Abstract
RMI1/BLAP75 (RecQ-Mediated Genome Instability 1/Bloom Associated protein 75) is an OB-fold protein highly conserved from yeast to human. Previous studies showed that RMI1 is required for the stability of the BLM/RMI1/TopIIIα complex and for the suppression of elevated sister chromatids exchange (SCE). The presence of RMI1 strongly stimulates the Holliday dissolution activity of the Bloom helicase in vitro. The in vivo function of RMI1, however, remains largely undefined. To address this question, we generated RMI1 knockout mice through homologous replacement targeting. We found that, while RMI1+/− mice showed no obvious developmental phenotype, deletion of both mRMI1 alleles resulted in early embryonic lethality before implantation. To determine whether RMI1 plays a role in tumorigenesis, we generated RMI1/p53 double heterozygous mice and analyzed their onset of ionizing radiation-induced tumor development. RMI1+/−/p53+/− mice succumbed to tumor with a higher frequency and exhibited a substantially shortened survival when compared to the wild type, RMI1+/− and p53+/− cohorts. These results demonstrated a dual-role of RMI1 in embryonic development and tumor suppression.
Keywords: Tumor development, Radiation, RMI1, Bloom Syndrome, genomic instability
Introduction
Bloom syndrome (BS) is a rare autosomal recessive disorder featured by sun sensitivity, male infertility, immune deficiency, and predisposition of a broad spectrum of cancers [1]. BS is caused by defects in the BLM helicase gene, a member of the RecQ helicase family conserved from bacteria to human [2]. Cells derived from BS patients are characterized by drastically elevated Sister Chromatin Exchange (SCE) level, which indicates an important function of BLM in controlling homologous recombinations between sister chromatids [3]. BLM is also suggested to be involved in the activation of S-phase checkpoint under replication stress [4,5], and sister chromatid decatenation in anaphase [6]. Biochemical and genetic studies showed that BLM is able to prevent strand crossing caused during repair of DNA double strand breaks (DSB). Upon formation of a double Holliday junction (HJ) intermediate during DSB repair, the BLM/RMI1/TopIIIα complex acts to facilitate the migration and dissolution of HJs in a non-crossover manner [7,8].
RMI1/BLAP75 is an OB-fold-containing protein identified as a BLM-interacting protein through biochemical purification [9]. RMI1 forms an evolutionarily conserved complex (BTB complex) with BLM and Topo3. Mutations in the RMI1 gene in yeast result in genomic instability and elevated SCE [9–11]. This complex is involved in the BLM-dependent dissolution of double Holliday junctions during homologous recombination [11–14]. Recently, RMI2, an additional component of the BLM-RMI1-Topo3α complex, was identified [11,15]. RMI1 interacts with RMI2 through its C-terminal OB2 (RPA1-C) domain, whereas it binds to BLM and Topo3α through its N-terminal region, which contains the OB1 (RecG) domain [11].
RMI1 is required for the BTB complex stability. Knocking down of RMI1 diminished protein levels of other components of the complex, especially Topo3α and RMI2 [9,11]. It is also involved in BLM function. Depletion of RMI1 led to increased SCE level similar to BLM knockdown [9]. Biochemical studies indicated that RMI1 regulates BTB complex activity. RMI1 stimulates branch migration of BLM protein on single HJs, promotes DNA-binding and catalytic activity of Topo3α, and facilitate BLM-Topo3α complex in double HJ dissolution [11,13,15,16].
Knockout mouse strains of BLM and Topo3α have been established. Complete disruption of either gene leads to embryonic lethality [17–19] [20]. Whereas Topo3α mice exhibited no haploinsufficiency, BLM+/− mice die early than wild type controls after murine leukemia virus (MLV) injection, and develop enhanced gastrointestinal tumor formation when crossed with APC+/− mice [17]. To explore the function of RMI1 in vivo and its potential impact on the maintenance of genomic stability and tumorigenesis, we generated RMI1 knockout mice via homologous targeting. We show that the RMI1 gene is required for early embryonic development as embryos die before implantation. Moreover, RMI1+/−/p53+/− double-heterozygous mice developed a phenotype of accelerated tumor formation when mice were exposed to ionizing radiation. These results indicate that RMI1 function in both early embryonic development and tumor suppression.
Material and Methods
Generation of RMI1 +/− mice
The mouse RMI1 gene is located on chromosome 13 (spanning from 57519450 to 57528410). A full-length cDNA sequence (BC037694, purchased from MGC) was used as a probe to screen a mouse BAC library RPCI-22 generated by Pieter De Jong. Three BAC clones (444B20, 369A5, and 444P22) were identified and further verified for containing undisrupted RMI1 genomic sequences by Southern blotting. To generate the knockout construct, a 4.5kb fragment upstream of RMI1 exon3 released by BamHI digesting was cloned into pBS-PGK-neo-bpA-LoxP vector [21] (a kind gift from Richard Behringer, MDACC). A 1.7kb fragment down stream of exon 3 served as the short arm was cloned by PCR with embedded Pvu II site at 5′, and ligated into the pMCI-TK-pA vector [22]. The final targeting vector was constructed by joining the 5′ arm-Neo and the 3′ arm-TK fragments. Transfection of ES (129 G4 and TC1) cells was carried out by the Genetically Engineered Mouse Facility (GEMF) at MDACC. RMI1+/− ES cell clones were selected for blastocyst injection to produce chimeras. These mice were then backcrossed with c57/BL6 for 5 generations.
