Abstract
Background
Breast cancers can be classified by hierarchical clustering using an “intrinsic” gene list into one of at least five molecular subtypes: basal-like, HER2, luminal A, luminal B, and normal breast-like. Five different intrinsic gene lists composed of varying numbers of genes have been used for molecular subtype identification and classification of breast cancers. The aim of this study was to determine the objectivity and interobserver reproducibility of the assignment of molecular subtype classes by hierarchical cluster analysis.
Methods
Three publicly available breast cancer datasets (n = 779) were subjected to two-way average-linkage hierarchical cluster analysis using five distinct intrinsic gene lists. We used free-marginal Kappa statistics to analyze interobserver agreement among five breast cancer researchers for the whole classification and for each molecular subtype separately according to each intrinsic gene list for each breast cancer dataset.
Results
None of the classification systems tested produced almost perfect agreement (Kappa ≥ 0.81) among observers. However, substantial interobserver agreement (70.8% to 76.1% of the samples and free-marginal Kappa scores from 0.635 to 0.701) was consistently observed in all datasets for four molecular subtypes (luminal, basal-like, HER2, and normal breast-like). When luminal cancers were subdivided (luminal A, B, and C), none of the classification systems produced substantial agreement (Kappa ≥ 0.61) in all the datasets analyzed. Analysis of each subtype separately revealed that only two (basal-like and HER2) could be reproducibly identified by independent observers (Kappa ≥ 0.81).
Conclusions
Assignment of molecular subtype classes of breast cancer based on the analysis of dendrograms obtained with hierarchical cluster analysis is subjective and shows modest interobserver reproducibility. For the development of a molecular taxonomy, objective definitions for each molecular subtype and standardized methods for their identification are required.
CONTEXTS AND CAVEATS
Prior knowledge
Hierarchical cluster analysis is used to classify tumors into subtypes identified through microarray-based gene expression profiling. These approaches are considered more objective and reproducible than histopathologic and immunohistochemical methods, but the subjectivity involved in assigning the molecular subtypes through dendrogram analysis has not been systematically analyzed.
Study design
The interobserver reproducibility among five breast cancer researchers experienced in molecular subtype assignment was determined using dendrograms and heatmaps generated by hierarchical clustering methods of three breast cancer datasets and five intrinsic gene lists.
Contribution
The identification of subgroups of luminal cancers and normal breast-like cancers by visual inspection of dendrograms obtained from hierarchical cluster analysis shows suboptimal levels of interobserver agreement, even when the molecular subtypes are known a priori and guidelines for the identification of these subtypes are provided.
Implications
The assignment of molecular subtypes of breast cancer based on the visual inspection of dendrograms obtained with hierarchical cluster analysis is subjective and shows only modest interobserver reproducibility, particularly when subclassification of luminal cancers into two or more groups is required. Class discovery studies need to take into account both the stability of the clusters and the reproducibility of the classification system.
Limitations
The datasets used were retrospectively accrued; hence, they may not have a balanced distribution of the different molecular subtypes and do not include samples of normal breast tissue. Publicly available microarray results were used for the hierarchical cluster analyses, and the data were not renormalized. A small proportion of genes from the intrinsic gene lists could not be reannotated in all datasets owing to different microarray platforms and changes in gene annotation.
From the Editors
The use of high-throughput methods for the analysis of cancers has provided new opportunities for understanding the diversity and heterogeneity of cancers and to devise classification systems that better recapitulate the biology and clinical behavior of human tumors. Class discovery studies have led to the identification of molecular subgroups of prognostic significance in multiple types of cancer, including lymphomas (1), sarcomas (2), pediatric malignancies (3), melanomas (4) and carcinomas (5, 6). There is a perception that these approaches may be more objective and reproducible than histopathologic and immunohistochemical methods (7,8).
Microarray-based gene expression profiling has highlighted the existence of breast cancer subtypes with distinct biology and clinical behavior (9,10). Expression profiling class discovery studies have led to a working model for a breast cancer molecular taxonomy (5,11–14), which has become widely used and recently adopted for the design of clinical trials (eg, NCT00546156).
Breast cancers can be classified by hierarchical cluster analysis using an “intrinsic” gene list [ie, list of “genes with significantly greater variation in expression between different tumours than between paired samples from the same tumour” (5)] into at least one of five molecular subtype classes: luminal A, luminal B, basal-like, HER2, and normal breast-like (5,10–14). Hierarchical clustering algorithms aggregate samples based on the similarity of their gene expression patterns and produce dendrograms, which are two-dimensional representations of the similarity between the samples and genes analyzed (ie, for each of two samples, the smaller the distance in the dendrogram arm or branch, the more similar the expression profiles of the samples). Five different intrinsic gene lists composed of varying numbers of genes have been reported (5,11–14). It has been assumed that molecular subtypes identified in different studies using different intrinsic gene lists are equivalent and reproducible with regard to their clinical, biological, and prognostic characteristics (ie, luminal A cancers in study “A” identified by the intrinsic gene list “a” are synonymous with luminal A cancers in study “B” identified by the intrinsic gene list “b”) (7,12,13).
Although hierarchical clustering has been widely used to identify molecular subtypes of breast cancer, this approach can only be applied retrospectively to sufficiently sized cohorts of patients (10,15) but not prospectively to individual samples. Therefore, three microarray-based “Single Sample Predictors” (SSPs) based on centroids (ie, the mean expression profile of each subtype) were developed (12–14). To define the SSPs, each molecular subtype was initially identified by hierarchical clustering based on the intrinsic gene list, and then the centroids of each molecular subtype (ie, luminal A, luminal B, HER2, basal-like, and normal breast-like) were derived. These SSPs, which can be applied to individual samples based on the correlations between the expression profile of a given sample and each of the centroids, recapitulate the classification obtained with hierarchical cluster analysis. We (16) and others (17) have recently demonstrated that the agreement between these SSPs is modest and that they can only reliably identify basal-like breast cancers.
Previous studies have highlighted the biostatistical limitations of hierarchical cluster analysis of microarray expression profiles for the identification of molecular subtypes of breast cancer and the relative instability of some of the molecular subtypes identified by this type of approach (15,18–21). One fundamental aspect of microarray-based class discovery studies, which has not been systematically analyzed, is the subjectivity involved in assigning the molecular subtypes through the analysis of dendrograms generated with hierarchical clustering methods.
