Abstract
As the prevalence of obesity continues to rise, rapid and accurate tools for assessing abdominal body and organ fat quantity and distribution are critically needed to assist researchers investigating therapeutic and preventive measures against obesity and its comorbidities. Magnetic resonance imaging (MRI) is the most promising modality to address such need. It is non-invasive, utilizes no ionizing radiation, provides unmatched 3D visualization, is repeatable, and is applicable to subject cohorts of all ages. This article is aimed to provide the reader with an overview of current and state-of-the-art techniques in MRI and associated image analysis methods for fat quantification. The principles underlying traditional approaches such as T1-weighted imaging and magnetic resonance spectroscopy as well as more modern chemical-shift imaging techniques are discussed and compared. The benefits of contiguous 3D acquisitions over 2D multi-slice approaches are highlighted. Typical post-processing procedures for extracting adipose tissue depot volumes and percent organ fat content from abdominal MRI data sets are explained. Furthermore, the advantages and disadvantages of each MRI approach with respect to imaging parameters, spatial resolution, subject motion, scan time, and appropriate fat quantitative endpoints are also provided. Practical considerations in implementing these methods are also presented.
Keywords: MRI, fat imaging, organ fat, body fat quantification, adipose tissue
INTRODUCTION
The prevalence of obesity has risen steadily (1). Literature evidence has attributed obesity-related health risks to the accumulation of subcutaneous and visceral adipose tissue (SAT, VAT) and ectopic organ fat (2–4). Non-invasive assessment of body fat has become an important component in obesity research and fat measures are useful as biomarkers to stratify risks and evaluate the efficacy of therapies.
Many methods are available for body fat assessment (5,6). Anthropometry, hydrodensitometry, air displacement plethysmography (ADP), bioelectric impedance (BIA), and dual energy X-ray absorptiometry (DEXA) have been widely used. The first four are indirect techniques because they measure body density or resistance, which are then converted into percent body fat using generalized equations (7). Hydrodensitometry and ADP only estimates total body fat. BIA and DEXA are limited to total and regional body fat measures. Indirect methods are not able to further differentiate between SAT and VAT or identify ectopic fat.
Recent years has seen an increased utilization of computed tomography (CT) (8–10), quantitative magnetic resonance (QMR) (11,12), and particularly magnetic resonance imaging (MRI) for body fat assessment (13–24). Like DEXA, these are direct techniques because they identify fat based on the tissue's unique properties. QMR does not yield images and is limited to measuring total body fat mass. In contrast, CT and MRI can differentiate SAT and VAT with multi-dimensional images. However, only MRI, with its many sensitive and flexible contrast mechanisms, can quantify ectopic fat. MRI involves no ionizing radiation, which allows for indefinite repeatability in longitudinal studies and in children. It is emerging as a comprehensive tool for fat quantification (25).
This article reviews state-of-the-art MRI methods and image analysis tools for body and organ fat quantification. A summary of MRI of fundamental concepts that underpin T1-weighted imaging, magnetic resonance spectroscopy (MRS), and chemical-shift imaging (CSI) is given. Relevant examples are shown using data acquired from a 3 Tesla General Electric scanner (GE Healthcare, Signa HDx 14M5), unless otherwise stated. General procedures for extracting adipose tissue volumes and organ fat measures from abdominal data are discussed and illustrated using commercial software (SliceOmatic, Tomovision, Inc.). The advantages and disadvantages of MRI approaches with regards to imaging parameters, subject motion, scan time, and quantitative endpoints are highlighted, along with practical considerations in implementation. The article is presented in the context of abdominal imaging, but the content can be translated to other anatomies.
MRI METHODOLOGY
MRI Signal Formation
MRI signals arise from the magnetization of nuclei when placed in a BO magnetic field. Hydrogens, or spins, of water and fat are the most commonly measured nuclei. Today’s clinical MRI scanners utilize a BO of 1.5 or 3 Tesla to align spins and generate magnetization. The spins rotate at a characteristic resonant frequency that is proportional to BO. Radiofrequency (RF) pulses tuned to this frequency are then used to repetitively excite the magnetization by a specified flip angle. Signals are acquired from a RF pulse sequence by receivers as the excited magnetization decays, or relax, between successive pulses. The relaxation rates are known as T1 and T2 times (26).
T1 Weighting
MRI signal intensities are influenced by many factors including spin density, tissue-specific T1 and T2 rates, the flip angle, and the repetition time of the pulse sequence. The T1 of fat is one of the shortest in vivo, which indicates a very rapid recovery of the magnetization between successive RF excitations. By using a T1-weighted sequence, strong tissue contrast can be achieved between short-T1 fat and muscles and organs with longer T1 values (Figure 1). Thus bright fat can be easily identified and delineated by simple signal thresholding from darker structures. T1-weighted sequences are very common in clinical MRI. They are available as standard software on all commercial MRI scanners (GE, Philips, Siemens, Hitachi, and Toshiba) and can be easily implemented. Tissue T1 values typically increase with BO field strength, such that slight adjustments in pulse sequence parameters are needed to maintain comparable tissue contrast between 1.5 and 3 Tesla.
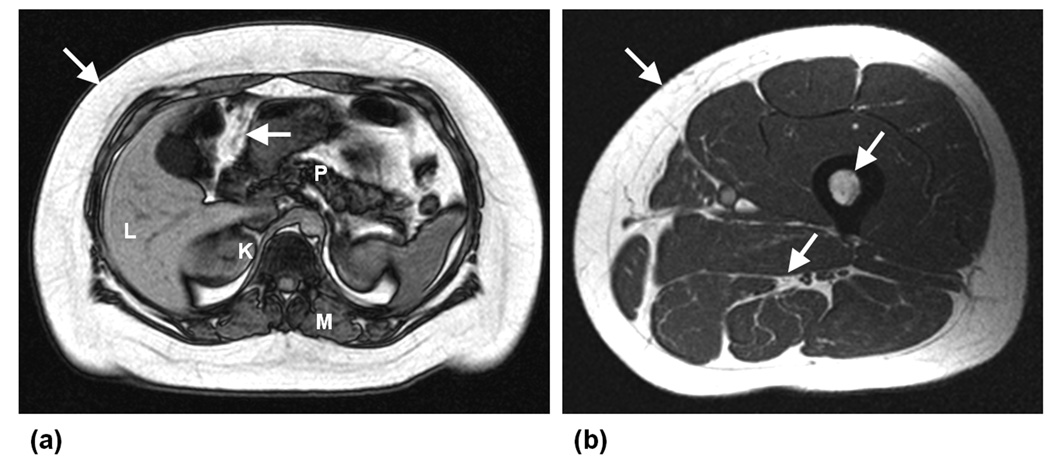
Figure 1.
Examples of T1-weighted axial MR images acquired in the (a) abdomen and (b) thigh, demonstrating the typical high signal intensities of fatty tissues (arrows) in contrast to other darker muscles and organs (L: liver, P: pancreas, K: kidneys, M: muscle). Data were acquired on a GE 1.5 Tesla scanner (Signa HD, 12M5).
Magnetic Resonance Spectroscopy
Single-voxel MRS has been the standard for ectopic fat quantification (27–32). MRS does not provide anatomical information. It instead yields a precise spectrum of chemical composition within one interrogated voxel. MRS relies on chemical shift, which refers to differences in the resonant frequencies of fat and water spins. Fat spins are characterized by a spectral peak that is offset from water spins. Due to differences in their chemical surroundings, lipid spins have a lower resonance. At 1.5 and 3 Teslas, the water-fat chemical shift is approximately 210 and 420 Hz, respectively (14,33–35). MRS provides an intuitive visualization of the presence and relative quantity of chemical species. It also benefits greatly from increasing BO fields due to larger chemical shift separations between peaks. MRS packages should be available on most commercial MRI scanners as standard or optional software but usually requires some expertise to implement and analyze.
Frequency-Selective MRI
Since MRI involves RF pulses tuned to specific frequencies, techniques that exploits the resonant frequency difference between fat and water to quantify SAT and VAT has been developed. One approach is to selectively excite fat spins by tuning the RF pulses to their resonant frequency while simultaneously suppress the water signals (36–38). The appearance of frequency-selective images resembles T1-weighted results and the sharp contrast between fat and musculature readily facilitates thresholding procedures. Frequency-selective techniques can be implemented on all MRI scanners though the capability may not be included as standard software. The technique is sensitive to spatially varying BO inhomogeneities, which worsens with increasing magnetic field. Despite greater chemical shift separation between fat and water at higher field strengths which allows for easier frequency selectivity, BO inhomogeneity can significantly degrade its performance.