Southern blot genotyping for ES cells
G418-resistant ES cell colonies were isolated and propagated in 96-well plates. Two probes, Probe1 and Probe3, were used in Southern blot genotyping from either 5′ or 3′ end, respectively. For 5′ end, Probe1 was amplified by PCR (primer 1A, 5′CCGCCTTTGGTCGTGACTGACAAC; primer1B, 5′GCTCGGCGGACCTGTTAACACCAG). Genomic DNA was digested with NheI, and transferred to membrane before hybridized with p32-labeled probe1 at 63°C overnight. The membrane was washed twice before exposed to film. For the 3′ end, probe3 was PCR-amplified (probe3A, 5′CTGTGGCTAGAAAGATGGTTCAGC; probe3, 5′GAGGGCATGGCCCAGACTTG), labeled, and hybridized overnight at 65°C with PvuII digested genomic DNA.
PCR genotyping for mice and embryos
Genomic DNA was extracted from tail or toe snips. Tissues were first incubated with 250uL lysis buffer (10 mM Tris-HCl pH8.0, 100mM NaCl, 2.5mM EDTA, 0.5%SDS) and 10uL Proteinase K (20ug/mL) at 55°C overnight. Then, proteins were removed by 100uL tail salt buffer (4.21M NaCl, 0.63M KCl, 10mMTrisHCl PH8.0). Genomic DNA then was precipitated by 400 μl EtOH and washed by 70% EtOH before dissolved in 200uL ddH2O. A PCR based RMI1 genotyping procedure was performed. Following primers were used: LWT3A, 5′ CTTTAAGTATCGCCCTCCGTTTTG; LWT3B, 5′CTTAACATGCCATGTTGCCAAAAG; NS3A, 5′ AGCAAGGGGGAGGATTGGGAAGACA; NS3B, 5′GCGGAGCTGCCCCAGCTTTAAGCT. Three primers were used in p53 genotyping, all were obtained from genetically Engineered Mouse Facility at MDACC: X6, 5′AGCGTGGTGGTACCTTATGAGC; 7, 5′GGATGGTGGTATACTCAGAGCC; neo19, 5′GCTATCAGGACATAGCGTTGGC. E3.5 embryos were flushed out of the uterus using the PBS buffer and genotyped by a nested PCR strategy. First round PCR primers were described below: LWT1A, 5′CTCATCCCAGAGTAAGGTGGCCGACTAT; LWT1B, 5′CACAAGCTTCCAGCCACATTGGAGGTAC; NS2A, 5′TTCGCAGCGCATCGCCTTCTATCG; NS2B, 5′AGCAGGGTCTCGCCTTTTGCCTCGAGAG. LWT3 and NS3 primer pairs were used in the second round of PCR. Statistical analyses for the P values were carried by Person’s Chi square test.
Cell culture and Clonogenic survival assay
HCT116 strains were maintained in 10% fetal bovine serum and grown in a humidified 5% CO2-incubator at 37°C. RMI1 shRNA transfection and clonogenic assay were performed as described previously [9]. Briefly, cells were transiently transfected with either RMI1 shRNA or control luciferase shRNA expressed from a NeoR and U6 promoter-driven vector [23], then counted and seeded onto 100 mm plates in the presence of G418. Colonies were allowed to grow for 2 weeks before counting.
Ionizing radiation treatment and tumor analyses
Littermates from RMI1+/−p53+/− intercrossing were subjected for IR irradiation at 4 Gy (Nasatron) at 6 weeks of age, followed by daily observation for at least 1 year. Mice with visible tumor phenotype or morbidity were sacrificed by CO2 asphyxiation. Tissue samples were preserved in 10% formalin and embedded in paraffin cassettes prior to slide cutting and staining with hematoxylin and eosin (H&E). Survival rate was estimated by the Kaplan-Meier method. P-value was derived from the log-rank test embedded in the GraphPad Prism package.
