I355K), which introduced a charged residue into the fourth of ten membrane-spanning domains (Fig. 6c, Supplementary Fig. 2). Compound heterozygosity for both alleles caused a B cell deficiency as severe as either parental strain (Fig. 6d,e), indicating that both emptyhive and spelling phenotypes were caused by X-linked recessive loss of function alleles of Atp11c (see Fig. 6f for protein schematic).
Figure 6.
The emptyhive phenotype is caused by a recessive mutation of Atp11c. Chromosomal (a) and fine (b) mapping of the emptyhive phenotype. (c) Sequence traces of the mutated nucleotides (C2113T, Q655*; T1214A, I355K) in Atp11c as detected in emptyhive and spelling hemizygous males. (d, e) Allelism test of spelling and emptyhive alleles. Progeny from an Atp11cspl/Y × Atp11cemp/+ mating were tested for phenotypic complementation by measuring the frequency of B cells in blood. Each symbol represents an individual mouse. (f) Predicted ATP11C protein domain structure. TM, transmembrane domain; P, phosphorylation domain; A, actuator domain; N, nucleotide binding domain.
Atp11c is a previously uncharacterized gene encoding a P4-type ATPase, expressed most prominently in the liver, but also by many other cell types28 (Supplementary Fig. 3). Little is known about mammalian P4-type ATPases in general, although mutation of one (ATP8B1) can cause progressive intrahepatic cholestasis in man29. While P-type ATPases are typically involved in cation transport30, the P4 subtype includes inwardly-translocating lipid flippases thought to enrich aminophospholipids at the cytoplasmic leaflet of lipid bilayers31,32. Because P4-type ATPases have previously been implicated in phosphatidylserine (PS) asymmetry in yeast31, we hypothesized that increased surface PS exposure and consequent PS-mediated phagocytosis could account for diminished numbers of B cells in Atp11c mutant mice33. PS expression on λ5-expressing cells was examined using Annexin V, yet no increase in surface expression was observed (Supplementary Fig. 4). Another requirement for phospholipid asymmetry could conceivably occur at the level of lipid rafts: membrane domains specialized in signal transduction34 and potentially important for IL-7R or pre-BCR35 signaling. Total surface expression of the raft-associated ganglioside GM1 (a target of the B subunit of cholera toxin) did not differ between wild-type and mutant cells (Supplementary Fig. 4), although closer examination of membrane microdomains around the IL-7R or pre-BCR will be needed to assess assembly of the rafts themselves.
Although not an aminophospholipid, phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3) is a phospholipid vital for signaling initiated at the IL-7R and pre-BCR36. Mutation of Cd19 or Pik3cd cripples the function of phosphatidylinositol-3-kinase (PI3K) and generation of PtdIns(3,4,5)P3 at the cytoplasmic leaflet of the pre-BCR signalosome, resulting in an impairment of MZ and B-1 B cell development37,38. Conversely, in the absence of the PtdIns(3,4,5)P3 phosphatase PTEN, an excess of PtdIns(3,4,5)P3 leads to an expansion of MZ and B-1 subsets37. Since mutations of both positive (Pik3cd, Pik3r1, Btk, Pik3ap1, Plcg2 and Blnk) and negative (Pten and Ptpn6) regulators of PtdIns(3,4,5)P3 availability are known to affect B cell development39, and since phosphatidylinositol asymmetry may be disrupted in emptyhive progenitors, we combined the emptyhive mutation with mutations of Cd19, Cd45, Inpp5d and Ptpn6. Neither genetic enhancement (Inpp5d, Ptpn6 mutation) nor diminishment (Cd19, Cd45 mutation) of PtdIns(3,4,5)P3 availability altered the emptyhive phenotype (Supplementary Fig. 5), implying that a disruption of phosphatidylinositol asymmetry at the cytoplasmic leaflet of the pre-BCR was an unlikely cause of B cell deficiency in Atp11c mutant mice.
