Abstract
The results of investigations into performing DNA sequencing chemistry on a picoliter-scale electrowetting digital microfluidic platform are reported. Pyrosequencing utilizes pyrophosphate produced during nucleotide base addition to initiate a process ending with detection through a chemiluminescence reaction using firefly luciferase. The intensity of light produced during the reaction can be quantified to determine the number of bases added to the DNA strand. The logic-based control and discrete fluid droplets of a digital microfluidic device lend themselves well to the pyrosequencing process. Bead-bound DNA is magnetically held in a single location, and wash or reagent droplets added or split from it to circumvent product dilution. Here we discuss the dispensing, control, and magnetic manipulation of the paramagnetic beads used to hold target DNA. We also demonstrate and characterize the picoliter-scale reaction of luciferase with adenosine triphosphate to represent the detection steps of pyrosequencing and all necessary alterations for working on this scale.
Keywords: DNA sequencing, digital microfluidics, pyrosequencing, electrowetting, picoliter reactions
1 Introduction
With the continued drive to create fast, low-cost methods for sequencing DNA [1], there is a trend toward smaller and smaller devices [2–4]. The use of smaller devices, commonly microfluidic devices, significantly reduces the necessary amount of reagents, one of the main contributors to the overall cost of DNA sequencing. However, as the number of possible sequencing applications emerge, so too does the number of possible technologies, each of which has different requirements for throughput, speed, sensitivity, accuracy, robustness, operational skill, and other operational variables.
Pyrosequencing is a sequencing method, originally introduced in the late 1980’s [5], which makes use of the side products of sequential nucleotide addition to initiate a series of reactions that ultimately produce a chemiluminescent signal when a correct nucleotide is introduced. The nucleotides are individually added to a primed template strand and cycled as the sequencing progresses [6]. As a correct base is added to the DNA strand of interest, pyrophosphate (PPi) is released into solution:
| (1) |
The released pyrophosphate is then converted to adenosine triphosphate (ATP) via a reaction with adenosine 5′-phosphosulfate (APS) and ATP sulfurylase:
| (2) |
after which the ATP will react with firefly luciferase, luciferin and oxygen to produce light:
| (3) |
The intensity of the light produced is closely related to the amount of ATP present, and it can be used to quantitatively determine the number of a particular nucleotide added to the strand. Since its introduction, pyrosequencing has been used in a number of applications [7–10], ranging from analyzing single nucleotide polymorphisms [11, 12] and detecting rare mutations [13] to full genome or ribosomal analysis [14–16].
Several examples of microfluidics-based pyrosequencing have been introduced. Zhou et. al. introduced a pyrosequencing device which makes use of a series of capillaries to dispense nucleotides to a microchamber containing the DNA to be sequenced [17]. However, the read length on this device was very limited due to successive dilution, and reading of relatively long homopolymer nucleotide additions was unreliable. Russom et. al. describe a microfluidic flow-through device in which DNA is bound to microbeads that are then trapped in a microfabricated filter structure within a nanoliter-scale reaction chamber [18, 19]. Reagents are continuously supplied to the captured beads and any luminescence signal is observed for real-time sequencing. While the flow-through device itself was relatively simple, it required a fairly complex reagent injection system. Margulies et. al. introduced a massively parallel system that makes use of an array of microfabricated picoliter-scale wells [20]. The device demonstrated high accuracy and throughput, but again required the use of large and cumbersome pumping systems to supply reagents to the flow chamber holding the picowell array.
Unlike continuous-flow style microfluidics, digital microfluidics produces discrete fluid droplets that can be directed and controlled to a greater degree. These droplets can in turn act as micro-, nano-, or even picoliter-sized reactors, with the necessary reagents delivered as needed. The process of electrowetting is based on the principle of electrocapillarity, in which an electric field is used to induce a change in the effective surface energy at a solid insulator-liquid interface [21]. On an electrowetting digital microfluidic device, voltage is applied to such an interface where the liquid involved is polarizable or conductive. Often, the device is enclosed and filled with oil to prevent evaporation of the aqueous fluid. As the applied electric field alters the internal charge of the fluid, it exerts a force on the fluid relative to the surface of the device. Combined with the change in contact angle due to the altered surface energy, this force deforms the fluid and causes it to increase the surface contact area between the two phases [21–23]. When using electrowetting to manipulate droplets on a digital microfluidic device, the forward and reverse processes are combined to translocate the droplet [21]. During actuation of a droplet, individually operated electrodes are successively activated and deactivated in series. As the previous electrode is deactivated and releases voltage-induced forces, the next is activated and applies the forces to pull the droplet forward. Variations in how the electrodes are activated can be used to combine, mix, split, or dispense droplets from a large fluid reservoir [24–26]. Combined with programmable control units, these devices can be adapted to countless applications, including polymerase chain reaction (PCR) [27], clinical diagnostics [28], proteomic sample preparation [29], simple separations [30], and other complex biochemical techniques [31, 32], and they have been integrated with a number of detection and analysis techniques [29, 33, 34].
Previous work has been done to perform pyrosequencing on an electrowetting digital microfluidic device capable of producing droplets several hundred nanoliters in volume [35]. To initiate sequencing on this device, the target DNA is bound to paramagnetic beads, which are dispensed and directed to a magnetic region. By magnetically holding the DNA in one location [36–38], reagents can be added and excess split away without loss of the DNA, thereby eliminating all difficulties caused by dilution. The DNA beads can also be rinsed with a few successive droplets of buffer to prevent cross-contamination. Steps for nucleotide addition and detection can be separated to obtain more quantitative signal readings than those observed in flow-through systems, providing more accuracy for longer homopolymer additions. While the entire process has been relatively slow to date, reagent consumption has been reduced to a few hundred microliters for each individual solution, and the device and controller are easily transportable, making the system ideal for use in point of care or other field situations. Scaling similar devices to produce droplets of tens of picoliters has been demonstrated [23]. The reduction in size produces a device that requires only a few microliters of reagent volume and can be run on as little power as a nine volt battery. In this article, we explore the logistics of paramagnetic bead manipulation and characterize the behavior of the luciferase reaction to provide the groundwork for performing a pyrosequencing process similar to what has been done on larger electrowetting devices instead on a picoliter-scale electrowetting digital microfluidic device.