Results
Generation of an RMI1 Knockout Mouse Model
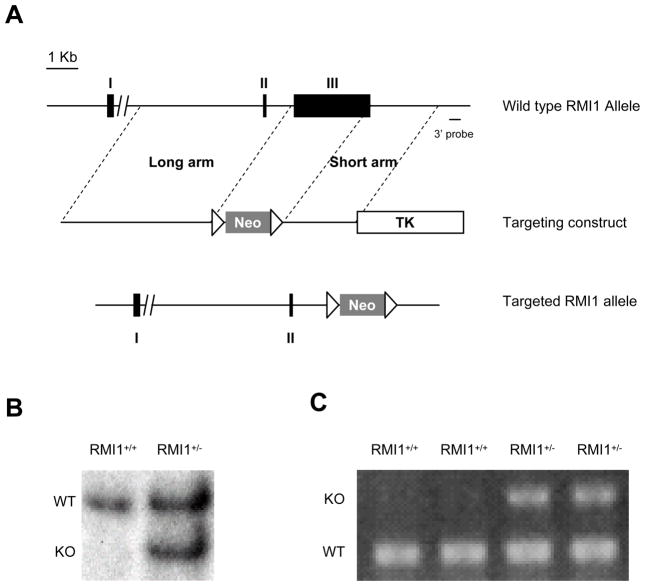
To study the physiological function of RMI1 in vivo, we generated a knockout mouse strain of RMI1 by replacement targeting of exon 3, which contains the entire RMI1 coding sequence. RMI1 exon 3 was replaced by a Cre-floxed neomycin selective cassette to create a null-allele (Fig. 1A). ES cells carrying the targeted allele were selected by southern blot using probes located both 5′ and 3′ to the targeted region, respectively (Fig. 1B). RMI1+/− ES cells were used to generate chimeras through standard blastocyst injection. Germline transmission was confirmed by PCR genotyping (Fig. 1C). Mice heterozygous for RMI1 were obtained by backcrossing the chimeras with C57BL/6 strain for 5 generations
Figure 1.
Targeting of the RIM1 locus. A. RMI1 replacement targeting strategy. Solid boxes depict the three RMI1 exons (I, II, and III) with exon III being the only coding exon. Neo: Neomycin selection marker (positive marker). Open triangles: CreLoxP sites. TK: TK cassette (negative selection marker). B. Southern blot of Pvu II-digested RMI1+/+ and RMI1+/− ES cells with the 3′ probe. WT: wild type allele. KO: Targeted allele. C. typical PCR genotyping results of RMI1+/+ and RMI1+/− mice using genomic DNA extracted from tail clips.
RMI1−/− embryos failed to pass implantation stage
Mice with a RMI1+/− genotype were healthy and fertile throughout the duration of this study (24 months). Long-term observation showed no sign of shortened life span or increased spontaneous tumor formation. However, crossing between RMI1+/− mice yielded no RMI1−/− offspring, which indicates that deletion of both RMI1 alleles leads to embryonic lethality. To further investigate this, we performed embryo dissection at various stages of development. Initially, genotyping of embryos from RMI1+/− intercrossing recovered neither RMI1−/− embryo nor absorbed residuals between 8.5 to 13.5 day post coitum, an indication that RMI1−/− embryos may die at an early stage of embryonic development. Subsequently, we harvested blastocysts at 3.5 dpc. Again, neither RMI1−/− nor abnormal blastocysts were present in the blastocyst stage (Table 1). On the other hand, RMI1 wild type and heterozygous embryos were recovered at or near Mendelian ratio. Collectively, these results suggest that RMI1 is essential for early embryonic development and viability. Homozygous deletion of RMI1 most likely leads to pre-implantation lethality.
Table 1.
Genotype analyses of Rmi1+/− intercross. Embryos were harvested from day E8.5 to E10.5
| Date | RMI+/+ | RMI+/− | RMI−/− |
|---|---|---|---|
| E3.5 | 7 | 11 | 0 |
| E8.5 | 11 | 18 | 0 |
| E9.5 | 6 | 12 | 0 |
| E10.5 | 2 | 6 | 0 |
| Combined | 19 | 36 | 0 |
p53 deletion alleviated cellular lethality caused by RMI1 loss, but has no detectable effect on embryonic lethality
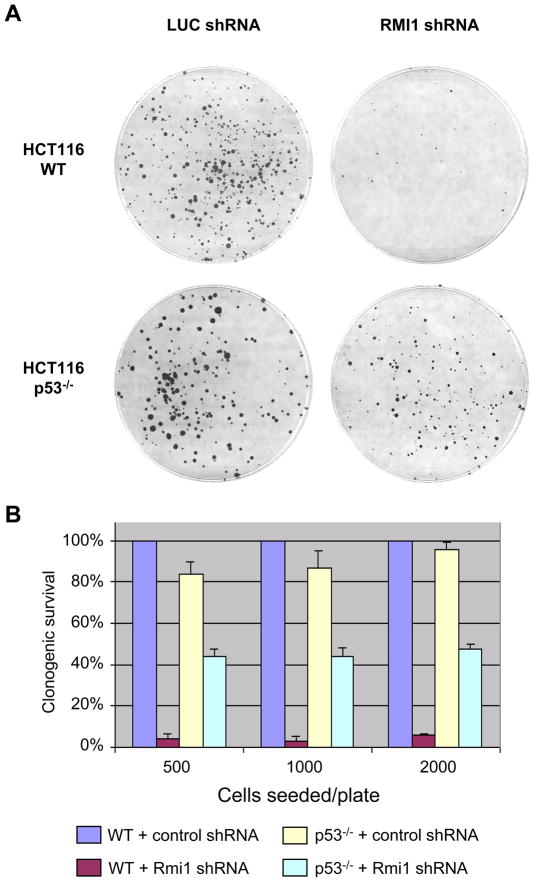
The early embryo lethality may reflect that RMI1 function is critical in the maintenance of genomic integrity in the context of the BLM/RMI1/TopIIIα complex. Previously, we have shown that cells with reduced RMI1 level led to the disruption of the BLM/RMI1/TopIIIα complex and display elevated SCE similar to BLM mutant cells. Complete knockdown of RMI1 resulted in severe inhibition of cell proliferation in tissue culture cells [9]. To determine whether increased genomic instability is a primary cause of cellular lethality, we tested whether p53 deficiency could rescue the cellular lethality of RMI1 knockdown cells. We transfected RMI1 and control (luciferase) shRNA vectors into isogenic p53+/+ and p53−/− HCT116 cells derived from somatic cellular targeting [24]. Cells were grown in G418-containing media to select for the presence of shRNA vectors in the clonogenic assay
As shown in Figure 2, p53+/+ cells transfected with RMI1 shRNA exhibited a drastic reduction in clonogenic survival compared with cells transfected with control shRNA, a result consistent with our previous observations [9]. In contrast, clonogenic survival increased markedly in p53−/− cells. This result indicates that RMI1 loss most likely compromises the integrity of genomic DNA and leads to cell growth inhibition, which can be alleviated by loss of the p53-dependent surveillance mechanisms.