A context-sensitive requirement for ATP11C
Having identified the causative mutation of the emptyhive phenotype, we were able to directly compare fetal B lymphopoiesis among wild-type and Atp11c mutant siblings in utero. Since IgM+ cells only appear after ~E16 (ref. 40), we examined E18.5 fetal liver cells, and found no difference in the number or frequency of B220+IgM− or B220+IgM+ cells in emptyhive embryos (Fig. 7a,b). This finding provided further evidence that fetal B cell development was intact in emptyhive mice, and that residual B cells in mutant mice were of pre-adult origin. Despite this result, mutant fetal liver cells were unable to reconstitute any B cell lineage in irradiated adult recipients (Fig. 3c), indicating that the B cell deficiency was cell-autonomous, yet microenvironment-dependent.
Figure 7.
Intact B lymphopoiesis in fetal liver. Percentages (a) and absolute numbers (b) of fraction A-D (B220+IgM−) and immature (B220+IgM+, fraction E) B cells in the fetal liver of E18.5 embryonic siblings from Atp11cemp/Y × Atp11c+/emp matings. Each symbol represents an individual embryo, and data are representative of two experiments with 8–14 embryos per experiment.
Discussion
Our results define a key role for Atp11c in adult, but not fetal B cell development. MZ and B-1 B cell compartments were both relatively intact in mutant mice, whereas follicular B cell numbers declined with age, closely resembling that seen after postnatal deletion of Rag2 (ref. 4). Most strikingly, fetal liver B cell progenitors developed normally in Atp11c mutant fetal liver, but not in the adult bone marrow, revealing a cell-intrinsic, but context-dependent requirement for ATP11C.
Few genes have been reported to act in such a way1, with Il7 and Il7r being the prototypical examples7. Il7, Il7r and Atp11c are all essential for adult, but not fetal B cell development, with a cell-intrinsic requirement for both Il7r and Atp11c to sustain Ebf1 transcription in pre-pro-B (B220+CD43+BP-1−CD24−) cells, and promote their differentiation to the pro-B (B220+CD43+BP-1−CD24+) stage9. But there are many key distinctions, the most notable being that T cell development proceeds in Atp11c mutants, but not in the absence of IL-7 or IL-7Rα8,41,42. Even among B cell progenitors, Igll1 transcription and λ5 expression are intact in Atp11c, but not Il7r mutants9. Ebf1−/− mice show a similar arrest in B cell development, yet they too fail to transcribe Igll1, and since these mice lack all peripheral IgM+ cells it does not appear to be an adult-restricted defect12. Atp11c mutant progenitors can clearly proliferate and differentiate ex vivo in the presence of IL-7, as well as during fetal development in the absence of FLT3 signaling6,43, and unlike Il7r mutant fetal liver cells (which can give rise to mature B cells in the bone marrow of irradiated adults7), have their developmental potential determined by their environment, rather than their source.
While ATP11C is clearly not essential for all IL-7-dependent phenotypes, it may still be quantitatively or qualitatively important, affecting only a given threshold or branch of IL-7R signaling, or migration of B cell progenitors to a stromal depot of IL-7. Another interpretation is that ATP11C has only a transiently important role in early B cell development. Temporal restriction of IL-7R expression appears to be critical, since retroviral overexpression arrests B cell development at the pre-pro-B stage and inhibits Ebf1 expression44, implying that transient downregulation of IL-7R is important for the progression of B cell development.
IL-7 is also necessary for sustaining expression of EBF and the progression of B cell development beyond the pre-pro-B stage, but not because of IL-7R signaling in pre-pro-B cells themselves. Instead, signaling during the common lymphoid precursor to pre-pro-B transition induces high and persistent expression of Ebf1, presumably via a series of feedback loops at the Ebf1 promoter45, and allows maturation of pre-pro-B cells even in the absence of IL-7 (refs. 9,10,46). Even though Ebf1 appears not to be expressed in Atp11c mutant precursors, an EBF target gene is (Igll1), suggesting that ATP11C may be important not for the initial induction of Ebf1, but instead to sustain Ebf1 expression through a feedback loop45. Whatever the cause, a failure to sustain Ebf1 expression has broad consequences for the regulation of early B cell development13.