Chemical-Shift MRI
Chemical-shift imaging (CSI) integrates fat-water spectral detection with imaging formation (39). CSI represents a family of approaches that have been developed over the past 25 years. Dixon was the first to demonstrate that by controlling the time when data was acquired after RF excitation (e.g. echo time), the net detected MRI signal can comprise either of fat and water in-phase (W+F, aligned) or out-of-phase (W-F, anti-aligned), as illustrated in Figure 2 (40). Using this two-point approach, separated fat and water images could be obtained by algebraic manipulation (34). Intuitively, one realizes that for a voxel containing only fat or water, its net signal will be the same on in-phase and out-of-phase acquisitions as one component’s signal will be zero. In contrast, a voxel containing fat and water will have different signals on the two acquisitions. With higher BO field strengths, the echo time separation between in-phase and out-of-phase signals decreases from every 2.4 milliseconds at 1.5 Tesla to every 1.2 milliseconds at 3 Tesla.
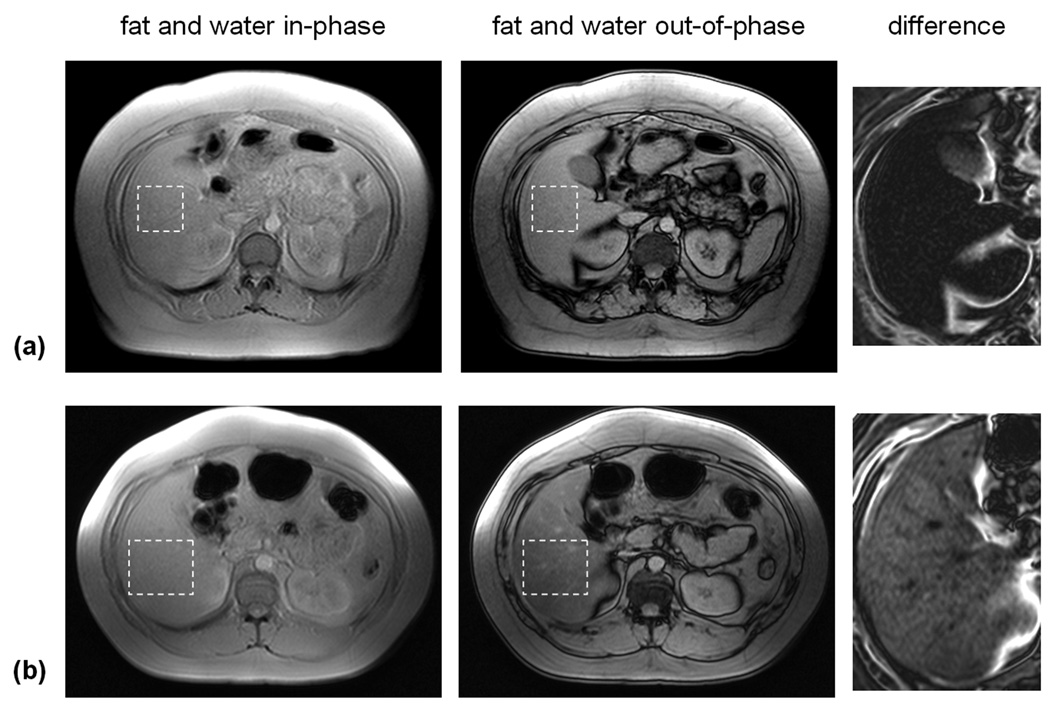
Figure 2.
Example of two-point CSI technique. In-phase (IP) and out-of-phase (OP) images from two subjects are shown. In the OP images, a dark line is present at all fat and lean tissue interfaces. This is caused by signal cancellation between fat and water species within the voxels. All IP and OP images are displayed on the same grayscale. In (a), signal intensities within the liver between the two images are similar, indicating very little presence of hepatic fat. However in (b), the liver signal intensity is markedly lower in the OP image (dashed region), indicative of fat presence. This is visualized in the zoomed difference images. The liver in (a) exhibits residual signal whereas that in (b) shows appreciable signal. As corroborated by MRS, the hepatic fat fractions were 3.4% and 21.2%, respectively
CSI has evolved over the past decade (34,41,42). More robust protocols have been developed to address BO inhomogeneity, which results from manufacturing imperfections, perturbations of the magnetic field by placement of a human body, and at air-tissue-bowel interfaces. Modern CSI approaches (43) also address fat-water signal ambiguity, which refers to the inability of previous approach to detect whether fat or water is the predominant specie present in a voxel. A voxel contain x% fat and (100-x)% water can be indiscernible from a voxel containing x% water and (100- x)% fat (e.g. there is ambiguity about the 50% fat fraction apex). Many literature reports have used CSI to measure organ fat fraction (29,43–48). While not difficult to implement, they often require technician involvement and manufacturer support in establishing appropriate imaging parameters. CSI pulse sequences and reconstruction software (GE – Lava Flex, IDEAL, Philips – mDixon, Siemens – Dixon, Hitachi – FatSep) may only be available on certain new commercial MRI models as standard clinical or optional research-dedicated software.
Recent advances in CSI have led to the development of comprehensive methods such as IDEAL (Iterative Decomposition with Echo Asymmetry and Least squares) (49). Along with several variants (50,51), IDEAL is a generalization of the multi-echo Dixon model and it has been demonstrated at both 1.5 and 3 Tesla. At both field strengths, IDEAL is robust to BO inhomogeneity, addresses fat-water signal ambiguity such that fat fractions can be measured uniquely across the 0–100% range (52,53), accounts for relaxation (54,55), and models the multiple spectral peaks of fat (56). A single IDEAL acquisition yields fat-only and water-only images and fat fraction maps (F/(F+W)) that are optimal in signal-to-noise ratio (57) (Figure 3), as well as in-phase and out-of-phase series. It has been validated against MRS and other CSI methods (29,48,58). IDEAL has also been used to study brown and white adipose tissues (59), spine (60), and knee cartilage (61,62). To the author’s knowledge, IDEAL is a patented and proprietary algorithm that is commercially available only from GE Healthcare on their Discovery 1.5 and 3 Tesla platforms and as a research software on the Signa HDx platforms.
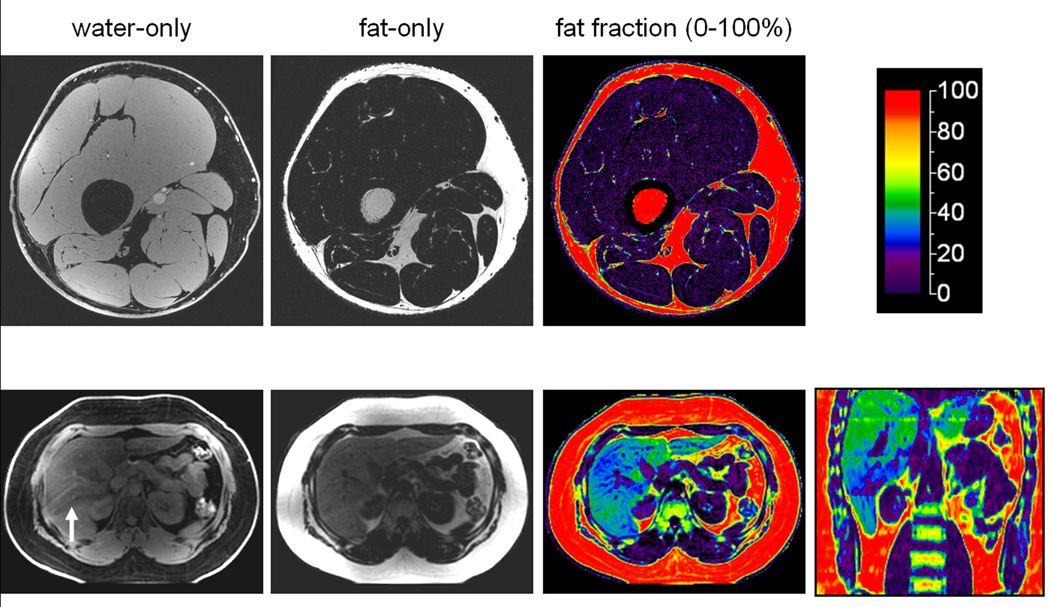
Figure 3.