Figure 2.
Clonogenic survival assay of HCT116 cells subjected to RMI1 shRNA expression. A. Colony formation in parental or p53−/− HCT116 cells transfected with RMI1 shRNA or control (Luciferase) shRNA. B. Colony numbers from each group were counted and normalized against the parental HCT116 transfected with control shRNA. Error bars were derived from three independent experiments.
Given that p53 deficiency was able to mitigate the cellular lethality caused by RMI1 loss, we asked whether the early mouse embryonic lethality could be similarly alleviated in a p53−/− background. We performed intercross between double heterozygous (RMI1+/−/p53+/−) male and female mice and collected 47 blastocysts for RMI1 genotyping. Of the 47 blastocysts, 19 were found to be wild type (RMI1+/+) and 28 were heterozygous (RMI1+/−). This result indicates that p53 deletion is unlikely sufficient in extending the viability of RMI1−/− embryos beyond the implantation stage (P<0.05).
Double heterozygosity of RMI1 and p53 led to shortened half life of radiation-induced foci
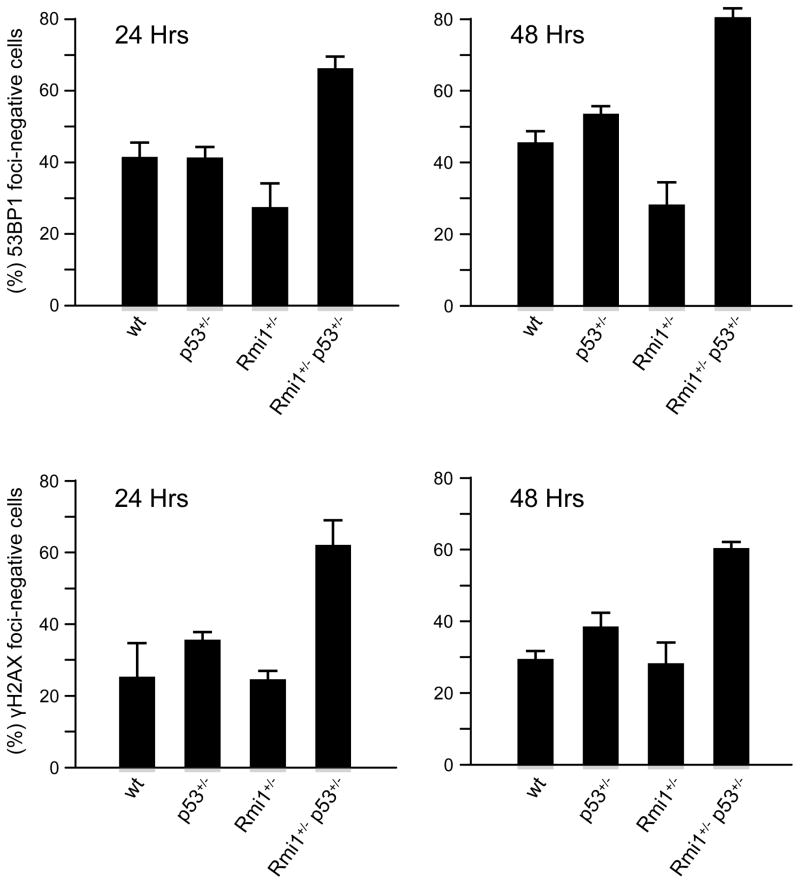
The Bloom helicase has been suggested to play a role in preventing genomic instability by promoting dissolution of double Holliday junctions during DNA double strand break repair [12,25]. Thus, interruption of the BLM/RMI1/Top3α complex function likely impacts radiation-induced DSB repair, and such impact may be exaggerated by p53 heterozygosity. We therefore measured the ionizing radiation-induced nuclear focus formation in MEF cells derived from litter-mates with the following genotypes, wt, RMI1+/+/p53+/−, RMI1+/−/p53+/+, and RMI1+/−/p53+/−.