Beyond the transcriptional consequences, what is the substrate of ATP11C, and how might it affect IL-7 responsiveness and the sustained expression of Ebf1? The aminophospholipids (phosphatidylserine and phosphatidylethanolamine) are primary candidate substrates based upon the study of ATP11C orthologs in yeast31,32, and lipid reconstitution experiments will be important to determine which specific variants are trafficked by ATP11C, and in which membrane compartments and/or domains. Further work will follow to define how this translates into an aberrant sensitivity to IL-7, and particularly why it does so only in the context of B cell development in the adult bone marrow. Presumably this outcome is related to phospholipid asymmetry during IL-7R signal transduction, or perhaps because of impaired migration to an IL-7-rich stromal niche. Identification of the substrate and affected pathway should expose a vital target for the specific regulation of adult B cell development, while Atp11c mutant strains will provide key experimental models to study membrane asymmetry and the origins of B cell immunity and malignancy.
Supplementary Material
Acknowledgments
We thank M. Gutierrez for animal care; T. Robinson, S. Kalina & C. Ross for genotyping; C. Goodnow (John Curtin School of Medical Research) & R. Brink (Garvan Institute) for mice; A. Feeney for discussions; D. La Vine for panel 5E; and reviewers for their valuable critiques. Supported by the Bill & Melinda Gates Foundation and National Institute of Allergy and Infectious Diseases Broad Agency Announcement Contract HHSN272200700038C (to B.B.), a General Sir John Monash Award (to O.M.S.), and an Irvington Institute Fellowship of the Cancer Research Institute (to C.N.A.).
Footnotes
Author Contributions
O.M.S. designed and performed experiments, analyzed data and wrote the paper under the guidance of B.B.; C.N.A. and E.P. identified the spelling phenotype, and assisted with immunization experiments; Y.X. and P.L. assisted with positional cloning and mutation identification, and C.H. measured immunoglobulin isotypes with reagents contributed by D.N..
References
- 1.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 2.Lopes-Carvalho T, Foote J, Kearney JF. Marginal zone B cells in lymphocyte activation and regulation. Curr Opin Immunol. 2005;17:244–250. doi: 10.1016/j.coi.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Tung JW, Herzenberg LA. Unraveling B-1 progenitors. Curr Opin Immunol. 2007;19:150–155. doi: 10.1016/j.coi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Hao Z, Rajewsky K. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J Exp Med. 2001;194:1151–1164. doi: 10.1084/jem.194.8.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho TL, Mota-Santos T, Cumano A, Demengeot J, Vieira P. Arrested B lymphopoiesis and persistence of activated B cells in adult interleukin 7(-/)- mice. J Exp Med. 2001;194:1141–1150. doi: 10.1084/jem.194.8.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sitnicka E, et al. Complementary signaling through flt3 and interleukin-7 receptor alpha is indispensable for fetal and adult B cell genesis. J Exp Med. 2003;198:1495–1506. doi: 10.1084/jem.20031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi K, Kondo M. Developmental switch of mouse hematopoietic stem cells from fetal to adult type occurs in bone marrow after birth. Proc Natl Acad Sci USA. 2006;103:17852–17857. doi: 10.1073/pnas.0603368103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Freeden-Jeffry U, et al. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dias S, Silva H, Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J Exp Med. 2005;201:971–979. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malin S, et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 13.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 14.Yabas M, et al. ATP11c is critical for phosphatidylserine internalization and B lymphocyte differentiation. Nat Immunol. 2011 doi: 10.1038/ni.2011. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelms KA, Goodnow CC. Genome-wide ENU mutagenesis to reveal immune regulators. Immunity. 2001;15 :409–418. doi: 10.1016/s1074-7613(01)00199-6. [DOI] [PubMed] [Google Scholar]