2 Materials and methods
2.1 Materials
All materials were used as provided by their respective suppliers. Tris(hydroxymethyl)aminomethane (Tris), ethylenediaminetetraacetic acid (EDTA), magnesium acetate, dithiothreitol (DTT), adenosine triphosphate (ATP), pyrophosphate (PPi), Tween 20, Triton X-15, and high conductivity grade poly(3,4-ethylenedioxythiophene) poly(styrenesulfonate) (PEDOT:PSS or PEDOT) were obtained from Sigma-Aldrich. Luciferin and adenosine 5′-phosphosulfate sodium salt (APS) were acquired from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). QuantiLum recombinant firefly luciferase was obtained from Promega Corporation, (Madison, WI), and ATP sulfurylase was purchased from New England Biolabs, Inc. (Ipswich, MA). Beads for statistical dispensing studies were obtained from Polysciences, Inc. (Warrington, PA) and Invitrogen Corp. (Carlsbad, CA). Norland optical adhesives NOA65 and NOA68 UV were purchased from ThorLabs (Newton, NJ). SU8 2015 was acquired from MicroChem (Newton, MA) and CYTOP fluorinated polymer from Bellex International Corp. (Wilmington, DE). Silicon wafers for the electrowetting substrate were purchased from Silicon Quest International, Inc. (Santa Clara, CA), and the 1 mm thick polycarbonate sheeting for top plates was purchased from McMaster-Carr (Elmhurst, IL).
2.2 Chip fabrication
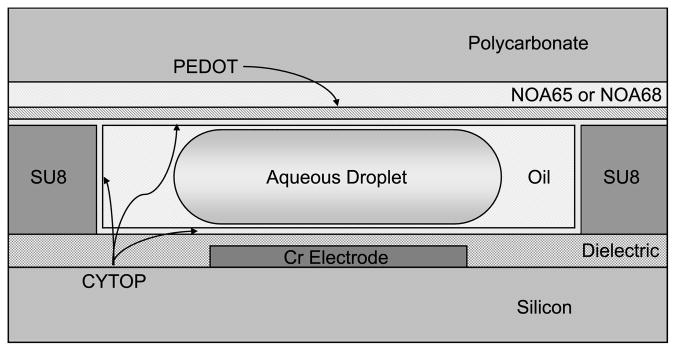
A diagram of the composition of the device used is shown in Figures 1. For this device, a silicon wafer, pre-coated with 1 μm of thermal oxide, is diced into 4–6 pieces. The electrodes are patterned by first laying down 100 nm of chromium, applying and developing the design with positive photoresist, and removing the excess metal using chromium etchant. The dielectric for the devices consists of a layer of tantalum pentoxide (Ta2O5) and a layer of Parylene C. As the SU8 used to form the gasket does not adhere well to the Parylene, the Ta2O5 is applied first, followed by SU8, which is then patterned, the substrate diced, and the Parylene C applied. A continuous film of Parylene C over the entire chip seals the underlying layers. A layer of CYTOP is spin-coated on the surface to provide the hydrophobicity necessary for electrowetting. This device, while somewhat limited in design flexibility, is very robust and is ideal for the simple, repetitive procedures used in the experiments described.
Figure 1.
Cross section of the electrowetting digital microfluidic device.
The substrate for all of the top plates used in these devices is a 1 mm thick piece of polycarbonate milled with a Roland MDX-20 mill (Irvine, CA) with a 1/32” bit. In order to allow multiple uses of the device, the chip is not bonded, and the top plate is held in place with pins. However, slight irregularities in the surface height of the SU8 gasket material can create gaps between the substrates and disrupt device operation. To achieve a better seal, a layer of Norland Optical Adhesive (NOA) 65 or 68 is applied to the polycarbonate substrate, spin-coated to a thickness of 50–100 μm [39]. The cured optical adhesive is relatively flexible and will deform around most imperfections in the SU8 without impinging on the reservoir or channels. A 150 nm layer of conductive PEDOT was applied to the top plate for grounding of the electrodes, followed by a layer of CYTOP as the hydrophobic layer. The new top plate composition has been determined to be nearly as robust as the polycarbonate-ITO-CYTOP composition used in the past, with no discernable loss of conductivity despite deformation of the chip-side layers. A diagram of the layers of this type of top plate is shown in Figure 1.
2.3 Feature geometry
An image of the reservoir, gasket, and electrode geometry is shown in Figure 2. In this device, the primary reservoir electrode is 2.5 mm wide and the internal reservoir electrode is 1 mm. Both are utilized during dispensing from the 140 nL-volume reservoir. The individual actuation electrodes consist of a body 100 μm2 with 2 rounded protrusions extending 30 μm at each side. The interlocking protrusions facilitate electrowetting and droplet movement by providing extra contact with droplets on neighboring electrodes. A gap of 5 μm separates the individual electrodes. This particular device is a single-level metal device with 10 μm wire connections for each electrode. The SU8 gasket is spin-coated to a thickness of 20 μm and provides boundaries for the reservoir and channels surrounding the electrodes. Channel widths defined by the gasket are approximately 250 μm wide and offset to cover the connection wires. The reservoir neck opening is approximately 100 μm wide, and the gasket tapers to a point on either side to facilitate “pinching” of the aqueous phase during dispensing. These features create a droplet volume of approximately 300 pL.
Figure 2.