Examples of 3 Tesla IDEAL. Reconstructed water-only, fat-only, and fat fraction images are illustrated for the thigh and upper abdomen. In the color fat fractions, the scale represents 0–100% percent fat content. In the thighs, note that subcutaneous and intramuscular adipose tissues and bone marrow are denoted by high (> 90% red) fat fractions. In the liver example, the color representation is indicative of very high 40% hepatic fat content. Coronal reformat illustrates the entire liver. The arrow in the water image denotes ripple-like artifacts from respiratory motion.
Summary
MRI and MRS approaches provide a vast array of sensitive methods for quantifying fat (Table 1). The interested reader is referred to three extensive review articles in recent literature for additional detailed technical explanations (34,41,42).
Table 1.
Summary of MR imaging and spectroscopy methods for fat quantification.
| Basis of Fat–Water Differentiation |
Typical Dimensionality |
Advantages | Disadvantages | Quantitative Endpoints |
|
|---|---|---|---|---|---|
| T1-weighted MRI | T1 relaxation | 2D and 3D | strong fat-water tissue contrast, standard on all scanners, easy to implement, rapid scans | T1 contrast varies with BO field strength, requires protocol optimization | SAT and VAT volumes |
| Frequency-Selective MRI | chemical shift | 2D and 3D | strong fat-water tissue contrast, option on most scanners, rapid scans | performance varies with BO field strength and magnet inhomogeneity | SAT and VAT volumes |
| MR Spectroscopy (MRS) | chemical shift | single voxel | high spectral resolution of chemical profile, very accurate measure of fat to water fractions | limited coverage, requires expertise in protocol prescription, data collection and spectral analysis | ectopic fat fraction |
| Chemical Shift MRI | chemical shift | 2D and 3D | complete fat-water tissue separation with accuracy comparable to MRS | not available on all scanners, requires special image reconstruction algorithms and protocol optimization for different BO field strengths | SAT and VAT volumes, ectopic fat fraction |
SAT subcutaneous adipose tissue. VAT visceral adipose tissue.
IMAGE ANALYSIS
The workflow for extracting fat volumes and organ fat fractions from MRI data remains a daunting and costly task. It requires substantial data post-processing and analysis by an experienced operator. The procedure involves the transfer of images to an offline workstation, followed by the use of dedicated software that requires manual intervention (63). Free packages such as ImageJ and Osirix and commercial programs such as SliceOmatic (Tomovision, Inc.), Analyze (AnalyzeDirect, Inc.) and Matlab (The Mathworks, Inc.) are commonly used for segmentation. The analysis of MRS data requires additional software (64). In our experience, post-processing of a 3D abdominal MRI volume takes 45–60 minutes.
Subcutaneous and Visceral Adipose Tissue
Since adipose tissue is composed primarily of lipids, their identification and quantification in T1-weighted, frequency-selective, and chemical-shift images is straightforward. MRS can not be used since it does not provide anatomical data. For T1 and frequency-selective approaches, a binary threshold is usually applied to the signal intensities of each voxel. Since fat appears brighter, those with intensities greater than a set threshold are labeled as fat while those with lower intensities are excluded. Due to intensity variations that may arise across the image, the local threshold may need to be adjusted accordingly (Figure 4). For CSI data, the analysis can utilize the fat-only images and apply a similar threshold procedure. Alternatively, the analysis can exploit the fat fraction map where the range is consistently fixed from 0–100%. Only voxels with fat fractions greater than a set value (e.g. 80%) are then labeled as fat.
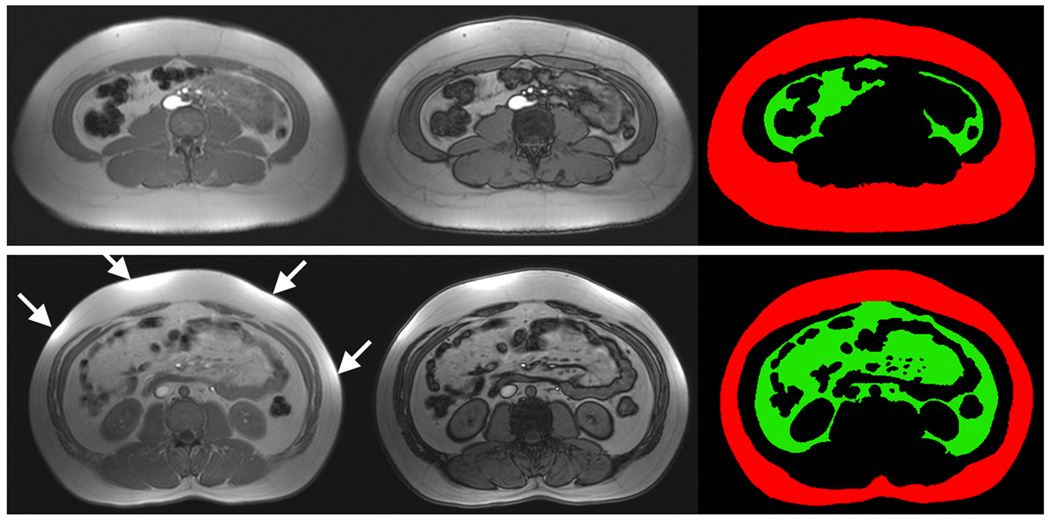
Figure 4.
Examples of SliceOmatic segmentation to delineate the subcutaneous (red) and visceral (green) adipose tissue depots. Note the evident difference in visceral adipose tissue quantity between the two subjects. T1-weighted in-phase (left column) and out-of-phase (middle column) images are shown. Labels (right column) were generated from in-phase images for volume accuracy, using the out-of-phase image for fat-muscle boundary guidance. Arrows highlight bright intensity regions caused by close proximity of the anatomy to receiver arrays. Note that bowels, intramuscular fat, blood vessels, bone, and spine-vertebrae structures are excluded from visceral fat segmentation.
Regardless of the MRI technique, the operator must also delineate the intra-abdominal boundary that separates SAT and VAT and the perimeter of the abdomen to exclude background. Additional exclusions of bowel contents, intramuscular fat (65), spine and vertebrae, blood vessels, and other non-relevant structures are also required since some of these can have signal intensities that mimic fat. An area or volume measure is then computed for 2D multi-slice and contiguous 3D data based on the identified fat voxels, respectively. With the exception of CSI fat fraction data that is always normalized between 0% and 100%, the operator must determine suitable thresholds on a subject-to-subject basis. This is necessary since signal intensities vary across anatomy, subjects, examinations, and equipment and is usually not consistent. In SliceOmatic and Analyze, the operator has control over a sliding threshold and an interface that provides visual feedback on the performance of the chosen threshold. Figure 4 and Figure 5 illustrate multiple examples.
Figure 5.
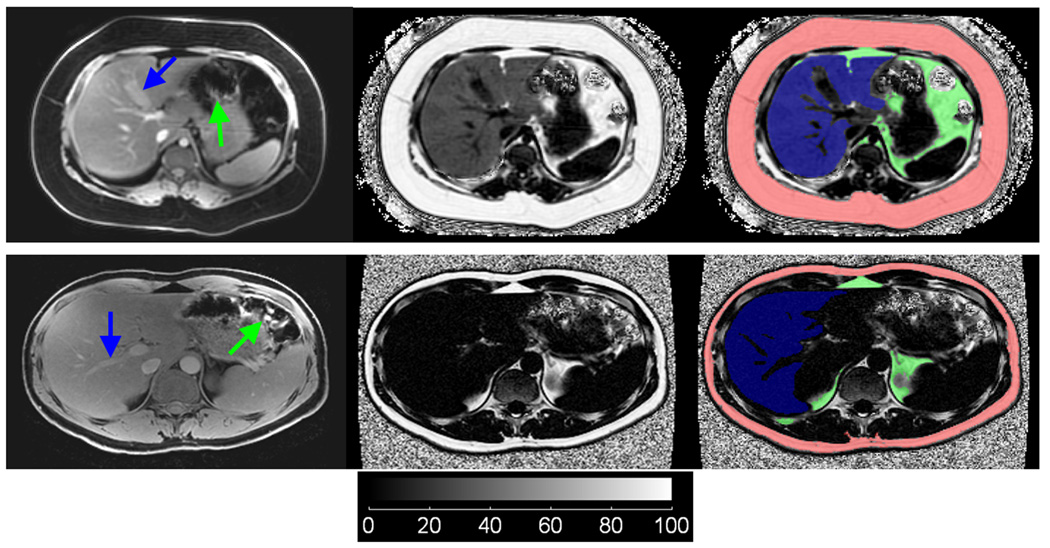
IDEAL image segmentation from two subjects, one with a fatty liver (top row) and one with a non-fatty liver (bottom row). Water-only, fat fraction, and overlay of segmentation labels (Red: subcutaneous adipose tissue, Green: visceral adipose tissue, Blue: liver) are shown in the left, middle, and right columns, respectively. For illustration purpose, the fuzzy noisy background in the fat fraction images has not been removed. In the fatty liver subject, note that hepatic vessels and the gall bladder (blue arrows) are well delineated in both water and fat fraction images, whereas in the non-fatty liver subject, such structures are only observed in the water image. Note that corresponding segmentation labels of the liver exclude these non-liver-tissue structures. Similarly, VAT labels do not include erroneous signals from empty bowels (green arrows).