The aim of this study was to determine the objectivity and interobserver reproducibility of the assignment of molecular subtype classes by hierarchical clustering (ie, do different observers assign the same patients to the same molecular subtype when they analyze the same dendrogram?). To address this question, we subjected three breast cancer datasets in the public domain [NKI-295 (22), TransBig (23), and Wang (24)] to hierarchical cluster analysis using five intrinsic gene lists from Perou et al. (5), Sorlie et al. (11), Sorlie et al. (12), Hu et al. (13), and Parker et al. (14). Subsequently, we determined the interobserver reproducibility among five breast cancer researchers who are experienced in molecular subtype assignment using the dendrograms and heatmaps generated by hierarchical clustering methods and the five intrinsic gene lists.
Material and Methods
Microarray Datasets
Microarray data from the publicly available breast cancer datasets, NKI-295 (22) (n = 295), Wang (24) (n = 286), and TransBig (23) (n = 198), were used for hierarchical cluster analysis. The normalized microarray-based gene expression data were retrieved from the internet or public repositories (NKI-295: http://microarray-pubs.stanford.edu/wound_NKI/explore.html; Wang: GEO:GSE2034; TransBig: GEO:GSE7390). Further details about the datasets and data acquisition are provided in Supplementary Table 1 (available online).
Hierarchical Cluster Analysis
The assignment of molecular subtypes of breast cancer based on hierarchical cluster analysis was essentially performed as previously described (5,11–14,25). The distinct intrinsic gene lists reported by Perou et al. (5) (496 probes corresponding to 349 unique Human Genome Organization [HUGO] [http://www.genenames.org/] gene symbols), Sorlie et al. (11) (456 probes corresponding to 395 unique HUGO gene symbols), Sorlie et al. (12) (552 probes corresponding to 492 unique HUGO gene symbols), Hu et al. (13) (1400 probes corresponding to 1176 unique HUGO gene symbols), and Parker et al. (14) (1918 probes and 1918 unique HUGO gene symbols) were retrieved (Supplementary Tables 2–6 and Supplementary Methods, available online).
A substantial proportion of the gene identifiers reported in the original publications have changed in more recent genome builds; therefore, annotations of intrinsic gene lists and breast cancer datasets were comprehensively updated and mapped to build 36 of the human genome (Ensembl assembly 54 [http://www.ensembl.org/index.html]) as described previously (16) (Supplementary Tables 2–6, available online). Of the identifiers tested (ie, HUGO gene symbols, Ensembl, and Unigene [http://www.ncbi.nlm.nih.gov/unigene]), the annotation with HUGO gene symbols allowed for the retrieval of the highest proportion of genes in the majority of intrinsic gene lists and datasets (Supplementary Table 7, available online). As observed in the original dendrograms and descriptions of the intrinsic gene lists (5,11–13), when multiple probes mapped to the same gene, all were included in the hierarchical cluster analysis. Analyses were performed in R version 2.9.0 (http://cran.rproject.org/).
Two-way average-linkage hierarchical clustering (median centered by feature and gene and Pearson correlation as the gene similarity metric) was applied to each dataset using Cluster 3.0 [http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm#ctv] as previously described (11–14,25), and results were visualized with Java Treeview [http://jtreeview.sourceforge.net/]. Additional details of the microarray data handling and hierarchical clustering are available in the Supplementary Methods (available online). Annotated datasets are available at http://rock.icr.ac.uk/collaborations/Mackay/centroid.correlations.Eset; annotated intrinsic gene lists used for hierarchical clustering, clustered data and Java Treeview files for each of the datasets presented are available at http://rock.icr.ac.uk/collaborations/Mackay/observer.clustering.
Molecular Subtype Assignment
To determine the reproducibility of microarray-based classification of breast cancers by hierarchical cluster analysis, the study curator (J. S. Reis-Filho) selected five researchers on the basis of experience in microarray-based expression profiling analysis, previous publications on the use of microarray-based molecular taxonomy of breast cancer, and having a first or senior author publication on microarrays in a journal with a 2008 Thompson ISI impact factor greater than 5.
The study curator created guidelines that described in detail how each molecular subtype should be identified by the visual analysis of the dendrograms obtained with hierarchical cluster analysis for each intrinsic gene list. These guidelines were based on extracts from the studies by Perou et al. (5), Sorlie et al. (11), Sorlie et al. (12), Hu et al. (13), and Parker et al. (14), and they summarized the characteristics of each molecular subtype according to each intrinsic gene list, provided graphical representations of the dendrograms and heatmaps obtained by applying each intrinsic gene list to a separate dataset of breast cancers, and additional details extracted from their respective original studies (for details see Supplementary Methods, available online). It should be emphasized that the original publications (5,11–14) did not provide clear guidelines of the levels at which the dendrogram branches should be cut to define the molecular subtypes.
The guidelines, the dendrograms, and color heatmaps obtained from hierarchical cluster analysis of the three breast cancer datasets (22–24) using the five intrinsic gene lists (5,11–14) (Supplementary Figures 1–15, available online) were sent to five of the authors (A. Mackay, B. Weigelt, A. Grigoriadis, B. Kreike, R. Natrajan) via email, together with a copy of the original studies (5,11–14) describing the intrinsic gene lists. Observers were requested to classify each dataset according to the methods described by Perou et al. (5), Sorlie et al. (11), Sorlie et al. (12), Hu et al. (13), and Parker et al. (14), identifying all molecular subtypes described in each publication. If samples in a dendrogram could not be assigned to a molecular subtype with confidence, the observers could opt for considering the sample as unclassifiable, as done in Sorlie et al. (12) and Parker et al. (14). A request to keep the correspondence strictly confidential was made, and no discussions with other researchers were permitted. The identity of each observer was kept confidential from the other study participants. Molecular subtype assignments were made by each observer blinded to the results reported by the other observers and sent directly to the study curator.
Analysis of Agreement
Statistical analysis of the molecular subtype assignments made by the five observers was performed by two of the authors (RA’H and JSR-F), without providing any feedback to the observers. The percentage of overall agreement and the multirater analysis of agreement was performed as previously described (26). We used the free-marginal Kappa statistics of Brennan and Prediger (26), which is optimal for the assessment of agreement among more than two observers (ie, raters) when categorical variables are used, and observers are not forced to assign a certain number of samples to each category. The choice of free-marginal Kappa score was based on the fact that this method minimizes prevalence-related biases and would be compatible with the choice of observers considering samples that could not be assigned to a molecular subtype as unclassifiable. Using this statistical method, Kappa values can range from −1.0 to 1.0, with −1.0 indicating perfect disagreement below chance, 0.0 agreement equal to chance, and 1.0 perfect agreement above chance. Kappa values can be interpreted as follows: 0.01–0.2 as slight agreement, 0.21–0.4 as fair agreement, 0.41–0.6 as moderate agreement, 0.61–0.8 as substantial agreement, and 0.81–1.0 as almost perfect agreement (26–28).