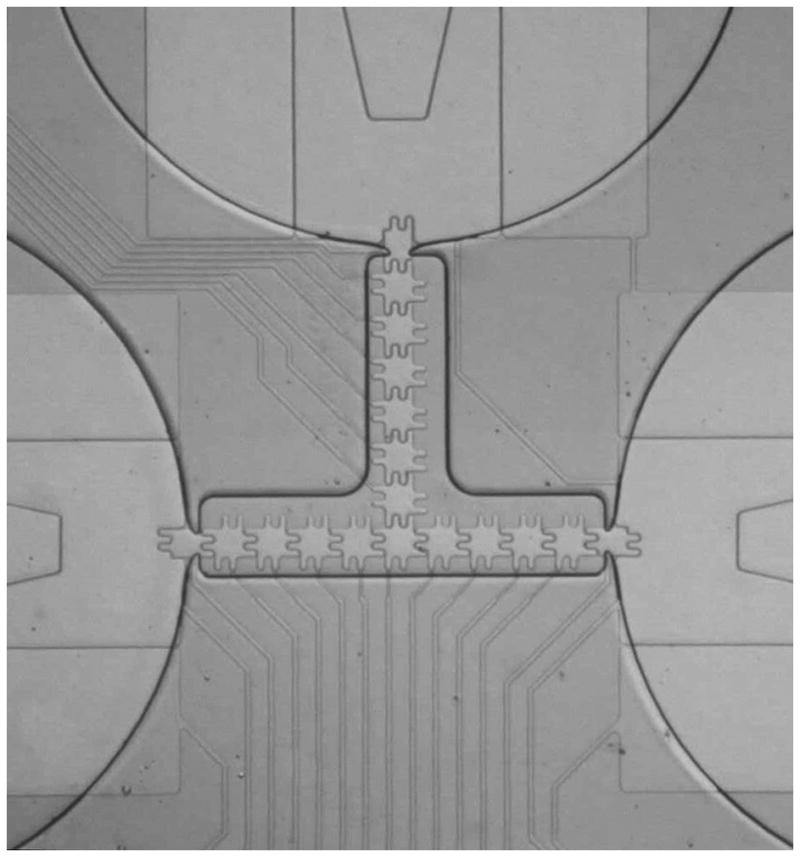
Image of the reservoirs and actuation electrodes on a model device.
2.4 Device control and detection
Contact to the electrowetting chips is made by a 22 pin clip connected to a 32-channel relay control board (National Control Devices, LLC). The relay board is connected via USB to a computer and controlled by a custom program. Electrode control can be performed both manually and automatically by simple text programs. Input voltage is supplied by an Agilent 33250A function generator (Agilent Technologies, Santa Clara, CA) amplified by a FLC F10A 10x voltage amplifier (FLC Electronics, Göteborg, Sweden). The grounding plane of the top plate is connected to the negative output of the amplifier with a mini alligator clip. All devices are operated at 20–30 Vpp. Observation for full-light tests is done with a Basler avA1000–120kc CCD camera (Basler Electronic, Highland, IL) mounted on an Optem Zoom 125 microscope lens with 10x objective (Qioptic, Paris, France). Low-light observations are performed with a Photometrics ES2 cooled CCD camera (Tucson, AZ) equipped with a 10x objective lens with 1 cm focal distance.
3 Results and Discussion
3.1 Bead dispensing
In the experimental outline for pyrosequencing on a large scale electrowetting device, the target DNA is bound to paramagnetic beads and magnets are used to manipulate the bead behavior. In larger-scale devices, the volume of fluid in the bead droplets relative to the bead size means that slight variations in the number of beads per droplet, and therefore the amount of DNA present, have little impact on the intensity of light produced during the luciferase reaction. However, at the picoliter scale, variations in the bead content per droplet can have a significant effect on the recorded intensity. To better understand the consequences of variations in bead concentration in a droplet-by-droplet basis, a series of experiments were performed using solutions with varying sizes and concentrations of beads. The beads used in these tests were Polysciences Fluoresbrite 2 μm and Polybead 10 μm carboxyl-functionalized polystyrene spheres and Invitrogen Dynabeads M-270 2.8 μm streptavidin-functionalized paramagnetic beads. Droplets of the bead suspensions were dispensed and the number of beads per droplet tabulated.
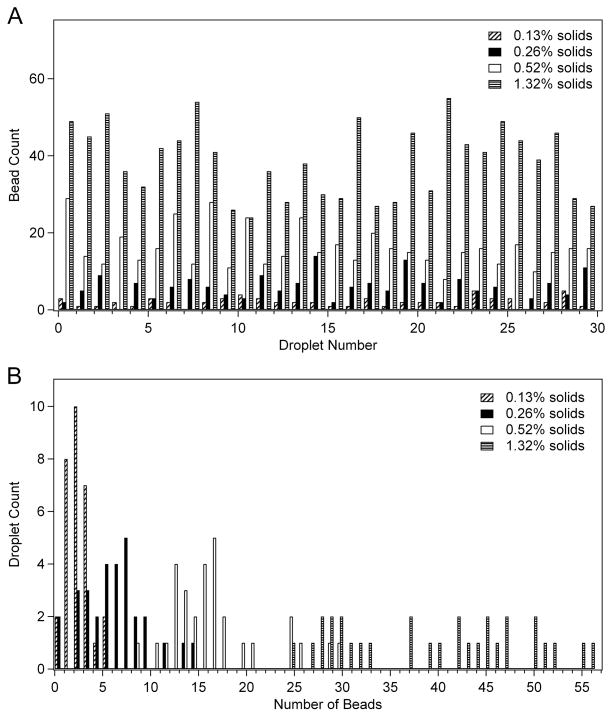
For all bead solutions, 30 droplets were dispensed and the beads counted. To prevent a buildup of beads at the edges of the reservoir fluid, the reservoirs were stirred by alternating activation of the two reservoir electrodes between each dispensed droplet. The droplets were dispensed to the point that the amount of fluid in the reservoir was significantly depleted by the end of the test to fully characterize bead count. In Figure 5A, the count of 10 μm beads in successive droplets is tabulated for several concentrations. This data indicates significant variation in the count, but no discernable trend was observed in the bead count over time or reduction of the volume of the fluid in the reservoir. Figure 5B displays the same data in a histogram. The average bead content was 2 beads ±1 for the lowest concentration of 0.13% solids (20x dilution of stock sample), 6 beads ±3 for 0.26% solids (10x dilution), 16 beads ±5 for 0.52% solids (5x dilution), and 39 beads ±9 for 1.32% solids (2x dilution). To determine whether the disparity in bead count per droplet was a result of any resistance to movement experienced by the large 10 μm beads, measurements were also performed using the 2 μm beads. While slightly less dispersed than the 10 μm beads, significant variation in bead count per droplet was also observed with the 2 μm beads (ex: 8.3 beads ±2.9).