- 16.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karasuyama H, et al. The expression of Vpre-B/lambda 5 surrogate light chain in early bone marrow precursor B cells of normal and B cell-deficient mutant mice. Cell. 1994;77:133–143. doi: 10.1016/0092-8674(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 18.Jensen CT, et al. FLT3 ligand and not TSLP is the key regulator of IL-7-independent B-1 and B-2 B lymphopoiesis. Blood. 2008;112:2297–2304. doi: 10.1182/blood-2008-04-150508. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe-Fukunaga R, Brannan CI, Copeland NG, Jenkins NA, Nagata S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature. 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- 20.Bouillet P, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 21.Strasser A, et al. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc Natl Acad Sci USA. 1991;88:8661–8665. doi: 10.1073/pnas.88.19.8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 23.Brink RA, et al. Immunoglobulin M and D antigen receptors are both capable of mediating B lymphocyte activation, deletion, or anergy after interaction with specific antigen. J Exp Med. 1992;176:991–1005. doi: 10.1084/jem.176.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phan TG, et al. B cell receptor-independent stimuli trigger immunoglobulin (Ig) class switch recombination and production of IgG autoantibodies by anergic self-reactive B cells. J Exp Med. 2003;197:845–860. doi: 10.1084/jem.20022144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelanda R, Schaal S, Torres RM, Rajewsky K. A prematurely expressed Ig(kappa) transgene, but not V(kappa)J(kappa) gene segment targeted into the Ig(kappa) locus, can rescue B cell development in lambda5-deficient mice. Immunity. 1996;5:229–239. doi: 10.1016/s1074-7613(00)80318-0. [DOI] [PubMed] [Google Scholar]
- 26.Spanopoulou E, et al. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1- deficient mice. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 27.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 28.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bull LN, et al. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat Genet. 1998;18:219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- 30.Kühlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 31.Tang X, Halleck MS, Schlegel RA, Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- 32.Holthuis JC, Levine TP. Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- 33.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 34.Dykstra M, Cherukuri A, Sohn HW, Tzeng SJ, Pierce SK. Location is everything: lipid rafts and immune cell signaling. Annu Rev Immunol. 2003;21:457–481. doi: 10.1146/annurev.immunol.21.120601.141021. [DOI] [PubMed] [Google Scholar]
- 35.Guo B, Kato RM, Garcia-Lloret M, Wahl MI, Rawlings DJ. Engagement of the human pre-B cell receptor generates a lipid raft-dependent calcium signaling complex. Immunity. 2000;13:243–253. doi: 10.1016/s1074-7613(00)00024-8. [DOI] [PubMed] [Google Scholar]
- 36.Fruman DA, Cantley LC. Phosphoinositide 3-kinase in immunological systems. Semin Immunol. 2002;14:7–18. doi: 10.1006/smim.2001.0337. [DOI] [PubMed] [Google Scholar]
- 37.Anzelon AN, Wu H, Rickert RC. Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat Immunol. 2003;4:287–294. doi: 10.1038/ni892. [DOI] [PubMed] [Google Scholar]
- 38.Janas ML, et al. The effect of deleting p110delta on the phenotype and function of PTEN-deficient B cells. J Immunol. 2008;180:739–746. doi: 10.4049/jimmunol.180.2.739. [DOI] [PubMed] [Google Scholar]
- 39.Miosge LA, Goodnow CC. Genes, pathways and checkpoints in lymphocyte development and homeostasis. Immunol Cell Biol. 2005;83:318–335. doi: 10.1111/j.1440-1711.2005.01353.x. [DOI] [PubMed] [Google Scholar]
- 40.Raff MC, Megson M, Owen JJ, Cooper MD. Early production of intracellular IgM by B-lymphocyte precursors in mouse. Nature. 1976;259:224–226. doi: 10.1038/259224a0. [DOI] [PubMed] [Google Scholar]
- 41.Peschon JJ, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grabstein KH, et al. Inhibition of murine B and T lymphopoiesis in vivo by an anti-interleukin 7 monoclonal antibody. J Exp Med. 1993;178:257–264. doi: 10.1084/jem.178.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vosshenrich CA, Cumano A, Müller W, Di Santo JP, Vieira P. Thymic stromal-derived lymphopoietin distinguishes fetal from adult B cell development. Nat Immunol. 2003;4:773–779. doi: 10.1038/ni956. [DOI] [PubMed] [Google Scholar]
- 44.Purohit SJ, et al. Determination of lymphoid cell fate is dependent on the expression status of the IL-7 receptor. EMBO J. 2003;22:5511–5521. doi: 10.1093/emboj/cdg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roessler S, et al. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kikuchi K, Kasai H, Watanabe A, Lai AY, Kondo M. IL-7 specifies B cell fate at the common lymphoid progenitor to pre-proB transition stage by maintaining early B cell factor expression. J Immunol. 2008;181:383–392. doi: 10.4049/jimmunol.181.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.