Ectopic Fat
Ectopic fat quantification requires approaches that can separate signals from lipid and non-lipid components within each imaging voxel. Therefore, the aforementioned binary threshold approach of “all fat” or “no fat” voxel classification is inadequate and only CSI methods are appropriate. Rather than measuring volume, a fractional estimate of fat and water is computed. For MRS, the areas under the fat and water spectral peaks represent the relative amounts of each species present. Software such as jMRUI allows the operator to determine each peak’s area efficiently. The MRS fat fraction is then defined as the ratio of area under the fat peaks to the combined area of the fat and water peaks (48).
CSI provides a fat fraction map across the imaging volume. The map is computed from the ratio of the reconstructed fat-only images to the sum of the fat and water images. To determine the fat fraction at a particular location, the operator needs to draw a region-of-interest (ROI). The statistical distribution of fat fractions within the ROI (e.g. mean, standard deviation) is then computed. To further quantify organ fat fraction, the operator needs to manually segment the entire organ. Once segmented, an overall average fat fraction across the organ can be computed. Unlike single-voxel MRS, a volumetric fat fraction map affords the operator immense flexibility. One can determine the average fat fraction across any arbitrary ROI, including different lobes of the liver, the head and the tail of the pancreas, and across muscles.
For accurate fat fractions, operator expertise is required for drawing ROIs and segmenting organs. In some cases, the fat fraction signal contrast surrounding abdominal organs may be poor, limiting an operator’s ability to directly segment structures from the fat fraction data series and visualize organ-muscle boundaries. This is particularly true for liver, pancreas, and kidney with low fat content that appear visually similar to surrounding muscles. In these instances, reference gray-scale image series, including water-only, fat-only, in-phase, and out-of-phase data, are needed to provide anatomical landmarks. The operator can then segment from these gray-scale images and transfer the ROIs to the corresponding fat fraction data (Figures 5 and 6). In our experience, manual analysis of organ fat accounts more than 70% of the post-processing time. The pancreas and the kidneys are also in close proximity to VAT depots. An inaccurately drawn ROI meant to enclose the organs may contain erroneous VAT voxels. In such cases, the high VAT fat fractions will significantly impact the apparent measured organ fat fraction.
Figure 6.
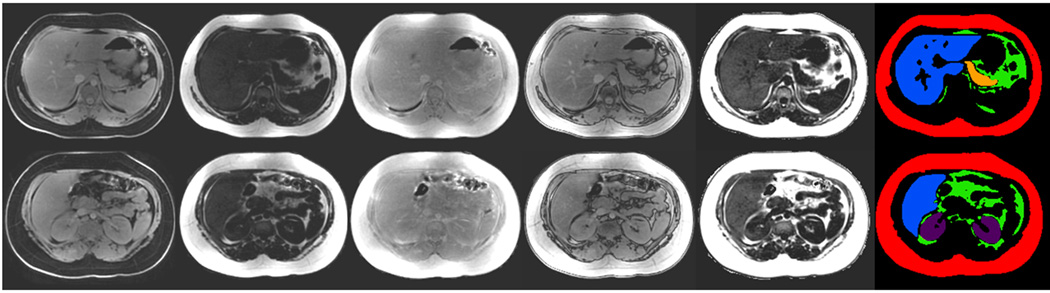
Manual image segmentation using SliceOmatic and the full spectrum of IDEAL data to further delineate abdominal organs (Red: subcutaneous adipose tissue, Green: visceral adipose tissue, Blue: liver, Yellow: pancreas, Purple: kidneys). Left to right: water-only, fat-only, in-phase, out-of-phase, fat fraction, and segmented labels. Labels were generated from the first four columns of source gray-scale images.
Summary
Post-processing by a trained operator is needed for computing quantitative fat endpoints. For SAT and VAT volumes, strong tissue contrast to can facilitate signal-intensity-based thresholding. With the aid of software, this procedure can be efficiently performed. The measurement of ectopic fat requires MRS and CSI. Manual segmentation of organs demands familiarity with software and anatomy, as well as significant operator input.
DISCUSSION
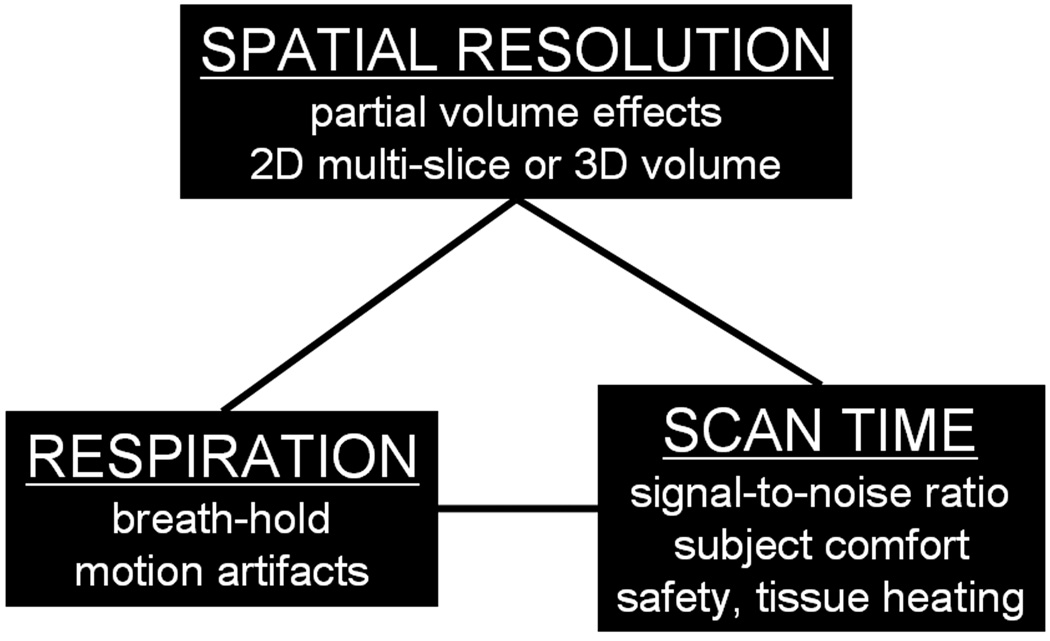
An investigator has several choices in MRI for body fat assessment. In addition to selecting the appropriate technique, one must also consider spatial resolution, scan time, and the need for breath-holds to minimize subject motion (Figure 7).
Figure 7.
Practical imaging parameters and considerations in MRI. In designing an appropriate protocol, one must consider the desired quantitative endpoints and leverage spatial resolution, scan time, patient safety, and the need for breath-holding.
Spatial resolution describes the dimensions of the imaging voxel. For MRS, 1–8 cm3 is commonly used. MRS requires a large voxel and several signal averages to yield a clean spectrum. Multiple scans are also needed to account for T1 and T2 relaxation (66). MRS voxel prescription requires operator expertise such that placement of the voxel does not contain any undesired tissues. Motion from the subject or shifting of organs due to respiration or peristalsis after voxel placement can lead to a spatial misalignments (48). Respiration should be monitored with a bellows transducer for data synchronization. Alternatively, multiple and consistent breath-holds can be used (31). In practice, these approaches may not be possible in subjects with a large abdominal circumference or who have difficulties with breath-holds.