Figure 5.
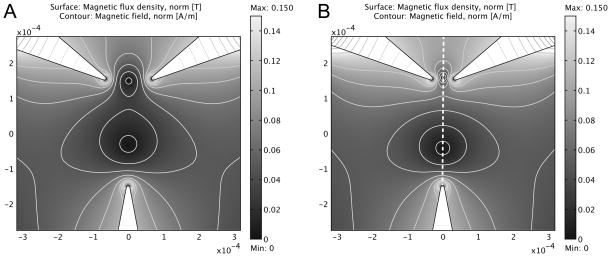
Simulation of magnetic split-well formation in a two-dimensional magnetic field simulation of three triangular sources. All magnetization sources are of equal magnitude, || M⇀i || = 1×105 A/m. Separation between the top paired electrodes is (A) 120 μm and (B) 50 μm. The dashed line represents the common axis of the magnetic wells.
3.2 Magnetic manipulation of paramagnetic beads
3.2.1 Experimental proof of concept
While some data manipulation can be used to compensate for the disparity in luminescence signal caused by varying bead counts (and thereby different DNA concentrations), greater consistency in signal would be preferable. A logical approach is the use of magnetic fields to control the behavior of the beads within the reservoir. In the literature, microfabricated electromagnets [40] and wire mesh [41–43] have been incorporated into devices to manipulate magnetic beads. When controlling the beads within the reservoir of a digital microfluidic device, either of these approaches can be used to create magnetic wells in the reservoir. The well locations and strengths can be altered to control the number of beads that can approach the reservoir exit.
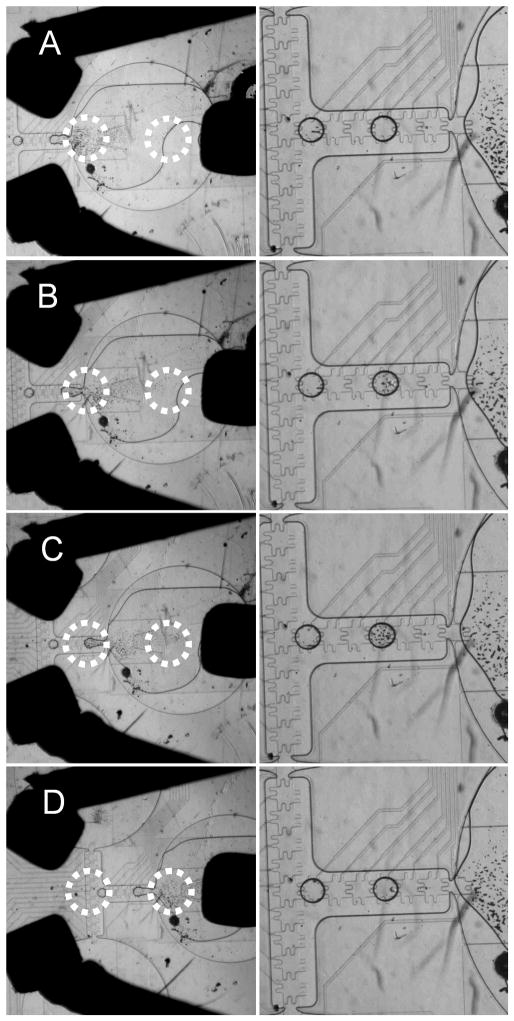
For experimental proof of concept, three 1 mm3 permanent magnets were mounted on wires with like poles pointed inward and positioned around a reservoir filled with a 1.0 mg/mL solution of paramagnetic beads. Figure 4 shows several images in which a micromanipulator was used to shift the position of the three mounted magnets to produce different concentrations of beads in the dispensed droplets. The approximate location of the magnetic energy wells is indicated by the red circles. In each image, the droplet sitting at the cross point of the electrode pathways was dispensed prior to introduction of the magnetic fields. In the first pair of images (Figure 4A), the magnetic well closest to the reservoir exit is located well within the reservoir. As a result, the beads are held in place and none are dispensed. Moving the magnets slightly to the left (Figure 4B) moves the well to the mouth of the reservoir. In this case, the beads are dispensed with the droplets in a concentration similar to the original bulk concentration. Further movement of the magnets (Figure 4C) shifts the magnetic well over the “gate” electrode, or the electrode furthest from the reservoir used during dispensing. This pulls most of the beads out of the reservoir while dispensing and produces a droplet with the highest number of beads. Movement of the magnetic well past the electrodes used for dispensing (Figure 4D) causes the second well to capture most of the beads within the reservoir, and no beads are dispensed. Each magnet placement was repeated, and the results were reproducible. In these experiments, the magnets were separated from the reservoirs by the full thickness of the top plate, so the fields experienced by the beads were relatively low, and several minutes were needed to allow the beads to reach their equilibrium point in the magnetic wells. Incorporation of the magnets or microfabrication of field producing features directly onto the device will produce faster response times, and further refinement of magnet placement and strength will provide even greater control.
Figure 4.
Paramagnetic bead manipulation of 1.0 mg/mL solution of Invitrogen Dynabeads M-270 2.8 μm with four magnet positions and the resulting bead droplet. The left-hand droplet in each image was dispensed prior to application of the magnetic field. Approximate locations of the field wells are circled with a dotted line.
3.2.2 Simulation for Magnet Optimization
While manipulation of magnetic beads while dispensing was successful, the process was difficult to control with the mounted magnets. Therefore, a series of simulations were performed with the Comsol Multiphysics electromagnetics modeling package, providing information on optimal magnet geometries appropriate for the formation of two localized magnetic wells, the effects of magnet spacing on split-well geometry, and the magnitude of forces obtainable with the dual-well magnet geometry. For simplicity, each simulation was performed in two dimensions and under magnetostatic conditions. In the computational domain, magnetic structures were defined with general, homogenous magnetization vectors, M⇀i, of units [A/m] and the constitutive relation,
| (4) |
where B⇀ is the magnetic flux density and H⇀ is the magnetic field. The medium surrounding the magnetic structures was considered to be water with unitary relative permeability, μr ≈ 1, and the constitutive relation,
| (5) |
where μ0 is the permeability of vacuum. A triangular mesh was generated with an element area ratio of approximately 2×10−4, affording each of the simulations 1.15×105 degrees of freedom. Given the magnetic sources, M⇀i; constitutive relations, relative medium permeability, and domain meshing, the magnetic vector potential field was computed in each simulation.