Each MEF cell culture was subjected to 1.5 Gy of ionizing radiation, the nuclear focus formation of γ-H2AX and 53BP1, two surrogate markers indicative of DSB processing, was scored at different time points to monitor the processing of DSBs. The initial onset of foci formation, as detected 30 minutes post IR, was similar among MEF cells with all four genotypes (data not shown). At 24 and 48 hours post IR (Figure 3), RMI1+/−/p53+/+ MEF cells showed similar percentage of foci negative cells as the wt cells. The RMI1+/+/p53+/− genotype also did not affect the loss of foci as previously reported [26]. However, double heterozygous RMI1+/−/p53+/− MEF cells showed much reduced foci number for both 53BP1 and γH2AX compared to wt, RMI1+/+/p53+/−, and RMI1+/−/p53+/+ MEF cells, as indicated by the percentages foci-negative cells. This result suggest that double heterozygosity in RMI1 and p53 impacted the processing of DSBs and is likely give rise to genomic instability.
Figure 3.
IR-induced foci exhibit shortened half life in RMI1+/− p53+/− MEF cells. MEF cells generated from litter mates were exposed to 1.5 Gy of gamma radiation. Cells were fixed at 0, 6, 24 and 48 hours and immunohistochemistry was performed to visualize and monitor the disappearance of 53BP1 and γH2AX foci. Foci-negative cells are defined by less than 2 foci per nuclei for 53BP1 and less than 5 foci per nuclei for γH2AX, respectively. Data points were derived from three independent experiments with duplicated samples. Error-bars represented standard deviations.
RMI1 heterozygosity enhanced radiation-induced tumor development in p53+/− mice
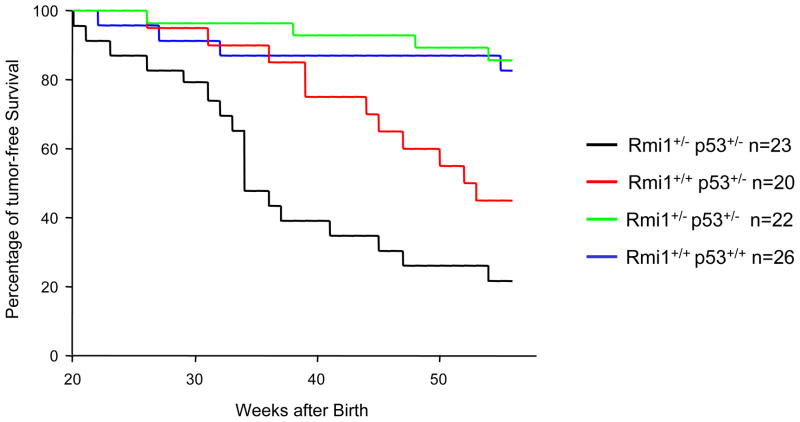
Since p53 deletion alleviates cellular lethality caused by RMI1 depletion and p53+/− heterozygosity facilitates IR-induced foci loss in RMI1+/− cells, we reasoned that reduced p53-dependent surveillance, in the form of p53 heterozygosity, may amplify the radiation-induced tumor development in RMI1+/− mice. To test this hypothesis, four cohorts of mice, RMI1+/−/p53+/−, RMI1+/−/p53+/+, RMI1+/+/p53+/−, and RMI1+/+/p53+/+, were exposed to 4 Gy of gamma radiation at the age of 6 weeks. Tumor development was monitored for 54 weeks after irradiation following standard observation procedures (supplementary material). Mice developed visible tumors or suffered from morbidity were sacrificed and examined for gross and histologic analyses. All mice were euthanized at the conclusion of the tumor monitoring.
As shown in Figure 4 and Table 2, RMI1+/−/p53+/+ mice displayed a tumorigenesis rate (23 %, 5/22 mice) similar to that of the RMI1+/+/p53+/+ wild type controls (31 %, 8/26), suggesting that loss of one RMI1 allele alone does not lead to a significant increase in radiation-induced tumorigenesis. In addition, the 50% survival periods of both the WT and RMI1+/− mice are beyond 55 weeks. Mice in the RMI1+/+/p53+/− control cohort more frequently succumbed to tumor (50%, 10/20 mice or 13 tumors in 20 mice) with an average tumor free rate of 45 weeks. Importantly, RMI1+/−/p53+/− double heterozygous mice exhibited a much higher tumor incidence (74%, 17/23 mice or 21 tumors in 23 mice) and a markedly decreased survival (average 27 weeks). The difference of average tumor free time between the RMI1+/−/p53+/− and RMI1+/+/p53+/− cohorts is statistically significant (P<0.02). Importantly, mice in the RMI1+/−/p53+/− group died of tumor much sooner (52% within 35 weeks) than the control groups (10% in p53+/−, 14% in the RMI1+/− and 4% in the WT groups, respectively). These results clearly indicate a cooperation of p53 and RMI1 deficiency.