We determined the interobserver agreement for the whole classification obtained from the analysis of the dendrogram produced with each intrinsic gene list for each breast cancer dataset [NKI-295 (22), Wang (24) and TransBig (23)]. In addition, we analyzed interobserver agreement for each molecular subtype according to each intrinsic gene list for each breast cancer dataset.
To test whether the free-marginal Kappa scores, when the luminal group was subdivided into the A, B, and C subgroups [Sorlie et al. (11)], were statistically significantly lower than the free-marginal Kappa scores when the luminal cancers were considered as a single group [Perou et al. (5)], we used a nonparametric test (Mann–Whitney U test).
Samples from these datasets were not previously classified by the proponents (5,11–14) of the molecular classification into the molecular subtypes by means of hierarchical clustering using all five intrinsic gene sets tested in this work. Thus, given that there is no available “gold standard” for the classification of samples from the three breast cancer datasets analyzed here into molecular subtypes by hierarchical clustering for all intrinsic gene lists, we also determined the percentage of samples with perfect agreement (samples for which all raters/observers agreed on the classification of the sample) and the percentage of samples with a “majority consensus” (samples for which three or more raters/observers agreed on the classification of a given sample into one of the molecular subtypes).
Results
Whole Classification Scheme
We first sought to define the reproducibility by different observers of the whole classification system according to Perou et al. (5), Sorlie et al. (11), Sorlie et al. (12), Hu et al. (13), and Parker et al. (14). None of the classification systems tested produced almost perfect agreement (free-marginal Kappa scores ≥ 0.81) among observers (Table 1, Supplementary Tables 8–10 available online).
Table 1.
Measures of agreement among five observers using five intrinsic gene lists in three breast cancer datasets*
| Gene list source | Overall agreement, % of samples | Free-marginal Kappa | Agreement 5/5, No. of samples (%) | Agreement 4/5, No. of samples (%) | Agreement 3/5, No. of samples | Majority consensus, % of samples |
| NKI-295 (n = 295)† | ||||||
| Perou et al. (5) | 70.8 | 0.635 | 125 (42.4) | 204 (69.2) | 295 | 100.0 |
| Sorlie et al. (11) | 62.4 | 0.561 | 136 (46.1) | 148 (50.2) | 233 | 79.0 |
| Sorlie et al. (12) | 68.7 | 0.624 | 119 (40.3) | 251 (85.1) | 251 | 85.1 |
| Hu et al. (13) | 78.9 | 0.754 | 182 (61.7) | 227 (76.9) | 295 | 100.0 |
| Parker et al. (14) | 75.7 | 0.708 | 172 (58.3) | 196 (66.4) | 295 | 100.0 |
| TransBig (n = 198)† | ||||||
| Perou et al. (5) | 76.1 | 0.701 | 121 (61.1) | 126 (63.6) | 194 | 98.0 |
| Sorlie et al. (11) | 64.1 | 0.582 | 82 (41.4) | 95 (48) | 193 | 97.5 |
| Sorlie et al. (12) | 65.2 | 0.582 | 99 (50) | 99 (50) | 164 | 82.8 |
| Hu et al. (13) | 78.4 | 0.748 | 115 (58.1) | 153 (77.3) | 195 | 98.5 |
| Parker et al. (14) | 56.2 | 0.475 | 58 (29.3) | 98 (49.5) | 173 | 87.4 |
| Wang (n = 286)† | ||||||
| Perou et al. (5) | 73.8 | 0.672 | 182 (63.6) | 185 (64.7) | 249 | 87.1 |
| Sorlie et al. (11) | 51.5 | 0.435 | 50 (17.5) | 136 (47.6) | 253 | 88.5 |
| Sorlie et al. (12) | 65.1 | 0.581 | 112 (39.2) | 175 (61.2) | 277 | 96.9 |
| Hu et al. (13) | 59.4 | 0.526 | 89 (31.1) | 152 (53.1) | 273 | 95.5 |
| Parker et al. (14) | 70.5 | 0.646 | 168 (58.7) | 173 (60.5) | 243 | 85.0 |
Free-marginal Kappa statistics were used to assess agreement among the five raters. Kappa values of 0.01–0.2 indicate slight agreement; 0.21–0.4, fair agreement; 0.41–0.6, moderate agreement; 0.61–0.8, substantial agreement; and 0.81–1.0, almost perfect agreement. Agreement 5/5 = perfect agreement among all five observers; agreement 4/5 = perfect agreement among four or five out of five observers; agreement 3/5 = perfect agreement among three of more observers out of five observers; majority consensus = three or more observers agreed on the classification of a given sample into one of the molecular subtypes.
For Perou et al. (5), four molecular subtypes were described, luminal, basal-like, HER2, and normal breast-like. The interobserver overall agreement for this classification system ranged from 70.8% to 76.1% of the samples according to the dataset analyzed, and the free-marginal Kappa scores ranged from 0.635 to 0.701 (ie, substantial agreement [Kappa scores ≥ 0.61] among observers in all datasets; Table 1; Figures 1, A, 2, A, and 3, A; Supplementary Figures 1–3, available online). Perfect interobserver agreement (five out of five observers) was found in 42.4% to 63.6% of samples. Importantly, interobserver disagreement in the classification system proposed by Perou et al. (5) was restricted to the assignment of luminal and normal breast-like subtypes (Table 2).
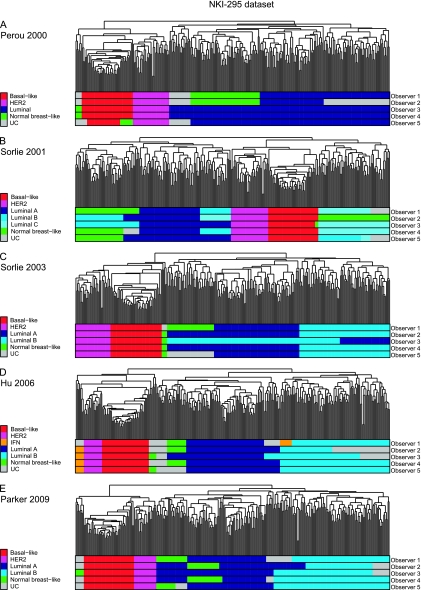
Figure 1.