With the magnetic vector potential known, the magnetic flux intensity, B⇀, was calculated and used to determine the force applied to a magnetic bead within the computed field. Electromagnetic field theory states that the force experienced by a paramagnetic bead in a magnetic field opposes the potential energy gradient of the field, but is also proportional to the magnetic moment of the bead, m⇀, and the gradient of the magnetic field,
| (6) |
The superparamagnetic nature of micron-sized beads, requires that m⇀ be proportional the applied magnetic field according to the equation,
| (7) |
where V is the volume of the bead, χ is the magnetic susceptibility of the bead. Thus, the force imparted by the field generated by the magnetic structures in each simulation can be defined as
| (8) |
Magnetic force calculations were based on beads measuring 2.8 μm in diameter with magnetic susceptibility of 0.165.
A three-magnet geometry was used to produce the desired field wells. The configuration utilizes paired triangular magnets juxtaposed at 120° with respect to one another and a third magnet separated by a set distance and oriented toward it’s two sister components. Figure 5 shows the results of two field simulations with different distances between the paired magnets. The tips of the triangular magnetic domains were drawn with a 5 μm radius of curvature. The vertical separation between the paired upper magnets and the lower magnet for each simulation was 300 μm. The horizontal separation of the paired tips in Figure 5A is 120 μm, and the spacing in Figure 5B is 50 μm. The difference in the field contours in Figure 5A and 5B displays a more concentrated gradient in the magnetic wells with smaller horizontal magnet spacing.
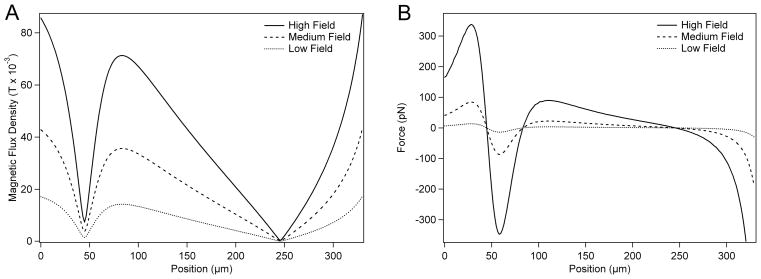
For variable magnetization strengths, magnetic forces were calculated for a single bead along the field cross-section indicated by the dashed line in Figure 5B. Figure 6A shows field profiles along the magnetic well common axis, taken with magnetization source magnitudes of 5×105 A/m, 2.5×105 A/m, and 1×105 A/m. These plots demonstrate how the extent of isolation in the wells varies directly with source magnetization strength. Figure 6B shows the force distribution along the common axis of the two magnetic wells. Stable points along the common axis occur where the magnetic force is equal to zero. A bead just before a stable point in Figure 6B will experience a positive force, oriented in the + direction (along the common axis), while a bead just past a stable point will experience a negative force, oriented in the - direction. The magnitude of the rectifying forces around the magnetic wells indicate the degree of their stability and varies directly with source magnetization strength, as demonstrated in Figure 6B.
Figure 6.
(A) Magnetic flux density profile along magnetic wells. Magnet spacing is 50 μm. (B) Magnetic force along the common axis of the magnetic split-wells. High, Medium and Low Field correspond to magnetization source strengths of 5×105 A/m, 2.5×105 A/m, and 1×105 A/m, respectively.
3.3 Representative pyrosequencing reactions
3.3.1 Reaction conditions
For all of the basic chemical reactions performed, a device was designed with three 3 mm wide reservoirs connected by a T-shaped array of actuation electrodes. This device was fabricated with the single-level metal process to ensure reliability of performance. The buffer solution used for the reactions contained 50 mM TRIS at pH 7.6, 5.0 mM magnesium acetate, 0.5 mM EDTA, 1.0 mM DTT, and 0.01–0.02% wt Tween 20. To determine detection limits and optimize the system, the chemistry was limited to the final step of the pyrosequencing process, as shown in Equation 3. The substrate solution consisted of 400 μg/mL luciferin and the specified amount of ATP in buffer, and the enzyme solution contained the specified concentration of luciferase in buffer. The two solutions were each injected via a micropipette into two of the three reservoirs, leaving the third to act as a waste reservoir. Droplets of approximately 300 pL were dispensed and moved with 20–30 Vpp applied to the electrodes, the exact value determined on a chip-by-chip basis. A droplet of luciferase solution was combined with a droplet of ATP solution, and the reaction was observed using a ES2 cooled CCD camera. The entire setup was housed in a dark box to prevent light contamination. Camera settings were 8x8 pixel binning, and 10 s exposure time. All reactions were started precisely at the beginning of the 10 s window and allowed to proceed to completion.
3.3.2 Dispensing enzyme-rich solutions
Dispensing of solutions with a high concentration of protein or enzyme, such as the luciferase solutions, was a challenge on the picoliter-scale device. Fouling of the chip surfaces over time causes the aqueous solutions to stick in the reservoir, and the surfactant-like behavior of some enzymes changes the contact angle and other fluidic properties, inhibiting dispensing and slowing droplet movement. In larger volume devices, these problems can be partially overcome by the momentum of greater fluid volumes, but in the smaller devices, dispensing solutions with high enzyme concentrations can be unreliable. Addition of an oil-based surfactant stabilized the oil films between the droplets and the chips, reducing the fouling effects of the enzymes, but did not completely eliminate the problem. As a result, enzyme concentrations were kept below a certain level, no more than 1 μg/μL luciferase when good repeatability was required and no more than 3 μg/μL luciferase during all experimentation.