Figure 4.
Exacerbation of IR-induced tumorigenesis in RMI1+/− p53+/− mice. Four cohorts with the indicated genotypes were exposed to 4 Gy of ionizing radiation at the age of 6 weeks. “n” indicates the final size of each cohort. Tumor-free survival was monitored for 54 weeks.
Table 2.
Tumor spectrum in various RMI1 heterozygous backgrounds. Mice with no obvious cause of death or death due to fight are listed as unknown. Four BP, three P and two WT mice showed two tumor types simultaneously
| Cohort size | Number of death | Number of death within 35 wks | lymphoma | sarcoma | carcinoma | Unknown | |
|---|---|---|---|---|---|---|---|
| RMI1+/−p53+/− | 23 | 19 | 12 | 12 | 5 | 4 | 1 |
| RMI1+/+p53+/− | 20 | 15 | 2 | 7 | 3 | 3 | 5 |
| RMI1+/−p53+/+ | 22 | 7 | 3 | 3 | 2 | 0 | 2 |
| RMI1+/+p53+/+ | 26 | 9 | 1 | 8 | 2 | 0 | 1 |
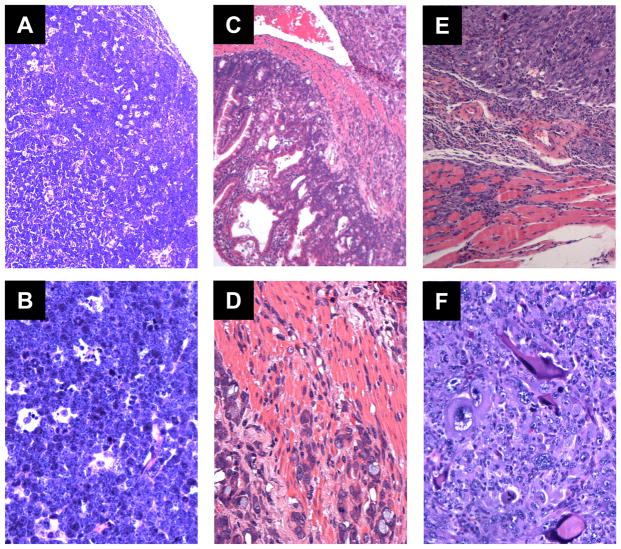
Lymphoma is known to be the primary tumor developed from IR irradiated p53+/− mice. The percentage of lymphoma incidences could vary from 48% to 90%, depending on different mouse inbred strains [27–30]. In the current study, we observed a similar trend. Tumors derived from p53+/− group consisted mainly of lymphomas (47%, 7/15), more than the combined percentage of carcinomas and sarcomas. Similarly, RMI1+/−p53+/− mice frequently developed lymphoma (57%, 12/23). Of note, four of the RMI1+/−/p53+/− mice and three of the p53+/− mice developed sarcoma or carcinoma in addition to lymphoma. In comparison to the control groups, RMI1+/−p53+/− mice seemed much more prone to aggressive tumor types, including high grade lymphoma, invasive carcinoma and invasive osteosarcoma (Figure 5). On the H & E sections, there is effacement of nodal architecture and lymphocytic infiltrate composed of intermediated to large sized lymphoid cells with distinct nuclei, brisk mitotic figures and frequent apoptotic debris inside histiocytes (Figure 5A and B). Figure 5C and D show small intestinal neoplasm, ranging from epithelial dysplasia to predominantly aggressive adenocarcinoma, infiltrating deeply into the intestinal wall. Invasive osteosarcoma is observed in a femur bone and infiltrating into adjacent skeletal muscle (Figure 5E and F). Frequent pleomorphic neoplastic cells are present, which frequently produce osteoid. Moreover, IR irradiated RMI1+/− or WT mice also developed lymphoma and sarcoma. However, the average survival rates from either RMI1+/− or WT groups were much longer than that of p53+/− or RMI1+−/p53+/− mice. Taking together, these results suggest RMI1 cooperate with p53 in suppressing tumor formation.
Figure 5.
Aggressive tumors derived from the RMI1+/− p53+/− mice, H & E stains. A and B, High grade lymphoma involving a lymph node (A, 100× ; B, 400× ). C and D, Aggressive and invasive adenocarcinoma of small intestine (C, 100× ; D, 400× ). E and F, Aggressive and invasive osteosarcoma involving skeletal muscle (E, 100× ; F, 400× ).
Discussion
The mouse model constructed in this study allowed us to demonstrate that RMI1 is essential for early embryo viability and attenuation of radiation induced tumor formation in p53 heterozygous background. These observations provide the first evidence in an animal model that RMI1 plays a role in the maintenance of genomic stability.