Spatial resolutions for imaging approaches are on the order of millimeters. While small voxels are desirable for visualizing anatomical detail, it leads to increased scan times and lowers image signal-to-noise ratio. Long scan times also increase the risk of motion and blurring artifacts. In CSI, blurring artifacts from respiration can further cause unrealistic fat fraction distributions within organs and at organ-fat interfaces. High spatial resolution scans also necessitate breath-holding, which may not be well-tolerated by children, uncooperative or obese subjects. Spatial resolution is determined by the field-of-view and the acquisition matrix, the number of slices, the slice thickness, and the inter-slice gap, if any. These parameters are freely chosen by the operator and are dependent on body habitus.
The operator must select between 2D multi-slice or 3D volumetric pulse sequences. With 2D techniques, data are acquired on a slice-by-slice basis such that data obtained for one slice is independent of any other slice. Subject motion and artifacts are generally not an issue. The time needed to acquire each slice is usually very short, requiring sub-seconds to a few seconds. The technique also allows the operator to specify a gap between non-contiguous adjacent slices. While this appears attractive in terms of scan efficiency, large inter-slice gaps and thick (~10 mm) slices can lead to partial volume errors and impact quantitative accuracy.
In contrast, 3D approaches acquire data that represent the entire volume. Volumetric imaging implies contiguous coverage and no gaps. Given similar imaging parameters and coverage, 3D protocols often contain more slices, have longer scan times, and are more susceptible to motion than 2D approaches. However, since signals arise from a volume rather than a slice, 3D methods exhibit greater signal-to-noise ratios. Breath-holding or respiratory gating is often required for abdominal 3D sequences. Three-dimensional approaches offers intrinsic advantages over 2D methods, such as exclusive parallel imaging acceleration, greater acceleration rates, and more robust parallel imaging performance to reduce scan time (67,68). Other 3D benefits include acquiring thinner slices and accommodating preparatory RF pulses that further enhances T1 tissue contrast. These advantages largely arise from hardware and RF tissue heating and safety limitations that are beyond the scope of this article.
Lastly, for a given set of resolution and parameters typically used in abdominal imaging, scan times in T1-weighted, frequency-selective, MRS, and CSI approaches should be comparable between 1.5 and 3 Tesla protocols. In some instances, safety and RF tissue heating limits may lengthen 3 Tesla protocol durations over their 1.5 Tesla counterparts. At higher BO field strengths, greater chemical shift differences can also cause CSI sequences to increase in scan time due to shorter in-phase and out-of-phase echo periodicities.
In the literature, 2–4 mm in-plane resolution and a slice thickness between 5–10 mm have been reported for SAT and VAT quantification. For 2D multi-slice approaches inter-slice gaps as wide as 10 mm have been reported. Since the determination of volumes rely on thresholding and an “all fat” or “no fat” labeling each voxel, any incorrect labeling can lead to quantification errors when using large voxels. Large voxels also generate partial volume effects where many can encompass both fat and lean tissues. These voxels exhibit ambiguous signal intensities that can cause threshold errors. Similarly, low spatial resolution and thick slices containing both organ and adjacent fat can exhibit unrealistic CSI fat fractions. There has been a transition in recent studies to adopt contiguous 2D multi-slice approaches with thin slices (22) and 3D protocols (24,69,70). While ectopic fat quantification with 3D IDEAL (Figure 8) has been described, its use in large cohorts has yet to be reported due to the method’s limited availability.
Figure 8.
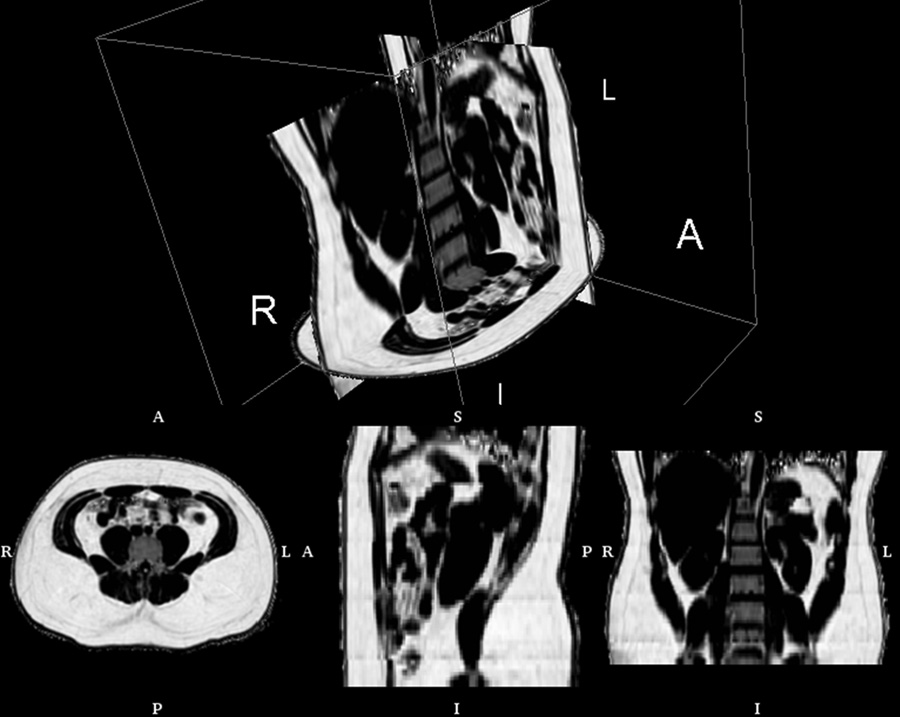
Abdominal fat fraction volume from IDEAL, illustrating the contiguous 3D nature of the data. The volume consists of more than 70 native axial slices and was obtained with five 15 second breath-holds (R: right, L: left, A: anterior, P: posterior, I: inferior, S: superior). Rendering created with 3DSlicer software.
Lastly, modern MRI platforms are equipped with receiver arrays designed specifically for abdominal parallel imaging (68). The array is strapped to the subject to provide high signal-to-noise ratios. However, they also create signal intensity variations across the imaging volume (Figure 4). If possible, these variations should be corrected with software from MRI vendors prior to image analysis. One attractive feature of fat fraction maps is that they are generated as a ratio of fat and water images. While the individual source images may exhibit signal variations, their ratio removes the bias. The use of arrays may be limited in obese subjects as they are unable to fit within the magnet bore with the receivers strapped.
Summary and Future Directions
The design of appropriate MRI protocols for fat assessment requires considerations of the quantitative endpoints and tradeoffs in the imaging parameters. Other practical issues include body sizes of the cohort and the cost and throughput efficiency of the study. The equipment cost associated with MRI can range from $300 – 600 per hour in a typical clinical radiology department. Regardless of the study, overhead time is always required for setup and for giving instructions to the subject. T1-weighted and frequency-selective approaches are very fast and typically require only a few seconds per slice for data acquisition. Whole-abdomen coverage with these sequences takes only a few minutes of actual scan time and is generally considered as a first option for SAT and VAT quantification. In our experience, utilizing only these sequences, an exam can be comfortably completed in 10 minutes. If ectopic fat fractions are needed, either a MRS or CSI protocol is then added. The MRS sequence necessitates significant user interaction, preparation work, and operator time. In our experience, MRS per single voxel takes approximately 5 minutes. The further addition of a 3D whole-abdomen CSI protocol such as IDEAL will require 5–7 more breath-holds, bringing the total examination time from setup to patient removal to 30–45 minutes.
Despite its rapid growth, MRI remains an untapped resource in obesity research and has not yet reached its full potential. Many opportunities remain for investigation. Several groups have recognized the time-consuming and labor-intensive work involved with manual image segmentation. This can be daunting in longitudinal studies, as the process must be performed for every scan at multiple time points across the same subject. Efficient semi-automated and automated algorithms have been proposed, but are usually limited to in-house usage (19,38,71–75). Techniques that can automatically segment adipose tissue and organs from a subject’s MRI data at subsequent time points while utilizing a priori information from the same subject’s baseline segmentations are being explored. The capability of achieving rapid intra-subject 3D registration, segmentation, and quantification will provide investigators with detailed person-specific information reflecting the temporal change in fat distribution and volumes in response to intervention. Such capability will also exploit the richness of 3D MRI that is unavailable in gapped 2D multi-slice data. Quality control also will be essential in assuring consistent accuracy. Fat measurements should be routinely validated against phantoms, between multiple blinded observers, and between manual and automated segmentations.