It should be noted that fouling problems are not observed when the molecule of interest is bound to the paramagnetic beads. Unlike in air-filled systems [44, 45], the aqueous droplet in an oil-filled device does not make direct contact with the chip surface. Instead, the oil completely encases the aqueous solution, even forming a film between the droplet and the electrode surface. While free molecules can partition across the oil-water interface to reach the chip surface, the hydrophilic paramagnetic beads with their bound molecules cannot.
3.3.3 ATP concentration variation
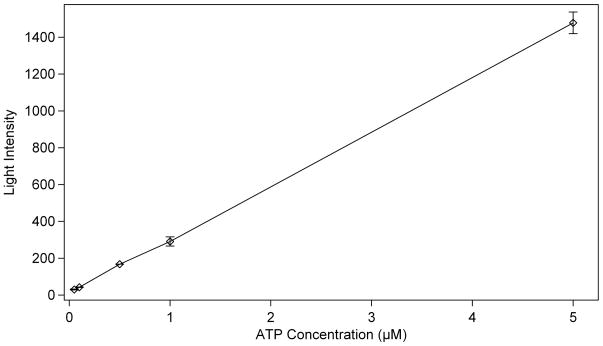
To determine the detection limits and response linearity of the reaction (Equation 3), a calibration curve was created by varying the concentration of ATP in the substrate solution. The reaction of ATP with luciferase represents the final reaction in the pyrosequencing process, producing the chemiluminescence used for detection. The enzyme concentration for these tests was held constant at 1.0 μg/μL luciferase, double the standard concentration used in the large volume devices in order to increase the initial light production. 400 ng/μL D-luciferin was added to each of the ATP solutions. Under the conditions described for the large scale device, an ATP concentration in the range of 10−1 μM represents the addition of a single base during pyrosequencing. The plot in Figure 7 shows the light intensity produced in the first 10 s of each reaction plotted versus the concentration of ATP. Concentrations are reported based on the amount of solute present in the droplet prior to combining the two droplets for the reaction. Intensity measurements were determined by taking an average of the cross-section of the droplet at its brightest point. The root mean square (rms) noise for each measurement was calculated, and the signal-to-noise ratio was determined for each reaction. The average rms noise was calculated to be 3.1 intensity units. From a plot of the signal-to-noise ratios, the limit of detection for these circumstances was calculated to be 7 nM ATP (S/N=5), which corresponds to the addition of a single nucleotide to 2 amol of DNA in a 300 pL droplet.
Figure 7.
Calibration curve of the intensity of light produced by the reaction of 1.0 μg/μL luciferase with a given concentration of ATP. All provided concentrations are prior to combining the two droplets to initiate the reaction. (n = 4)
3.3.4 Luciferase concentration variation
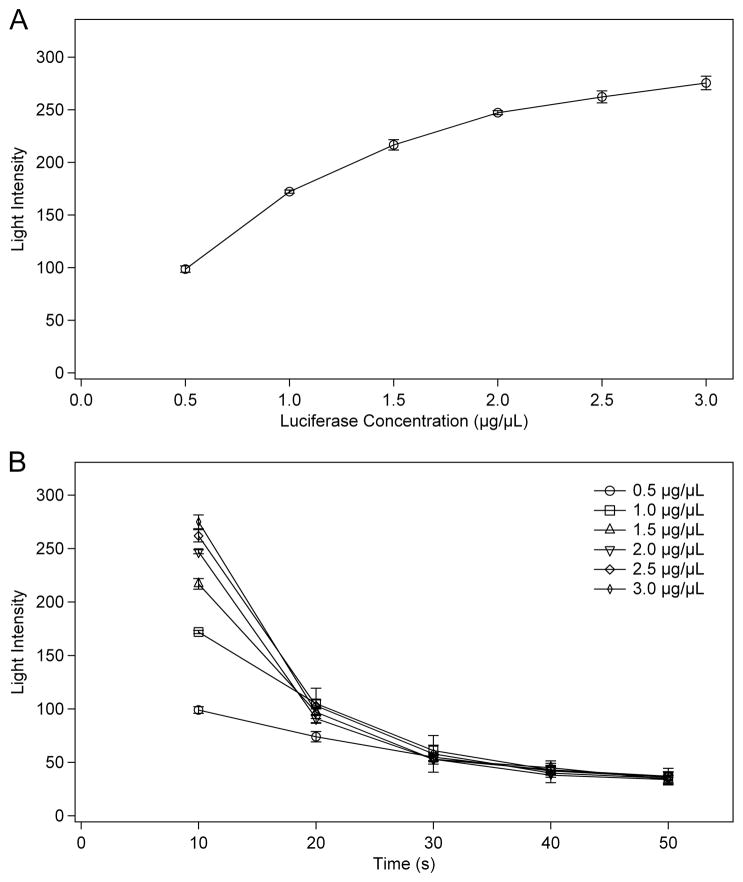
During initial testing of the chemiluminescence reaction for the ATP calibration curve, it was determined that the concentration of luciferase should be optimized to provide the maximum possible luminescence. Tests for larger volume devices were arranged such that the concentration of luciferase would control the rate of reaction, producing a long-term, relatively consistent emission of light. This was necessary so that any mixing or movement of the droplet would not prevent observation of a majority of the luminescence. However, as the light production scales directly with volume for a given concentration, the intensity was very low for the picoliter-scale device. In order to determine the timing and light production of the reaction on the smaller scale, a series of experiments were run with varying concentrations of luciferase. The concentration of ATP was 0.5 μM for all reactions, similar to that produced as a result of pyrosequencing on a larger electrowetting device. Figure 8 shows two plots of the luminescence intensity for reactions with luciferase concentrations between 0.5 μg/μL and 3.0 μg/μL. Figure 8A plots intensity versus luciferase concentration for the first 10 s of each reaction. In this plot, the intensity increases rapidly from 0.5 to 1.5 μg/μL luciferase, but begins to reach a maximum above 2.0 μg/μL luciferase. Figure 8B plots the same reactions as intensity versus time for all 5 concentrations. Since the amount of ATP is the same in each case, measuring the total light produced over the extent of the reaction would result in the same intensity for all reactions. The difference is how fast the complete reaction takes place, which is determined by how much luciferase is present relative to the amount of ATP. Higher concentrations of luciferase result in a brighter initial burst of light as much of the ATP is consumed at once, after which the intensity drops quickly. Conversely, lower concentrations of luciferase produce a “glow” reaction that is regulated primarily by diffusion and the release kinetics of the enzyme. For optimization of the reaction, higher concentrations of luciferase will provide better limits of detection. However, as discussed earlier, such high enzyme concentrations can be difficult to dispense in a regular fashion. During pyrosequencing reproducibility is vital, so luciferase concentrations between 1.0 and 2.0 μg/μL, high enough for good intensity and low enough for good dispensing, will be used.