Molecular subtype classification of the NKI-295 (22) dataset by five observers based on hierarchical cluster analysis using the “intrinsic” gene lists described by A) Perou et al. (5), B) Sorlie et al. (11), C) Sorlie et al. (12), D) Hu et al. (13), and E) Parker et al. (14). HER2 = human epidermal growth factor receptor 2; UC = unclassified.
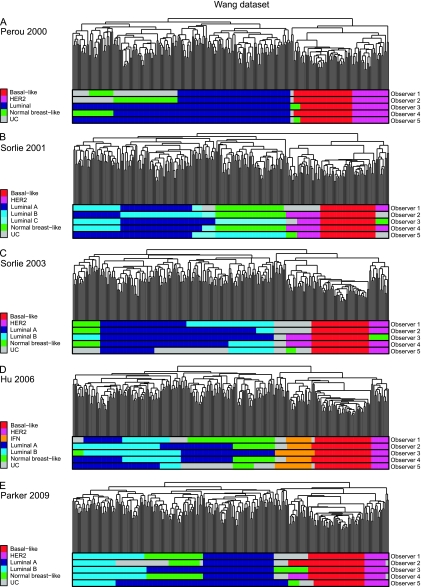
Figure 2.
Molecular subtype classification of the Wang (24) dataset by five observers based on hierarchical cluster analysis using the “intrinsic” gene lists described by A) Perou et al. (5), B) Sorlie et al. (11), C) Sorlie et al. (12), D) Hu et al. (13), and E) Parker et al. (14). HER2 = human epidermal growth factor receptor 2; UC = unclassified.
Figure 3.
Molecular subtype classification of the TransBig (23) dataset by five observers based on hierarchical cluster analysis using the “intrinsic” gene lists described by A) Perou et al. (5), B) Sorlie et al. (11), C) Sorlie et al. (12), D) Hu et al. (13), and E) Parker et al. (14). HER2 = human epidermal growth factor receptor 2; UC = unclassified.
Table 2.
Measures of agreement among five observers for each molecular subtype individually using five intrinsic gene lists in three breast cancer datasets*
| Molecular subtype | Breast cancer datasets† |
|||||||||||
| NKI-295 (n = 295) |
TransBig (n = 198) |
Wang (n = 286) |
||||||||||
| OA, % | F-M Kappa | PA, No. | MAX, No. | OA, % | F-M Kappa | PA, No. | MAX, No | OA, % | F-M Kappa | PA, No. | MAX, No. | |
| Perou et al. (5) | ||||||||||||
| Basal-like | 97.7 | 0.954 | 31 | 48 | 97.6 | 0.952 | 36 | 44 | 98.7 | 0.975 | 47 | 53 |
| Luminal | 74.3 | 0.486 | 60 | 207 | 80.6 | 0.612 | 73 | 137 | 80.1 | 0.601 | 102 | 197 |
| HER2 | 100 | 1.000 | 34 | 34 | 99 | 0.980 | 12 | 17 | 100 | 1.000 | 33 | 33 |
| Normal breast-like | 83.9 | 0.679 | 0 | 65 | 80 | 0.600 | 0 | 64 | 83.5 | 0.670 | 0 | 58 |
| UC | 85.6 | 0.713 | 0 | 88 | 94.9 | 0.899 | 0 | 18 | 85.2 | 0.705 | 0 | 76 |
| Sorlie et al. (11) | ||||||||||||
| Basal-like | 99.6 | 0.992 | 44 | 47 | 96.6 | 0.931 | 28 | 42 | 100 | 1.000 | 50 | 50 |
| Luminal A | 91.1 | 0.821 | 57 | 101 | 95.4 | 0.907 | 42 | 65 | 78.3 | 0.566 | 0 | 108 |
| Luminal B | 77.5 | 0.550 | 0 | 60 | 73.6 | 0.473 | 0 | 74 | 71.7 | 0.434 | 0 | 74 |
| Luminal C | 83.1 | 0.661 | 0 | 67 | 84.2 | 0.685 | 0 | 33 | 86.6 | 0.733 | 0 | 64 |
| HER2 | 100 | 1.000 | 35 | 35 | 99 | 0.980 | 12 | 17 | 92.4 | 0.849 | 0 | 43 |
| Normal breast-like | 79.3 | 0.586 | 0 | 67 | 85.6 | 0.711 | 0 | 33 | 83.5 | 0.670 | 0 | 64 |
| UC | 94.3 | 0.886 | 0 | 18 | 93.9 | 0.879 | 0 | 20 | 90.5 | 0.810 | 0 | 45 |
| Sorlie et al. (12) | ||||||||||||
| Basal-like | 100 | 1.000 | 48 | 48 | 95.9 | 0.917 | 39 | 54 | 100 | 1.000 | 52 | 52 |
| Luminal A | 73.8 | 0.477 | 0 | 124 | 77 | 0.539 | 48 | 115 | 80.5 | 0.610 | 49 | 157 |
| Luminal B | 76.8 | 0.536 | 38 | 162 | 78.7 | 0.574 | 0 | 67 | 83.7 | 0.674 | 0 | 79 |
| HER2 | 100 | 1.000 | 33 | 33 | 100 | 1.000 | 12 | 12 | 92.7 | 0.853 | 0 | 41 |
| Normal breast-like | 93.4 | 0.867 | 0 | 44 | 88.5 | 0.770 | 0 | 34 | 91 | 0.820 | 0 | 25 |
| UC | 93.4 | 0.867 | 0 | 44 | 90.3 | 0.806 | 0 | 24 | 82.3 | 0.646 | 11 | 117 |
| Hu et al. (13) | ||||||||||||
| Basal-like | 100 | 1.000 | 44 | 44 | 98.8 | 0.976 | 37 | 43 | 99.4 | 0.987 | 51 | 54 |
| Luminal A | 94.5 | 0.890 | 73 | 91 | 94.2 | 0.885 | 46 | 65 | 68 | 0.359 | 0 | 92 |
| Luminal B | 87.3 | 0.745 | 38 | 103 | 82.4 | 0.648 | 0 | 66 | 83.1 | 0.663 | 0 | 88 |
| HER2 | 100 | 1.000 | 17 | 17 | 97.8 | 0.956 | 12 | 23 | 100 | 1.000 | 16 | 16 |
| Normal breast-like | 94.9 | 0.898 | 0 | 18 | 87.5 | 0.749 | 0 | 26 | 86.2 | 0.724 | 0 | 79 |
| IFN | 97.4 | 0.948 | 0 | 11 | 100 | 1.000 | 20 | 20 | 97.3 | 0.947 | 22 | 36 |
| UC | 83.8 | 0.676 | 10 | 71 | 96.1 | 0.921 | 0 | 10 | 84.7 | 0.694 | 0 | 69 |
| Parker et al. (14) | ||||||||||||
| Basal-like | 100 | 1.000 | 47 | 47 | 99.8 | 0.996 | 40 | 41 | 96.8 | 0.936 | 46 | 69 |
| Luminal A | 82.1 | 0.642 | 41 | 110 | 74.7 | 0.495 | 0 | 73 | 83.6 | 0.671 | 64 | 156 |
| Luminal B | 91.5 | 0.831 | 63 | 109 | 68.4 | 0.368 | 0 | 73 | 94.1 | 0.883 | 39 | 67 |
| HER2 | 100 | 1.000 | 21 | 21 | 99.4 | 0.988 | 18 | 20 | 96.9 | 0.937 | 19 | 37 |
| Normal breast-like | 87.3 | 0.745 | 0 | 33 | 82.8 | 0.657 | 0 | 39 | 84 | 0.680 | 0 | 53 |
| UC | 90.4 | 0.809 | 0 | 32 | 87.3 | 0.745 | 0 | 46 | 85.7 | 0.715 | 0 | 67 |
Free-marginal Kappa statistics were used to assess agreement among the five raters. Kappa values of 0.01–0.2 indicate slight agreement; 0.21–0.4, fair agreement; 0.41–0.6, moderate agreement; 0.61–0.8, substantial agreement; and 0.81–1.0, almost perfect agreement. OA = overall agreement; F-M Kappa = free-marginal Kappa scores; PA = number of samples with perfect agreement; MAX = maximum number of samples; HER2 = human epidermal growth factor receptor 2; IFN = interferon-regulated molecular subtype; UC = unclassified.