In conclusion, MRI is the most powerful and comprehensive imaging tool for fat quantification. With CSI methods, recent studies are beginning to demonstrate their usefulness and explore new fat biomarkers in obesity. This article has provided introductory materials for investigators to survey and integrate MRI into their body composition studies.
Acknowledgements
The authors gratefully thank Huanzhou Yu and Ann Shimakawa from GE Healthcare for providing MRI technical support, and acknowledge grant support from the National Institutes of Health (Grant numbers: U54CA116848, R21DK081173, K25DK087931). The authors also thank Rosa Rangel and Sherryl Esplana from the University of Southern California for research assistance, as well as Mark Punyanitya from Columbia University for guidance with image segmentation.
Footnotes
Conflicts of Interests
None – all authors
REFERENCE
- 1.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999–2008. JAMA. 2010;303(3):235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 2.Bjorntorp P. Metabolic implications of body fat distribution. Diabetes Care. 1991;14(12):1132–1143. doi: 10.2337/diacare.14.12.1132. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary AK, Donnelly LF, Racadio JM, Strife JL. Diseases associated with childhood obesity. AJR Am J Roentgenol. 2007;188(4):1118–1130. doi: 10.2214/AJR.06.0651. [DOI] [PubMed] [Google Scholar]
- 4.Despres JP, Lemieux I, Bergeron J, Pibarot P, Mathieu P, Larose E, Rodes-Cabau J, Bertrand OF, Poirier P. Abdominal obesity and the metabolic syndrome: contribution to global cardiometabolic risk. Arterioscler Thromb Vasc Biol. 2008;28(6):1039–1049. doi: 10.1161/ATVBAHA.107.159228. [DOI] [PubMed] [Google Scholar]
- 5.Ellis KJ. Human body composition: in vivo methods. Physiol Rev. 2000;80(2):649–680. doi: 10.1152/physrev.2000.80.2.649. [DOI] [PubMed] [Google Scholar]
- 6.Mattsson S, Thomas BJ. Development of methods for body composition studies. Phys Med Biol. 2006;51(13):R203–R228. doi: 10.1088/0031-9155/51/13/R13. [DOI] [PubMed] [Google Scholar]
- 7.Jackson AS, Pollock ML. Steps toward the development of generalized equations for predicting body composition of adults. Can J Appl Sport Sci. 1982;7(3):189–196. [PubMed] [Google Scholar]
- 8.Kvist H, Chowdhury B, Grangard U, Tylen U, Sjostrom L. Total and visceral adipose-tissue volumes derived from measurements with computed tomography in adult men and women: predictive equations. Am J Clin Nutr. 1988;48(6):1351–1361. doi: 10.1093/ajcn/48.6.1351. [DOI] [PubMed] [Google Scholar]
- 9.Seidell JC, Bakker CJ, van der Kooy K. Imaging techniques for measuring adipose-tissue distribution--a comparison between computed tomography and 1.5-T magnetic resonance. Am J Clin Nutr. 1990;51(6):953–957. doi: 10.1093/ajcn/51.6.953. [DOI] [PubMed] [Google Scholar]
- 10.Goodpaster BH, Thaete FL, Kelley DE. Composition of skeletal muscle evaluated with computed tomography. Ann N Y Acad Sci. 2000;904:18–24. doi: 10.1111/j.1749-6632.2000.tb06416.x. [DOI] [PubMed] [Google Scholar]
- 11.Taicher GZ, Tinsley FC, Reiderman A, Heiman ML. Quantitative magnetic resonance (QMR) method for bone and whole-body-composition analysis. Anal Bioanal Chem. 2003;377(6):990–1002. doi: 10.1007/s00216-003-2224-3. [DOI] [PubMed] [Google Scholar]
- 12.Napolitano A, Miller SR, Murgatroyd PR, Coward WA, Wright A, Finer N, De Bruin TW, Bullmore ET, Nunez DJ. Validation of a quantitative magnetic resonance method for measuring human body composition. Obesity (Silver Spring) 2008;16(1):191–198. doi: 10.1038/oby.2007.29. [DOI] [PubMed] [Google Scholar]
- 13.Lancaster JL, Ghiatas AA, Alyassin A, Kilcoyne RF, Bonora E, DeFronzo RA. Measurement of abdominal fat with T1-weighted MR images. J Magn Reson Imaging. 1991;1(3):363–369. doi: 10.1002/jmri.1880010315. [DOI] [PubMed] [Google Scholar]
- 14.Brix G, Heiland S, Bellemann ME, Koch T, Lorenz WJ. MR imaging of fat-containing tissues: valuation of two quantitative imaging techniques in comparison with localized proton spectroscopy. Magn Reson Imaging. 1993;11(7):977–991. doi: 10.1016/0730-725x(93)90217-2. [DOI] [PubMed] [Google Scholar]
- 15.Abate N, Garg A, Coleman R, Grundy SM, Peshock RM. Prediction of total subcutaneous abdominal, intraperitoneal, and retroperitoneal adipose tissue masses in men by a single axial magnetic resonance imaging slice. Am J Clin Nutr. 1997;65(2):403–408. doi: 10.1093/ajcn/65.2.403. [DOI] [PubMed] [Google Scholar]
- 16.Thomas EL, Hamilton G, Patel N, O'Dwyer R, Dore CJ, Goldin RD, Bell JD, Taylor-Robinson SD. Hepatic triglyceride content and its relation to body adiposity: a magnetic resonance imaging and proton magnetic resonance spectroscopy study. Gut. 2005;54(1):122–127. doi: 10.1136/gut.2003.036566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross R, Goodpaster B, Kelley D, Boada F. Magnetic resonance imaging in human body composition research. From quantitative to qualitative tissue measurement. Ann N Y Acad Sci. 2000;904:12–17. doi: 10.1111/j.1749-6632.2000.tb06415.x. [DOI] [PubMed] [Google Scholar]
- 18.Schick F, Machann J, Brechtel K, Strempfer A, Klumpp B, Stein DT, Jacob S. MRI of muscular fat. Magn Reson Med. 2002;47(4):720–727. doi: 10.1002/mrm.10107. [DOI] [PubMed] [Google Scholar]
- 19.Wilhelm Poll L, Wittsack HJ, Koch JA, Willers R, Cohnen M, Kapitza C, Heinemann L, Modder U. A rapid and reliable semiautomated method for measurement of total abdominal fat volumes using magnetic resonance imaging. Magn Reson Imaging. 2003;21(6):631–636. doi: 10.1016/s0730-725x(03)00093-6. [DOI] [PubMed] [Google Scholar]
- 20.Goodpaster BH, Wolf D. Skeletal muscle lipid accumulation in obesity, insulin resistance, and type 2 diabetes. Pediatr Diabetes. 2004;5(4):219–226. doi: 10.1111/j.1399-543X.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 21.Machann J, Thamer C, Schnoedt B, Haap M, Haring HU, Claussen CD, Stumvoll M, Fritsche A, Schick F. Standardized assessment of whole body adipose tissue topography by MRI. J Magn Reson Imaging. 2005;21(4):455–462. doi: 10.1002/jmri.20292. [DOI] [PubMed] [Google Scholar]
- 22.Siegel MJ, Hildebolt CF, Bae KT, Hong C, White NH. Total and intraabdominal fat distribution in preadolescents and adolescents: measurement with MR imaging. Radiology. 2007;242(3):846–856. doi: 10.1148/radiol.2423060111. [DOI] [PubMed] [Google Scholar]
- 23.Kullberg J, Brandberg J, Angelhed JE, Frimmel H, Bergelin E, Strid L, Ahlstrom H, Johansson L, Lonn L. Whole-body adipose tissue analysis: comparison of MRI, CT and dual energy X-ray absorptiometry. Br J Radiol. 2009;82(974):123–130. doi: 10.1259/bjr/80083156. [DOI] [PubMed] [Google Scholar]
- 24.Berglund J, Johansson L, Ahlstrom H, Kullberg J. Three-point dixon method enables whole-body water and fat imaging of obese subjects. Magnetic Resonance in Medicine. 2010;63(6):1659–1668. doi: 10.1002/mrm.22385. [DOI] [PubMed] [Google Scholar]