Figure 8.
The light intensity of a reaction of 0.5 μM ATP with a given concentration of luciferase plotted versus (A) luciferase concentration or (B) time. All provided concentrations are prior to combining the two droplets to initiate the reaction. (n = 4)
4 Concluding Remarks
The preceding data and observations provide the groundwork for performing pyrosequencing on a picoliter-scale electrowetting device. Dispensing of solutions with different concentrations of microspheres was demonstrated, but the observed number of beads per droplet varies widely. Since the concentration of DNA that will be used for pyrosequencing depends on the concentration of beads to which it is bound, the intensity of light produced may vary widely under the current conditions. Use of magnets to create magnetic wells that control the concentration of beads dispensed has been successful, and the results are reproducible and improving. As a representation of the full pyrosequencing process, droplets containing ATP and luciferase have been combined and the resulting light intensity recorded. By varying the concentration of ATP in the substrate droplet, a detection limit of approximately 7 nM was calculated, which is well below the expected concentration for the addition of a single nucleotide in this device. Changing the concentration of luciferase in the enzyme droplet was also useful in optimizing the reaction conditions. Higher concentrations of luciferase than those used in larger-scale devices resulted in a brighter signal, which is helpful in the picoliter-scale devices that inherently produce lower light levels. With these results and further developments in the immediate future, pyrosequencing of DNA on a picoliter-scale digital electrowetting device is well within sight.
Figure 3.
Statistical analysis of bead dispensing plotted (A) in the order of droplet dispensing and (B) as a histogram of beads per droplet. Thirty droplets of each suspension were dispensed.
Acknowledgments
This research was supported by grant # R01HG004354-01 from the National Institute of Health.
List of abbreviations
- APS
adenosine 5′-phosphosulfate
- ATP
adenosine triphosphate
- NOA
Norland Optical Adhesive
- PEDOT
PSS or PEDOT, poly(3,4-ethylenedioxythiophene) poly(styrenesulfonate)
- Ppi
pyrophosphate
- rms
root mean square
Footnotes
Conflict of interest statement
The authors have declared no conflict of interest.
References
- 1.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2009;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 2.Leamon JH, Rothberg JM. Cramming More Sequencing Reactions onto Microreactor Chips. Chem Rev. 2007;107:3367–3376. doi: 10.1021/cr068297s. [DOI] [PubMed] [Google Scholar]
- 3.Voelkerding KV, Dames SA, Durtschi JD. Next-Generation Sequencing: From Basic Research to Diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 4.Wheeler DA, Srinivasan M, Egholm M, Shen Y, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–877. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 5.Hyman ED. A New Method of Sequencing DNA. Anal Biochem. 1988;174:423–436. doi: 10.1016/0003-2697(88)90041-3. [DOI] [PubMed] [Google Scholar]
- 6.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-Time DNA Sequencing Using Detection of Pyrophosphate Release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 7.Ahmadian A, Ehn M, Hober S. Clin Chim Acta. 2006:83–94. doi: 10.1016/j.cccn.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 8.Diggle MA, Clarke SC. Pyrosequencing Sequence Typing at the Speed of Light. Mol Biotechnol. 2004;28:129–137. doi: 10.1385/mb:28:2:129. [DOI] [PubMed] [Google Scholar]
- 9.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronaghi M. Pyrosequencing Sheds Light on DNA Sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 11.Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, et al. Single-Nucleotide Polymorphism Analysis by Pyrosequencing. Anal Biochem. 2000;280:103–110. doi: 10.1006/abio.2000.4493. [DOI] [PubMed] [Google Scholar]
- 12.Fakhrai-Rad H, Pourmand N, Ronaghi M. Pyrosequencing: An Accurate Detection Platform for Single Nucleotide Polymorphisms. Hum Mutat. 2002;19:479–485. doi: 10.1002/humu.10078. [DOI] [PubMed] [Google Scholar]
- 13.Thomas RK, Nickerson E, Simons JF, Janne PA, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 14.Dowd SE, Sun Y, Secor PR, Rhoads DD, et al. Survey of bacterial diversity in chronic wounds using Pyrosequencing, DGGE, and full ribosome shotgun sequencing BMC Microbiol 20088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabriel C, Danzer M, Hackl C, Kopal G, et al. Rapid high-throughput human leukocyte antigen typing by massively parallel pyrosequencing for high-resolution allele identification. Hum Immunol. 2009;70:960–964. doi: 10.1016/j.humimm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou G, Kamahori M, Okano K, Harada K, Kambara H. Miniaturized pyrosequencer for DNA analysis with capillaries to deliver deoxynucleotides. Electrophoresis. 2001;22:3497–3504. doi: 10.1002/1522-2683(200109)22:16<3497::AID-ELPS3497>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 18.Russom A, Tooke N, Andersson H, Stemme G. Single nucleotide polymorphism analysis by allele-specific primer extension with real-time bioluminescence detection in a microfluidic device. J Chromatogr A. 2003;1014:37–45. doi: 10.1016/s0021-9673(03)01033-1. [DOI] [PubMed] [Google Scholar]