With the introduction of subdivisions of the luminal molecular subtype into luminal A, luminal B, and luminal C in Sorlie et al. (11), the overall agreement rates (51.5% to 64.1%) and the free-marginal Kappa scores were substantially reduced (0.435–0.582—only moderate agreement among observers; Mann–Whitney U test one-tailed P = .05) (Table 1, Figures 1, B, 2, B, and 3, B, and Supplementary Figures 4–6, available online). Perfect interobserver agreement was 17.5% to 46.1%, and a majority consensus (three or more observers agreeing on the classification of a given sample) was found in 79% (NKI-295 dataset) to 97.5% (TransBig dataset) of the samples.
Of the remaining classification systems, including subdivisions of luminal cancers into luminal A and luminal B (12–14) (Supplementary Figures 7–15, available online), none produced free-marginal Kappa scores of at least 0.61 in all datasets (Table 1). It should be noted, however, that the Hu et al. (13) classification system had better overall agreement and free-marginal Kappa scores than the other classification systems with five or more subtypes in the NKI-295 and TransBig datasets (11,12,14) (Table 1, Figures 1, D, 2, D, and 3, D, Supplementary Figures 10–12, available online). Importantly, more than 95% of samples with a majority consensus were found in only two or more datasets when the Perou et al. (5) and Hu et al. (13) classifications were used, but not with Sorlie et al. (11), Sorlie et al. (12) or Parker et al. (14) (Table 1).
Analysis of Agreement of Each Molecular Subtype
Analysis of the interobserver agreement for the identification of each molecular subtype separately revealed that basal-like cancers could be reproducibly identified by independent observers in all datasets regardless of the classification system used, with overall agreement rates consistently greater than 95%, free-marginal Kappa scores of at least 0.81, and a majority consensus greater than 90% in all datasets (Table 2 and Supplementary Table 11, available online).
HER2-positive cancers also consistently displayed free-marginal Kappa scores of at least 0.81 and overall agreement rates greater than 90%; a majority consensus was found in more than 95% of samples in the NKI-295 (22) and TransBig (23) datasets, regardless of the classification used; however, in the Wang dataset, a majority consensus was found in more than 90% of samples only when Perou et al. (5) or Hu et al. (13) intrinsic gene lists were used (Table 2 and Supplementary Table 11, available online); with the other intrinsic gene lists, discordances were observed in the assignment of HER2-positive tumors as unclassifiable (Figure 2 and Supplementary Figures 2 and 14, available online).
The identification of luminal cancers and their subgroups and normal breast-like cancers failed to show acceptable levels of overall agreement or to consistently display free-marginal Kappa scores of at least 0.81 in all datasets (Table 2 and Supplementary Table 11, available online). When the Sorlie et al. (11) classification system comprising three categories of luminal cancers (ie, luminal A, B, and C) was used, a majority consensus for the classification of samples was found in less than 50% of luminal cancers [46.5% in NKI-295 (22), 50% in TransBig (23), and 48.8% in the Wang (24) dataset; Supplementary Table 11]. With the use of two categories of luminal cancers (ie, luminal A and luminal B) (12–14), a majority consensus for more than 50% of luminal cancers in all datasets was only found when hierarchical clustering using the intrinsic gene list of Hu et al. (13) was used. Notably, the interferon-rich subtype, only identified in the Hu et al. (13) intrinsic gene list, displayed almost perfect agreement levels among observers (overall agreement >97% and free-marginal Kappa scores ≥ 0.81; Table 2).
Discussion
The results presented in this study provide direct evidence that the identification of subgroups of luminal cancers and normal breast-like cancers by visual inspection of dendrograms obtained from hierarchical cluster analysis shows suboptimal levels of interobserver agreement, even when the molecular subtypes are known a priori, and guidelines for the identification of these subtypes are provided. The identification of basal-like and HER2 cancers showed almost perfect interobserver agreement rates regardless of the intrinsic gene list used.