- 25.Shen W, Liu H, Punyanitya M, Chen J, Heymsfield SB. Pediatric obesity phenotyping by magnetic resonance methods. Curr Opin Clin Nutr Metab Care. 2005;8(6):595–601. [PMC free article] [PubMed] [Google Scholar]
- 26.Stanisz GJ, Odrobina EE, Pun J, Escaravage M, Graham SJ, Bronskill MJ, Henkelman RM. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn Reson Med. 2005;54(3):507–512. doi: 10.1002/mrm.20605. [DOI] [PubMed] [Google Scholar]
- 27.Barac-Nieto M, Gupta RK. Use of proton MR spectroscopy and MR imaging to assess obesity. J Magn Reson Imaging. 1996;6(1):235–238. doi: 10.1002/jmri.1880060141. [DOI] [PubMed] [Google Scholar]
- 28.Kamba M, Meshitsuka S, Iriguchi N, Koda M, Kimura K, Ogawa T. Measurement of relative fat content by proton magnetic resonance spectroscopy using a clinical imager. J Magn Reson Imaging. 2000;11(3):330–335. doi: 10.1002/(sici)1522-2586(200003)11:3<330::aid-jmri13>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 29.Kim H, Taksali SE, Dufour S, Befroy D, Goodman TR, Petersen KF, Shulman GI, Caprio S, Constable RT. Comparative MR study of hepatic fat quantification using single-voxel proton spectroscopy, two-point dixon and three-point IDEAL. Magn Reson Med. 2008;59(3):521–527. doi: 10.1002/mrm.21561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weis J, Johansson L, Ortiz-Nieto F, Ahlstrom H. Assessment of lipids in skeletal muscle by high-resolution spectroscopic imaging using fat as the internal standard: Comparison with water referenced spectroscopy. Magn Reson Med. 2008;59(6):1259–1265. doi: 10.1002/mrm.21601. [DOI] [PubMed] [Google Scholar]
- 31.Lingvay I, Esser V, Legendre JL, Price AL, Wertz KM, Adams-Huet B, Zhang S, Unger RH, Szczepaniak LS. Noninvasive quantification of pancreatic fat in humans. J Clin Endocrinol Metab. 2009;94(10):4070–4076. doi: 10.1210/jc.2009-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Werven JR, Hoogduin JM, Nederveen AJ, van Vliet AA, Wajs E, Vandenberk P, Stroes ES, Stoker J. Reproducibility of 3.0 Tesla magnetic resonance spectroscopy for measuring hepatic fat content. J Magn Reson Imaging. 2009;30(2):444–448. doi: 10.1002/jmri.21837. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton G, Middleton MS, Bydder M, Yokoo T, Schwimmer JB, Kono Y, Patton HM, Lavine JE, Sirlin CB. Effect of PRESS and STEAM sequences on magnetic resonance spectroscopic liver fat quantification. J Magn Reson Imaging. 2009;30(1):145–152. doi: 10.1002/jmri.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassidy FH, Yokoo T, Aganovic L, Hanna RF, Bydder M, Middleton MS, Hamilton G, Chavez AD, Schwimmer JB, Sirlin CB. Fatty liver disease: MR imaging techniques for the detection and quantification of liver steatosis. Radiographics. 2009;29(1):231–260. doi: 10.1148/rg.291075123. [DOI] [PubMed] [Google Scholar]
- 35.Ren J, Dimitrov I, Sherry AD, Malloy CR. Composition of adipose tissue and marrow fat in humans by 1H NMR at 7 Tesla. J Lipid Res. 2008;49(9):2055–2062. doi: 10.1194/jlr.D800010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaldoudi E, Williams SC, Barker GJ, Tofts PS. A chemical shift selective inversion recovery sequence for fat-suppressed MRI: theory and experimental validation. Magn Reson Imaging. 1993;11(3):341–355. doi: 10.1016/0730-725x(93)90067-n. [DOI] [PubMed] [Google Scholar]
- 37.Machann J, Bachmann OP, Brechtel K, Dahl DB, Wietek B, Klumpp B, Haring HU, Claussen CD, Jacob S, Schick F. Lipid content in the musculature of the lower leg assessed by fat selective MRI: intra- and interindividual differences and correlation with anthropometric and metabolic data. J Magn Reson Imaging. 2003;17(3):350–357. doi: 10.1002/jmri.10255. [DOI] [PubMed] [Google Scholar]
- 38.Peng Q, McColl RW, Ding Y, Wang J, Chia JM, Weatherall PT. Automated method for accurate abdominal fat quantification on water-saturated magnetic resonance images. J Magn Reson Imaging. 2007;26(3):738–746. doi: 10.1002/jmri.21040. [DOI] [PubMed] [Google Scholar]
- 39.Hood MN, Ho VB, Smirniotopoulos JG, Szumowski J. Chemical shift: the artifact and clinical tool revisited. Radiographics. 1999;19(2):357–371. doi: 10.1148/radiographics.19.2.g99mr07357. [DOI] [PubMed] [Google Scholar]
- 40.Dixon WT. Simple proton spectroscopic imaging. Radiology. 1984;153(1):189–194. doi: 10.1148/radiology.153.1.6089263. [DOI] [PubMed] [Google Scholar]
- 41.Ma J. Dixon techniques for water and fat imaging. J Magn Reson Imaging. 2008;28(3):543–558. doi: 10.1002/jmri.21492. [DOI] [PubMed] [Google Scholar]
- 42.Bley TA, Wieben O, Francois CJ, Brittain JH, Reeder SB. Fat and water magnetic resonance imaging. J Magn Reson Imaging. 2010;31(1):4–18. doi: 10.1002/jmri.21895. [DOI] [PubMed] [Google Scholar]
- 43.Hussain HK, Chenevert TL, Londy FJ, Gulani V, Swanson SD, McKenna BJ, Appelman HD, Adusumilli S, Greenson JK, Conjeevaram HS. Hepatic fat fraction: MR imaging for quantitative measurement and display--early experience. Radiology. 2005;237(3):1048–1055. doi: 10.1148/radiol.2373041639. [DOI] [PubMed] [Google Scholar]
- 44.Yokoo T, Bydder M, Hamilton G, Middleton MS, Gamst AC, Wolfson T, Hassanein T, Patton HM, Lavine JE, Schwimmer JB, Sirlin CB. Nonalcoholic fatty liver disease: diagnostic and fat-grading accuracy of low-flip-angle multiecho gradient-recalled-echo MR imaging at 1.5 T. Radiology. 2009;251(1):67–76. doi: 10.1148/radiol.2511080666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guiu B, Petit JM, Loffroy R, Ben Salem D, Aho S, Masson D, Hillon P, Krause D, Cercueil JP. Quantification of liver fat content: comparison of triple-echo chemical shift gradient-echo imaging and in vivo proton MR spectroscopy. Radiology. 2009;250(1):95–102. doi: 10.1148/radiol.2493080217. [DOI] [PubMed] [Google Scholar]
- 46.O'Regan DP, Callaghan MF, Wylezinska-Arridge M, Fitzpatrick J, Naoumova RP, Hajnal JV, Schmitz SA. Liver fat content and T2*: simultaneous measurement by using breath-hold multiecho MR imaging at 3.0 T--feasibility. Radiology. 2008;247(2):550–557. doi: 10.1148/radiol.2472070880. [DOI] [PubMed] [Google Scholar]
- 47.Kellman P, Hernando D, Shah S, Zuehlsdorff S, Jerecic R, Mancini C, Liang ZP, Arai AE. Multiecho dixon fat and water separation method for detecting fibrofatty infiltration in the myocardium. Magn Reson Med. 2009;61(1):215–221. doi: 10.1002/mrm.21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu HH, Kim HW, Nayak KS, Goran MI. Comparison of fat-water MRI and single-voxel MRS in the assessment of hepatic and pancreatic fat fractions in humans. Obesity. 2010;18(4):841–847. doi: 10.1038/oby.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeder SB, McKenzie CA, Pineda AR, Yu H, Shimakawa A, Brau AC, Hargreaves BA, Gold GE, Brittain JH. Water-fat separation with IDEAL gradient-echo imaging. J Magn Reson Imaging. 2007;25(3):644–652. doi: 10.1002/jmri.20831. [DOI] [PubMed] [Google Scholar]