- 19.Russom A, Tooke N, Andersson H, Stemme G. Pyrosequencing in a Microfluidic Flow-Through Device. Anal Chem. 2005;77:7505–7511. doi: 10.1021/ac0507542. [DOI] [PubMed] [Google Scholar]
- 20.Margulies M, Egholm M, Altman WE, Attiya S, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollack MG, Fair RB. Electrowetting-based actuation of liquid droplets for microfluidic applications. Appl Phys Lett. 2000;77:1725–1726. [Google Scholar]
- 22.Fair RB. Digital microfluidics: is a true lab-on-a-chip possible? Microfluid Nanofluid. 2007;3:245–281. [Google Scholar]
- 23.Lin YY, Evans RD, Welch E, Hsu BN, et al. Actuators, B. 2010;105:465–470. doi: 10.1016/j.snb.2010.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren H, Fair RB, Pollack MG. Automated on-chip droplet dispensing with volume control by electro-wetting actuation and capacitance metering. Sens Actuators, B. 2004;98:319–327. [Google Scholar]
- 25.Paik P, Pamula VK, Fair RB. Rapid droplet mixers for digital microfluidic systems. Lab Chip. 2003;3:253–259. doi: 10.1039/b307628h. [DOI] [PubMed] [Google Scholar]
- 26.Cho SK, Moon H, Kim CJ. Creating, transporting, cutting, and merging liquid droplets by electrowetting-based actuation for digital microfluidiccircuits. J Microelectromech Syst. 2003;12:70–80. [Google Scholar]
- 27.Zhang T, Chakrabarty K, Fair RB. Behavioral Modeling and Performance Evaluation of Microelectrofluidics-Based PCR Systems Using SystemC. IEEE Transactions on Computer-aided Design of Integrated Circuits and Systems. 2004;23:843–858. [Google Scholar]
- 28.Srinivasan V, Pamula VK, Fair RB. An integrated digital microfluidic lab-on-a-chip for clinical diagnostics on human physiological fluids. Lab Chip. 2004;4:310–315. doi: 10.1039/b403341h. [DOI] [PubMed] [Google Scholar]
- 29.Moon H, Wheeler AR, Garrell RL, Loo JA, Kim CJC. An integrated digital microfluidic chip for multiplexed proteomic sample preparation and analysis by MALDI-MS. Lab Chip. 2006;6:1213–1219. doi: 10.1039/b601954d. [DOI] [PubMed] [Google Scholar]
- 30.Cho SK, Zhao Y, Kim CJC. Concentration and binary separation of micro particles for droplet-based digital microfluidics. Lab Chip. 2007;7:490–498. doi: 10.1039/b615665g. [DOI] [PubMed] [Google Scholar]
- 31.Su F, Chakrabarty K, Fair RB. Microfluidics-Based Biochips: Technology Issues, Implementation Platforms, and Design-Automation Challenges. IEEE Transactions on Computer-aided Design of Integrated Circuits and Systems. 2006;25:211–223. [Google Scholar]
- 32.Fair RB, Khlystov A, Tailor TD, Ivanov V, et al. Chemical and Biological Applications of Digital-Microfluidic Devices. IEEE Design & Test of Computers 2007. 2007 Jan-Feb;:10–24. [Google Scholar]
- 33.Luan L, Evans RD, Jokerst NM, Fair RB. Integrated Optical Sensor in a Digital Microfluidic Platform. IEEE Sensors Journal. 2008;8:628–635. [Google Scholar]
- 34.Shah GJ, Ohta AT, Chiou EPY, Wu MC, Kim CJC. EWOD-driven droplet microfluidic device integrated with optoelectronic tweezers as an automated platform for cellular isolation and analysis. Lab Chip. 2009;9:1732–1739. doi: 10.1039/b821508a. [DOI] [PubMed] [Google Scholar]
- 35.Thwar PK, Eckhardt AE, Rouse JL, Fair RB, et al. unpublished work. [Google Scholar]
- 36.Sista RS, Eckhardt AE, Srinivasan V, Pollack MG, et al. Heterogeneous immunoassays using magnetic beads on a digital microfluidic platform. Lab Chip. 2008;8:2188–2196. doi: 10.1039/b807855f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beyor N, Seo TS, Liu P, Mathies RA. Immunomagnetic bead-based cell concentration microdevice for dilute pathogen detection. Biomed Microdevices. 2008;10:909–917. doi: 10.1007/s10544-008-9206-3. [DOI] [PubMed] [Google Scholar]
- 38.Huang H, Zheng XL, Zheng JS, Pan J, Pu XY. Rapid analysis of alpha-fetoprotein by chemiluminescence microfluidic immunoassay system based on super-paramagnetic microbeads. Biomed Microdevices. 2009;11:213–216. doi: 10.1007/s10544-008-9226-z. [DOI] [PubMed] [Google Scholar]
- 39.Dupont EP, Luisier R, Gijs MAM. NOA 63 as a UV-curable material for fabrication of microfluidic channels with native hydrophilicity. Microelectron Eng. 2010;87:1253–1255. [Google Scholar]
- 40.Rong R, Choi JW, Ahn CH. An on-chip magnetic bead separator for biocell sorting. J Micromech Microeng. 2006;16:2783–2790. [Google Scholar]
- 41.Lee H, Purdon AM, Westervelt RM. Manipulation of biological cells using a microelectromagnet matrix. Appl Phys Lett. 2004;85:1063–1065. [Google Scholar]
- 42.Lee H, Purdon AM, Chu V, Westervelt RM. Controlled Assembly of Magnetic Nanoparticles from Magnetotactic Bacteria Using Microelectromagnets Arrays. Nano Lett. 2004;4:995–998. [Google Scholar]
- 43.Lee H, Liu Y, Hamb D, Westervelt RM. Integrated cell manipulation system—CMOS/microfluidic hybrid. Lab Chip. 2007;7:331–337. doi: 10.1039/b700373k. [DOI] [PubMed] [Google Scholar]
- 44.Shah GJ, Kim CJC. Meniscus-Assisted High-Efficiency Magnetic Collection and Separation for EWOD Droplet Microfluidics. J Microelectromech Syst. 2009;18:363–375. [Google Scholar]
- 45.Wang Y, Zhao Y, Cho SK. Efficient in-droplet separation of magnetic particles for digital microfluidics. J Micromech Microeng. 2007;17:2148–2156. [Google Scholar]