Microarray-based expression profiling analysis has led to a paradigm shift in the way breast cancer is perceived (9,10). Class discovery studies have demonstrated the existence of five main molecular subtypes, namely basal-like, HER2, luminal A, luminal B, and normal breast-like, but luminal C and interferon-regulated subtypes have also been described (5,9–14). These five main subtypes have been reported to have distinct clinical presentations (29), sites of relapse (30), histological features (31), responses to chemotherapy (14,32), and outcomes (10,11,13,15). Despite being derived from unsupervised approaches for class discovery, this molecular classification to some extent recapitulates the clinical subgroups of breast cancer identified in clinical practice. In fact, there is evidence to suggest a strong association between the molecular subtype classes (luminal A, luminal B, HER2, and basal-like) and the clinical categories of breast cancer (tamoxifen-sensitive estrogen receptor positive [ER+], tamoxifen-resistant ER+, trastuzumab-sensitive, and other) (9,10).
It has been argued that microarray expression profiling is the gold standard for breast cancer classification (7); however, several lines of evidence suggest that there are major limitations in our ability to assign samples consistently to specific molecular subtypes (10,15,33). We and others have recently demonstrated that SSPs fail to assign individual samples reproducibly into molecular subtypes (16,17). Here, we demonstrate that apart from basal-like and HER2 breast cancer subtypes, the interobserver reproducibility of breast cancer molecular subtype assignment using the methods and approaches originally used for this purpose is modest, in particular for the identification of luminal A, luminal B, and normal breast-like subtypes. None of the intrinsic gene lists concurrently provided almost perfect agreement (ie, free-marginal Kappa scores ≥0.81) for the luminal A, luminal B, and normal breast-like subtypes in any of the datasets. In comparison, for example, the interobserver agreement of ER and HER2 immunohistochemical staining of breast cancers using tissue microarrays has been reported to be high (Kappa scores ≥ 0.81) (34–37). Furthermore, the Kappa scores observed in this study overlap with those observed in analyses of interobserver agreement of histological grade (Kappa scores ranging from 0.43 to 0.83) (38). It should be noted that similar Kappa scores have been considered by many as inadequate and as evidence for the subjective nature of histological grade (39,40).
It is plausible that the limited interobserver agreement for the subclassification of luminal cancers may stem from attempting to identify distinct groups within a continuum (41). The distinction between luminal A and luminal B tumors is reported to be principally driven by the expression of proliferation-related genes (7,13); however, several studies have recently demonstrated that proliferation in ER+ breast cancers is a continuum rather than a bimodal distribution (10,41,42). Therefore, allocation of specific subgroups (eg, luminal A and B) by hierarchical cluster analysis is likely to be arbitrary and to depend on the population of samples subjected to the analysis (15,43), which may explain why the luminal B cluster was identified in the ER+ arm of the cluster dendrogram in three studies (11,13,14) and in the ER− arm in one study (12).
The lack of agreement in the identification of normal breast-like tumors should perhaps not come as a surprise, given that these tumors may constitute an artifact of gene expression profiling analysis [ie, analysis of tumor specimens with a disproportionately high percentage of normal tissue “contamination” (7,13,14)]. The normal breast-like gene cluster in the heatmaps has been either represented by only a few genes [ie, Sorlie et al. (12)] or not even specified [ie, Hu et al. (13)]. Moreover, this gene cluster was composed of different genes in studies in which it was reported (5,11,12). Notably, normal breast-like tumors have been reported in both the ER− (5,11,12) and the ER+ branch of the cluster dendrogram (13,14), which may have contributed to the poor reproducibility of the normal breast-like subtype assignment among different observers in this study.
Hierarchical cluster analysis was the method of choice for the development of the current working model of microarray-based breast cancer taxonomy (5,11–14) and of the SSPs, which can be applied prospectively to single samples for molecular subtype assignment. However, previous studies (15,19) (and references therein) have demonstrated that hierarchical cluster analysis has several limitations for the identification of subtypes of breast cancer, that is, relevant features and distance measures have to be selected a priori, the actual number of clusters is unknown, and clusters are always generated even when random data are used (21). Therefore, the emergence of “clusters” does not necessarily equate with biological significance. The interpretation of dendrograms resulting from the analysis of breast cancers is by no means trivial, as illustrated here (Figures 1–3; Supplementary Figures 1–15, available online); however, it becomes even more complex when different dendrograms are cut at different levels and different methods and approaches are used. In fact, in different studies, dendrograms obtained from hierarchical clustering using different intrinsic gene lists were cut at different levels (5,11,12,14), and sometimes molecular subtypes were defined in the same dendrogram by cutting the branches at different levels [eg, molecular subtype assignments described in Sorlie et al. (11)]. In the most recent publication by Parker et al. (14), the cluster dendrogram was analyzed using “SigClust” (44), a tool for assessing statistical significance of a cluster. This method was used to identify “prototypic tumor samples” from each of the molecular subtypes, which were used to derive a minimized gene set for the development of a 50-gene set for quantitative reverse transcription polymerase chain reaction for sample subtype prediction (PAM50). Conceivably, this method might lead to a more consistent assignment of clusters; it should be noted, however, that three different SSPs, two of which were generated with subtypes initially identified without the use of SigClust, showed equivalent associations with outcome in three distinct datasets (16).
The interpretation of the clusters identified by Perou et al. (5) was based on the relationship between the genes over- or underexpressed in samples classified into each cluster, and clinical and biological characteristics of breast cancers that were already known. Surprisingly, some of the genes that defined the initial subtypes were not present in subsequent versions of the intrinsic gene lists [eg, keratin 8/18 (KRT8/18), one of the defining features of luminal cancers, is not present in Sorlie et al. (12); keratin 17 (KRT17), but not keratin 5 (KRT5), the defining features of basal-like cancers, is not present in Hu et al. (13); aquaporin 7 (AQP7), integrin alpha 7 (ITGA7), thrombospondin receptor (CD36) [Sorlie et al. (11), aldo-keto reductase family 1, member C1 (AKR1C1) and phosphoinositide-3-kinase, regulatory subunit 1(α) (PIK3R1) (Sorlie et al. (12)], genes pertaining to the normal breast-like cluster in Sorlie et al. (11) and Sorlie et al. (12), respectively, are not present in Hu et al. (13)]. Another important limitation of hierarchical cluster analysis, as elegantly illustrated by Pusztai et al. (15), is the lack of stability of some of the subgroups identified. Although there are algorithms to determine the stability of clusters generated by hierarchical clustering (21), they do not provide an assessment of interobserver variability in molecular subtype assignment via inspection of the dendrograms. In this study, we systematically analyzed the ability of experienced observers to identify the molecular subtypes through the analysis of dendrograms and demonstrated that even when clear guidelines are provided, the assignment of samples is subjective and not entirely reproducible.