Studies by Chester et al showed that complete inactivation of the BLM loci resulted in embryonic growth retardation and lethality at day 13.5 [18]. Compared to BLM−/− mice, embryonic lethality of RMI1 homozygous mice occurred at a much earlier stage since no RMI1−/− blastocysts could be recovered. This phenotype is reminiscent of the Topo3α mice which exhibit a pre-implantation embryonic lethality [19]. Several factors could contribute to the early death of RMI1−/− embryos. RMI1 is found in two independent complexes, one consists of BLM, RMI1, RMI2, and Topo3α, the other contains RMI1, RMI2, and Topo3α without the Bloom helicase [11]. The presence of the second RMI1-containing complexes suggests that these OB-fold proteins may play an important role in Topo3α function, which is believed to carry out chromosome separation during mitosis. Thus it is plausible that the lethality of RMI1−/− embryos could arise from mitotic dysfunction. This notion is supported by studies of yeast Topo3 mutants, which displayed reduced sporulation and ineffective chromosome separations during mitosis [31–34].
Our previous studies indicated that the entire RMI1-RMI2 module is important for the integrity of the BLM-RMI1-RMI2-Topo3α complex and the stability of other components in this complex. Loss of RMI1 leads to significant reductions of BLM and Topo3α at the protein level [9,11]. Therefore, it is likely that RMI1−/− mouse embryos have insufficient levels of Topo3α and BLM. This would implicate that the resulting embryonic lethality could also reflect the compounded outcome of both functional and physical deficits of Topo3α and BLM.
We showed that the cellular lethality of RMI1 knockdown cells could be significantly alleviated by the absence of p53. This genetic interaction served as a key rationale for examining tumor development in RMI1+/−/p53+/− mice. It also prompted us to examine whether loss of p53 could rescue the embryonic lethality. Multiple attempts at isolating RMI1−/−/p53−/− embryos failed to yield any surviving blastocysts from intercrossing between RMI1+/−/p53+/− mice (P<0.05). Thus, it appears that the embryonic lethality could not be effectively mitigated by the loss of p53, further suggesting that cells deprived of RMI1 may have severely compromised genomic integrity and/or cell cycle transition difficulties. It also suggests that RMI1 function is more strictly required during embryonic development than in cultured cells.
We found that radiation-induced foci were resolved with an accelerated rate in RMI1+/−/p53+/− double heterozygous cells. This observation is unlikely attributed to cell death as apoptotic analysis showed no significant difference among the 4 different MEF cells when treated with the same dose of IR (1.5 Gy) as in the foci staining experiment. One plausible mechanism for the reduced foci is that the intermediate joint molecules during DSB repair is resolved more readily due to the compromised BLM/RMI1/TopIIIα complex function, which normally suppresses cross over resolution of Holliday junctions. This may in turn causes elevated translocations and gives rise to genomic instability. However, the exact role of p53 in promoting this process is unclear.
Our tumorigenesis study demonstrates that loss of one RMI1 allele exerts a significant impact on radiation-induced tumorigenesis in a p53+/− background, whereas RMI1 heterozygosity alone did not show a detectable enhancement of radiation-induced tumor development or progression. In particular, the median tumor-free survive age is 35 weeks from RMI1+/−p53+/− mice, comparing to 53 weeks from p53+/− Despite the earlier onset, the tumor spectrum of RMI1+/−p53+/− mice showed no major alteration from that of the p53+/− mice. This result suggests a recessive haploinsufficiency of RMI1 as a tumor suppressor, which impacts tumor development when coupled with p53 heterozygosity or other prevailing genetic backgrounds. Most likely, the reduced function of the BLM/RMI1/TopoIIIα complex creates a low level of genetic instability which is augmented by the partial loss of p53 function. Consistently, SNPs in the RMI1 gene and the BLM/RMI1/TopIIIα complex have been linked to the elevated risks of several malignancies [35,36]. However, the exact molecular mechanism of the RMI1-p53 interaction depends on further delineation of the molecular nature of genomic instability arisen from BLM/RMI1/TopIIIα defects. Recently, RMI1 heterozygosity has been found to be sensitive to high-fat diet-induced obesity [37]. But the link between RMI1 and metabolic control remains to be established.
Pathological analyses revealed a trend that RMI1+/−p53+/− mice were more susceptible to aggressive forms of lymphoma, osteosarcoma and adenocarcinoma (Table 2). This observation suggests that RMI1 heterozygosity may facilitate the progression of malignancies in additional to promoting early tumor onset. However, additional studies with larger cohort sizes are required to firmly establish this notion. Collectively, our results indicate a dual-role of RMI1 in vivo, its essentiality for early embryo viability and a potential tumor suppressor.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Gigi Lozano (Department of Genetics, UT MDACC) for critical suggestions to this work. The GEF Core facility provided ES cell selection and blastocyst injection services for the generation of the knockout mice. This work was supported, in whole or in part, by grants from the National Cancer Institute CA127945 and CA097175 (to L. L) and UT MDACC Physician Scientist Award, American Cancer Society IRG, UT MDACC IRG and Ladies Leukemia League (to M. J.Y). This work was also supported in part by the Intramural Research Program of the National Institute on Aging (Z01 AG000657-08), National Institute of Health (to W.W.)