- 50.Yokoo T, Shiehmorteza M, Bydder M, Hamilton G, Kono Y, Kuo A, Lavine JE, Sirlin CB. Spectrally-modeled hepatic fat quantification by multi-echo gradient-recalled-echo magnetic resonance imaging at 3.0T; 17th Meeting of the International Society of Magnetic Resonance in Medicine; 2009. p. 209. [Google Scholar]
- 51.Xiang QS, An L. Water-fat imaging with direct phase encoding. J Magn Reson Imaging. 1997;7(6):1002–1015. doi: 10.1002/jmri.1880070612. [DOI] [PubMed] [Google Scholar]
- 52.Bernard CP, Liney GP, Manton DJ, Turnbull LW, Langton CM. Comparison of fat quantification methods: a phantom study at 3.0T. J Magn Reson Imaging. 2008;27(1):192–197. doi: 10.1002/jmri.21201. [DOI] [PubMed] [Google Scholar]
- 53.Hines CD, Yu H, Shimakawa A, McKenzie CA, Brittain JH, Reeder SB. T1 independent, T2* corrected MRI with accurate spectral modeling for quantification of fat: validation in a fat-water-SPIO phantom. J Magn Reson Imaging. 2009;30(5):1215–1222. doi: 10.1002/jmri.21957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu CY, McKenzie CA, Yu H, Brittain JH, Reeder SB. Fat quantification with IDEAL gradient echo imaging: correction of bias from T1 and noise. Magn Reson Med. 2007;58(2):354–364. doi: 10.1002/mrm.21301. [DOI] [PubMed] [Google Scholar]
- 55.Bydder M, Yokoo T, Hamilton G, Middleton MS, Chavez AD, Schwimmer JB, Lavine JE, Sirlin CB. Relaxation effects in the quantification of fat using gradient echo imaging. Magn Reson Imaging. 2008;26(3):347–359. doi: 10.1016/j.mri.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu H, Shimakawa A, McKenzie CA, Brodsky E, Brittain JH, Reeder SB. Multiecho water-fat separation and simultaneous R2* estimation with multifrequency fat spectrum modeling. Magn Reson Med. 2008;60(5):1122–1134. doi: 10.1002/mrm.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pineda AR, Reeder SB, Wen Z, Pelc NJ. Cramer-Rao bounds for three-point decomposition of water and fat. Magn Reson Med. 2005;54(3):625–635. doi: 10.1002/mrm.20623. [DOI] [PubMed] [Google Scholar]
- 58.Reeder SB, Robson PM, Yu H, Shimakawa A, Hines CD, McKenzie CA, Brittain JH. Quantification of hepatic steatosis with MRI: the effects of accurate fat spectral modeling. J Magn Reson Imaging. 2009;29(6):1332–1339. doi: 10.1002/jmri.21751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu HH, Smith DL, Jr, Nayak KS, Goran MI, Nagy TR. Identification of brown adipose tissue in mice with fat-water IDEAL-MRI. J Magn Reson Imaging. 2010;31(5):1195–1202. doi: 10.1002/jmri.22162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss KL, Sun D, Cornelius RS, Weiss JL. Iterative Decomposition of Water and Fat with Echo Asymmetric and Least-Squares Estimation (IDEAL) Automated Spine Survey Iterative Scan Technique (ASSIST) Magnetic Resonance Insights. 2008;1(4):1–6. [Google Scholar]
- 61.Kijowski R, Woods MA, Lee KS, Takimi K, Yu H, Shimakawa A, Brittain JH, Reeder SB. Improved fat suppression using multipeak reconstruction for IDEAL chemical shift fat-water separation: application with fast spin echo imaging. J Magn Reson Imaging. 2009;29(2):436–442. doi: 10.1002/jmri.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kijowski R, Tuite M, Passov L, Shimakawa A, Yu H, Reeder SB. Cartilage imaging at 3.0T with gradient refocused acquisition in the steady-state (GRASS) and IDEAL fat-water separation. J Magn Reson Imaging. 2008;28(1):167–174. doi: 10.1002/jmri.21414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonekamp S, Ghosh P, Crawford S, Solga SF, Horska A, Brancati FL, Diehl AM, Smith S, Clark JM. Quantitative comparison and evaluation of software packages for assessment of abdominal adipose tissue distribution by magnetic resonance imaging. Int J Obes (Lond) 2008;32(1):100–111. doi: 10.1038/sj.ijo.0803696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naressi A, Couturier C, Castang I, de Beer R, Graveron-Demilly D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med. 2001;31(4):269–286. doi: 10.1016/s0010-4825(01)00006-3. [DOI] [PubMed] [Google Scholar]
- 65.Gallagher D, Kuznia P, Heshka S, Albu J, Heymsfield SB, Goodpaster B, Visser M, Harris TB. Adipose tissue in muscle: a novel depot similar in size to visceral adipose tissue. Am J Clin Nutr. 2005;81(4):903–910. doi: 10.1093/ajcn/81.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma P, Martin DR, Pineda N, Xu Q, Vos M, Anania F, Hu X. Quantitative analysis of T2-correction in single-voxel magnetic resonance spectroscopy of hepatic lipid fraction. J Magn Reson Imaging. 2009;29(3):629–635. doi: 10.1002/jmri.21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weiger M, Pruessmann KP, Boesiger P. 2D SENSE for faster 3D MRI. Magma. 2002;14(1):10–19. doi: 10.1007/BF02668182. [DOI] [PubMed] [Google Scholar]
- 68.Margolis DJ, Bammer R, Chow LC. Parallel imaging of the abdomen. Top Magn Reson Imaging. 2004;15(3):197–206. doi: 10.1097/01.rmr.0000136557.27727.79. [DOI] [PubMed] [Google Scholar]
- 69.Bornert P, Keupp J, Eggers H, Aldefeld B. Whole-body 3D water/fat resolved continuously moving table imaging. J Magn Reson Imaging. 2007;25(3):660–665. doi: 10.1002/jmri.20861. [DOI] [PubMed] [Google Scholar]
- 70.Kullberg J, Johansson L, Ahlstrom H, Courivaud F, Koken P, Eggers H, Bornert P. Automated assessment of whole-body adipose tissue depots from continuously moving bed MRI: a feasibility study. J Magn Reson Imaging. 2009;30(1):185–193. doi: 10.1002/jmri.21820. [DOI] [PubMed] [Google Scholar]
- 71.Kullberg J, Angelhed JE, Lonn L, Brandberg J, Ahlstrom H, Frimmel H, Johansson L. Whole-body T1 mapping improves the definition of adipose tissue: consequences for automated image analysis. J Magn Reson Imaging. 2006;24(2):394–401. doi: 10.1002/jmri.20644. [DOI] [PubMed] [Google Scholar]
- 72.Armao D, Guyon JP, Firat Z, Brown MA, Semelka RC. Accurate quantification of visceral adipose tissue (VAT) using water-saturation MRI and computer segmentation: preliminary results. J Magn Reson Imaging. 2006;23(5):736–741. doi: 10.1002/jmri.20551. [DOI] [PubMed] [Google Scholar]
- 73.Kullberg J, Ahlstrom H, Johansson L, Frimmel H. Automated and reproducible segmentation of visceral and subcutaneous adipose tissue from abdominal MRI. Int J Obes (Lond) 2007;31(12):1806–1817. doi: 10.1038/sj.ijo.0803671. [DOI] [PubMed] [Google Scholar]
- 74.Positano V, Gastaldelli A, Sironi AM, Santarelli MF, Lombardi M, Landini L. An accurate and robust method for unsupervised assessment of abdominal fat by MRI. J Magn Reson Imaging. 2004;20(4):684–689. doi: 10.1002/jmri.20167. [DOI] [PubMed] [Google Scholar]
- 75.Demerath EW, Ritter KJ, Couch WA, Rogers NL, Moreno GM, Choh A, Lee M, Remsberg K, Czerwinski SA, Chumlea WC, Siervogel RM, Towne B. Validity of a new automated software program for visceral adipose tissue estimation. Int J Obes (Lond) 2007;31(2):285–291. doi: 10.1038/sj.ijo.0803409. [DOI] [PMC free article] [PubMed] [Google Scholar]