Several groups, including ours, have previously attempted to define the molecular subtypes in breast cancer datasets using hierarchical cluster analysis (30,45–49). Given the lack of interobserver agreement and stability of some of the molecular subtypes, as discussed above, our findings indicate that breast cancer molecular subtype classifications performed by other investigators may not have necessarily reproduced those originally described and that molecular subtypes identified by the same intrinsic gene list in different cohorts analyzed by different observers are not necessarily equivalent.
This study had several limitations. First, the datasets used were retrospectively accrued; hence, they may not have a balanced distribution of the different molecular subtypes and do not include samples of normal breast tissue. Second, we used the publicly available processed microarray for the hierarchical cluster analyses; no attempts to renormalize the data were made. Third, although we endeavored to reannotate all genes from each gene list, a small proportion of genes from the intrinsic gene lists could not be annotated in all datasets owing to different microarray platforms and changes in gene annotation (see Supplementary Methods, available online). Fourth, the datasets included in this study derive from different microarray platforms. Fifth, one cannot rule out that if five observers from the groups of the proponents of the breast cancer taxonomy (5,11–13,36) were asked to assign the molecular subtypes based on the visual inspection of dendrograms and gene clusters, a better interobserver agreement would be found. Finally, this study focused on the interobserved reproducibility of the assignment of molecular subtypes by inspection of dendrograms and gene clusters; we have not investigated whether bioinformatic methods to define the statistical robustness of clusters [eg, SigClust (44), Pvclust (50), or R and D indices (21)] would increase the reproducibility.
It is beyond the scope of this work to evaluate algorithms for cluster analysis, and the choices of distance metrics and linkage. Instead, we have focused on the human-dependent component of class discovery analysis and demonstrated that this subjective component leads to substantial variability in cluster assignment. Moreover, a direct comparison between the molecular subtypes identified by distinct intrinsic gene lists applied to the same datasets [eg, Perou et al. (5) vs Parker et al. (14), Hu et al. (13) vs Sorlie et al. (11)] would provide an inflated rate of disagreement, because different numbers of subclasses were reported in each classification, and there is no gold standard for each of the intrinsic gene lists. In fact, the reported agreement between the Sorlie et al. (12) and Hu et al. (13) intrinsic gene lists, when applied to the same dataset, was 78% when the samples classified as the interferon-rich subtype were excluded (13).
The subjectivity and modest reproducibility of the interpretation of histopathologic features and immunohistochemical stainings have been heavily criticized, and the need for more objective methods to guide the breast cancer patient in decision making is clearly justified. Hierarchical cluster analysis is undoubtedly a powerful tool for class discovery and a useful first step for the development of a molecular classification. However, hierarchical clustering may not be an ideal choice as a method for breast cancer classification because it is neither entirely objective nor are its results entirely reproducible. In fact, current molecular classification systems for breast cancer are similarly to histopathology, descriptive, and prognostic (10,16). Based on the available data and the limitations of our knowledge on the heterogeneity of breast cancers, it is still not possible to determine with absolute certainty how many molecular subtype classes do exist (15). Hence, with the increasingly more coherent information about the genetic (51) and transcriptomic features of breast cancer (9,10,52), mechanisms of action of chemotherapy agents, and availability of humanized monoclonal antibodies and small molecule inhibitors that target specific molecular pathways and networks, perhaps molecular classification should be designed to provide more direct functional information for clinicians to facilitate the treatment of breast cancer patients. For example, a certain breast cancer subtype could be classified based on the presence or absence of overexpressed or mutated genes that may serve as predictive biomarkers for specific therapeutic agents (9,10,53). The development of such a taxonomy is likely to require integrative approaches combining descriptive analysis of genomic, transcriptomic, and proteomic data from sufficiently statistically powered cohorts (54) with multidimensional data from global functional analyses of large panels of cancer models (eg, genome-wide RNA interference screens and chemical screens) (9,10,51,53,55,56).
In conclusion, we demonstrate that assignment of molecular subtypes of breast cancer based on the visual inspection of dendrograms obtained with hierarchical cluster analysis is subjective and shows modest interobserver reproducibility, in particular when subclassification of luminal cancers into two or more groups is required. These results suggest that class discovery studies, in which subtypes are identified by inspection of dendrograms [eg, (5,11,12)], need to take into account both the stability of the clusters and the reproducibility of the classification system (16,41). This is of paramount importance, as SSPs used for the prospective classification of breast cancer patients into specific molecular subtypes have been derived from the analysis of subtypes originally identified by hierarchical clustering methods (12–14). For the incorporation of the molecular taxonomy into clinical trials, routine clinical practice and treatment decision making, stringent standardization of methodologies for the identification of breast cancer molecular subtypes, and objective definitions for each molecular subtype are of utmost importance.
Funding
This work was supported by Breakthrough Breast Cancer. B.W. is funded by a Cancer Research UK postdoctoral fellowship. We also acknowledge NHS funding to the NIHR Biomedical Research Centre. Jorge S. Reis-Filho is the recipient of the 2010 Cancer Research UK Future Leaders Prize.