References
- 1.German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72(6):393–406. [PubMed] [Google Scholar]
- 2.Ellis NA, Groden J, Ye TZ, et al. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83(4):655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 3.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc Natl Acad Sci U S A. 1974;71(11):4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davalos AR, Kaminker P, Hansen RK, Campisi J. ATR and ATM-dependent movement of BLM helicase during replication stress ensures optimal ATM activation and 53BP1 focus formation. Cell Cycle. 2004;3(12):1579–1586. doi: 10.4161/cc.3.12.1286. [DOI] [PubMed] [Google Scholar]
- 5.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. Embo J. 2003;22(16):4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. Embo J. 2007;26(14):3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci U S A. 2000;97(12):6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39(47):14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 9.Yin J, Sobeck A, Xu C, et al. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. Embo J. 2005;24(7):1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu L, Davies SL, North PS, et al. The Bloom’s syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275(13):9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 11.Xu D, Guo R, Sobeck A, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22(20):2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115(4):401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Bachrati CZ, Ou J, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103(11):4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L, Chan KL, Ralf C, et al. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. Embo J. 2005;24(14):2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh TR, Ali AM, Busygina V, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22(20):2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. J Biol Chem. 2007;282(43):31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- 17.Goss KH, Risinger MA, Kordich JJ, et al. Enhanced tumor formation in mice heterozygous for Blm mutation. Science. 2002;297(5589):2051–2053. doi: 10.1126/science.1074340. [DOI] [PubMed] [Google Scholar]
- 18.Chester N, Kuo F, Kozak C, O’Hara CD, Leder P. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom’s syndrome gene. Genes Dev. 1998;12(21):3382–3393. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, Wang JC. Mammalian DNA topoisomerase IIIalpha is essential in early embryogenesis. Proc Natl Acad Sci U S A. 1998;95(3):1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo G, Santoro IM, McDaniel LD, et al. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26(4):424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 21.Wakamiya M, Lindsay EA, Rivera-Perez JA, Baldini A, Behringer RR. Functional analysis of Gscl in the pathogenesis of the DiGeorge and velocardiofacial syndromes. Hum Mol Genet. 1998;7(12):1835–1840. doi: 10.1093/hmg/7.12.1835. [DOI] [PubMed] [Google Scholar]
- 22.Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336(6197):348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 23.Bao S, Lu T, Wang X, et al. Disruption of the Rad9/Rad1/Hus1 (9–1–1) complex leads to checkpoint signaling and replication defects. Oncogene. 2004;23(33):5586–5593. doi: 10.1038/sj.onc.1207753. [DOI] [PubMed] [Google Scholar]
- 24.Bunz F, Dutriaux A, Lengauer C, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 25.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426(6968):870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 26.Goodarzi AA, Noon AT, Deckbar D, et al. ATM Signaling Facilitates Repair of DNA Double-Strand Breaks Associated with Heterochromatin. Molecular Cell. 2008;31(2):167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 27.Backlund MG, Trasti SL, Backlund DC, Cressman VL, Godfrey V, Koller BH. Impact of ionizing radiation and genetic background on mammary tumorigenesis in p53-deficient mice. Cancer Res. 2001;61(17):6577–6582. [PubMed] [Google Scholar]
- 28.Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet. 1994;8(1):66–69. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- 29.Mori N, Yamate J, Umesako S, Hong DP, Okumoto M, Nakao R. Preferential induction of mammary tumors in p53 hemizygous BALB/c mice by fractionated irradiation of a sub-lethal dose of X-rays. J Radiat Res (Tokyo) 2003;44(3):249–254. doi: 10.1269/jrr.44.249. [DOI] [PubMed] [Google Scholar]
- 30.Umesako S, Fujisawa K, Iiga S, et al. Atm heterozygous deficiency enhances development of mammary carcinomas in p53 heterozygous knockout mice. Breast Cancer Res. 2005;7(1):R164–170. doi: 10.1186/bcr968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Win TZ, Goodwin A, Hickson ID, Norbury CJ, Wang SW. Requirement for Schizosaccharomyces pombe Top3 in the maintenance of chromosome integrity. J Cell Sci. 2004;117(Pt 20):4769–4778. doi: 10.1242/jcs.01351. [DOI] [PubMed] [Google Scholar]
- 32.Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol. 2005;25(11):4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang M, Bellaoui M, Zhang C, et al. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. Embo J. 2005;24(11):2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14(12):8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broberg K, Hoglund M, Gustafsson C, et al. Genetic variant of the human homologous recombination-associated gene RMI1 (S455N) impacts the risk of AML/MDS and malignant melanoma. Cancer Lett. 2007;258(1):38–44. doi: 10.1016/j.canlet.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Broberg K, Huynh E, Engstrom K, et al. Association between polymorphisms in RMI1, TOP3A, and BLM and risk of cancer, a case-control study. BMC Cancer. 2009;9(1):140. doi: 10.1186/1471-2407-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akira S, Masayasu Y, Chihiro Y, et al. RMI1 deficiency in mice protects from diet and genetic-induced obesity. FEBS Journal. 277(3):677–686. doi: 10.1111/j.1742-4658.2009.07513.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.