Supplementary Material
Footnotes
A. Mackay and B. Weigelt contributed equally to this study. The authors have no conflict of interest to declare. The funders did not have any involvement in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
References
- 1.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen TO, West RB, Linn SC, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359(9314):1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- 3.Khan J, Wei JS, Ringner M, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7(6):673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bittner M, Meltzer P, Chen Y, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406(6795):536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 5.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98(24):13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peppercorn J, Perou CM, Carey LA. Molecular subtypes in breast cancer evaluation and management: divide and conquer. Cancer Invest. 2008;26(1):1–10. doi: 10.1080/07357900701784238. [DOI] [PubMed] [Google Scholar]
- 8.He YD, Friend SH. Microarrays—the 21st century divining rod? Nat Med. 2001;7(6):658–659. doi: 10.1038/89022. [DOI] [PubMed] [Google Scholar]
- 9.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 10.Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220(2):263–280. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 11.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pusztai L, Mazouni C, Anderson K, Wu Y, Symmans WF. Molecular classification of breast cancer: limitations and potential. Oncologist. 2006;11(8):868–877. doi: 10.1634/theoncologist.11-8-868. [DOI] [PubMed] [Google Scholar]
- 16.Weigelt B, Mackay A, A’Hern R, Natrajan R, Tan DS, Dowsett M, et al. Breast cancer molecular profiling: a retrospective analysis of molecular subtype assignment using single sample predictors. Lancet Oncol. 2010;11(4):339–349. doi: 10.1016/S1470-2045(10)70008-5. [DOI] [PubMed] [Google Scholar]
- 17.Haibe-Kains B, Culhane A, Desmedt C, Bontempi G, Quackenbush J, Sotiriou C. Robustness of breast cancer molecular subtypes identification. Ann Oncol. 2010;21(suppl 4):iv49–iv59. [Google Scholar]
- 18.Dobbin K, Simon R. Comparison of microarray designs for class comparison and class discovery. Bioinformatics. 2002;18(11):1438–1445. doi: 10.1093/bioinformatics/18.11.1438. [DOI] [PubMed] [Google Scholar]
- 19.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99(2):147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 20.Michiels S, Koscielny S, Hill C. Interpretation of microarray data in cancer. Br J Cancer. 2007;96(8):1155–1158. doi: 10.1038/sj.bjc.6603673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McShane LM, Radmacher MD, Freidlin B, Yu R, Li MC, Simon R. Methods for assessing reproducibility of clustering patterns observed in analyses of microarray data. Bioinformatics. 2002;18(11):1462–1469. doi: 10.1093/bioinformatics/18.11.1462. [DOI] [PubMed] [Google Scholar]
- 22.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 23.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 25.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennan RL, Prediger DJ. Coefficient kappa: some uses, misuses, and alternatives. Educ Psychol Meas. 1981;41:687–699. [Google Scholar]
- 27.Randolph JJ. Joensuu University Learning and Instruction Symposium. Joensuu, Finland: October 14-15, 2005. Free-marginal multirater kappa: an alternative to Fleiss’ fixed-marginal multirater kappa. [Google Scholar]
- 28.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- 29.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 30.Smid M, Wang Y, Zhang Y, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68(9):3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 31.Livasy CA, Karaca G, Nanda R, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19(2):264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 32.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11(16):5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 33.Gusterson B. Do ‘basal-like’ breast cancers really exist? Nat Rev Cancer. 2009;9(2):128–134. doi: 10.1038/nrc2571. [DOI] [PubMed] [Google Scholar]
- 34.Wasielewski R, Hasselmann S, Ruschoff J, Fisseler-Eckhoff A, Kreipe H. Proficiency testing of immunohistochemical biomarker assays in breast cancer. Virchows Arch. 2008;453(6):537–543. doi: 10.1007/s00428-008-0688-4. [DOI] [PubMed] [Google Scholar]
- 35.Turbin DA, Leung S, Cheang MC, et al. Automated quantitative analysis of estrogen receptor expression in breast carcinoma does not differ from expert pathologist scoring: a tissue microarray study of 3,484 cases. Breast Cancer Res Treat. 2008;110(3):417–426. doi: 10.1007/s10549-007-9736-z. [DOI] [PubMed] [Google Scholar]
- 36.Parker RL, Huntsman DG, Lesack DW, et al. Assessment of interlaboratory variation in the immunohistochemical determination of estrogen receptor status using a breast cancer tissue microarray. Am J Clin Pathol. 2002;117(5):723–728. doi: 10.1309/PEF8-GL6F-YWMC-AG56. [DOI] [PubMed] [Google Scholar]
- 37.Turashvili G, Leung S, Turbin D, et al. Inter-observer reproducibility of HER2 immunohistochemical assessment and concordance with fluorescent in situ hybridization (FISH): pathologist assessment compared to quantitative image analysis. BMC Cancer. 2009;9:165. doi: 10.1186/1471-2407-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rakha EA, Reis-Filho JS, Baehner F, et al. Breast cancer prognostic classification in the molecular era: the role of histological grade [published online ahead of print] Breast Cancer Res. 2010;12(4):207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 40.Liedtke C, Hatzis C, Symmans WF, et al. Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J Clin Oncol. 2009;27(19):3185–3191. doi: 10.1200/JCO.2008.18.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lusa L, McShane LM, Reid JF, et al. Challenges in projecting clustering results across gene expression-profiling datasets. J Natl Cancer Inst. 2007;99(22):1715–1723. doi: 10.1093/jnci/djm216. [DOI] [PubMed] [Google Scholar]
- 42.Wirapati P, Sotiriou C, Kunkel S, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10(4):R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popovici V, Chen W, Gallas BG, et al. Effect of training sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010;12(1):R5. doi: 10.1186/bcr2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Hayes DN, Nobel A, Marron JS. Statistical significance of clustering for high-dimension, low–sample size data. J Am Stat Assoc. 2008;103(483):1281–1293. [Google Scholar]
- 45.Weigelt B, Horlings HM, Kreike B, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216(2):141–150. doi: 10.1002/path.2407. [DOI] [PubMed] [Google Scholar]
- 46.Kreike B, van Kouwenhove M, Horlings H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9(5):R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertucci F, Finetti P, Cervera N, et al. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res. 2006;66(9):4636–4644. doi: 10.1158/0008-5472.CAN-06-0031. [DOI] [PubMed] [Google Scholar]
- 48.Bertucci F, Finetti P, Rougemont J, et al. Gene expression profiling identifies molecular subtypes of inflammatory breast cancer. Cancer Res. 2005;65(6):2170–2178. doi: 10.1158/0008-5472.CAN-04-4115. [DOI] [PubMed] [Google Scholar]
- 49.Van Laere SJ, Van den Eynden GG, Van der Auwera I, et al. Identification of cell-of-origin breast tumor subtypes in inflammatory breast cancer by gene expression profiling. Breast Cancer Res Treat. 2006;95(3):243–255. doi: 10.1007/s10549-005-9015-9. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 51.Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462(7276):1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desmedt C, Haibe-Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14(16):5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 53.Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6(12):718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- 54.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci U S A. 2006;103(15):5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iorns E, Lord CJ, Turner N, Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov. 2007;6(7):556–568. doi: 10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]
- 56.Teschendorff AE, Caldas C. The breast cancer somatic ‘muta-ome’: tackling the complexity. Breast Cancer Res. 2009;11(2):301. doi: 10.1186/bcr2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.