Summary
Centrioles play a crucial role in mitotic spindle assembly and duplicate precisely once per cell cycle. In worms, flies, and humans, centriole assembly is dependent upon a key regulatory kinase (ZYG-1/Sak/Plk4) and its downstream effectors SAS-5 and SAS-6. Here we report a role for protein phosphatase 2A (PP2A) in centriole duplication. We find that the PP2A catalytic subunit LET-92, the scaffolding subunit PAA-1, and the B55 regulatory subunit SUR-6, function together to positively regulate centriole assembly. In PP2A-SUR-6-depleted embryos, the levels of ZYG-1 and SAS-5 are reduced and the ZYG-1- and SAS-5-dependent recruitment of SAS-6 to the nascent centriole fails. We show that PP2A physically associates with SAS-5 in vivo, and that inhibiting proteolysis can rescue SAS-5 levels and the centriole duplication defect of PP2A-depleted embryos. Together our findings indicate that PP2A-SUR-6 promotes centriole assembly by protecting ZYG-1 and SAS-5 from degradation.
Keywords: centriole duplication, PP2A, ZYG-1, SZY-20, C. elegans
Introduction
In animal cells, the size and number of microtubule-organizing centers, or centrosomes, are determined by the centrioles, barrel-shaped organelles having ninefold rotational symmetry (Marshall, 2009). Centrioles organize centrosomes and thus, the number of centrosomes per cell generally correlates with the number of centrioles. A typical centrosome possesses an orthogonally arranged pair of centrioles surrounded by a halo of pericentriolar material (PCM), a substance from which the microtubules are nucleated and anchored.
Centrioles duplicate precisely once per cell cycle with a new daughter centriole assembling next to each preexisting centriole during S phase (Strnad and Gonczy, 2008). Defects in this process lead to abnormal numbers of centrioles, a state that has been linked to both chromosome missegregation and tumor formation (Basto et al., 2008; Castellanos et al., 2008; Ganem et al., 2009; Silkworth et al., 2009). Although the structural complexity of centrioles varies among species, studies in worms and humans suggest the existence of a common centriole assembly pathway that involves the activities of a small set of conserved duplication factors (Delattre et al., 2006; Kleylein-Sohn et al., 2007; Pelletier et al., 2006).
The centriole assembly pathway has been described most thoroughly in the C. elegans embryo where the kinase ZYG-1 plays a key role (O’Connell et al., 2001). Related to vertebrate Plk4 and Drosophila Sak, ZYG-1 localizes early to the nascent centriole in a step that requires the coiled-coil protein SPD-2 (Delattre et al., 2006; Pelletier et al., 2006). ZYG-1 then recruits a complex of two conserved coiled-coil proteins SAS-5 and SAS-6, that is required for formation of the central tube, the first known structural intermediate (Dammermann et al., 2004; Delattre et al., 2006; Delattre et al., 2004; Leidel et al., 2005; Pelletier et al., 2006). SAS-5 and SAS-6 are also required for the stable incorporation of SAS-4, a coiled-coil protein that promotes the addition of nine sets of microtubules that surround the central tube (Dammermann et al., 2008; Delattre et al., 2006; Kirkham et al., 2003; Leidel and Gonczy, 2003; Pelletier et al., 2006).
Recently, attention has focused on the role of phosphorylation in centriole assembly. Kitagawa et. al (2009) have found that ZYG-1 promotes stable association of SAS-6 with the nascent centriole through phosphorylation. Also, human Plk4 has been shown to prevent centriole overduplication by negatively regulating its own levels through autophosphorylation (Guderian et al., 2010; Holland et al., 2010). While roles for kinases in centriole assembly have been established, the possible involvement of phosphatases remains an open question. Like kinases, phosphatases regulate a myriad of cellular processes and do so with a high degree of specificity. The most abundant phosphatases are members of the PP1 and PP2A family. These multimeric enzymes contain a catalytic subunit characteristic of a specific phosphatase family and a regulatory subunit that determines substrate specificity (Virshup and Shenolikar, 2009).
Here we find that LET-92, the PP2A catalytic subunit (hereafter referred to as PP2AcLET-92), physically associates with SZY-20, a negative regulator of ZYG-1 (Song et al., 2008). We find that PP2AcLET-92 is required for centriole duplication in C. elegans, and that it functions in this context as a holoenzyme consisting of the catalytic subunit PP2AcLET-92, the scaffolding subunit PP2AaPAA-1, and the regulatory subunit SUR-6. Further, we show that the most critical role of PP2AcLET-92-SUR6 in centriole assembly is to protect ZYG-1 and SAS-5 from degradation. Our work thus identifies PP2AcLET-92-SUR-6 as a new component of the centriole assembly pathway, and suggests that centriole duplication is controlled by the coordinated action of kinases and phosphatases.
Results
SZY-20 physically interacts with protein phosphatase 2A
Previously, we identified the RNA-binding protein SZY-20 as a factor that negatively regulates ZYG-1 levels at the centrosome (Song et al., 2008). To understand how SZY-20 controls ZYG-1, we sought to identify proteins that physically interact with SZY-20. Using affinity-purified α-SZY-20 antibodies, we immunoprecipitated SZY-20 from whole worm extracts and analyzed the precipitated material by mass spectrometry. Among the proteins that specifically co-precipitated with SZY-20, we identified one, that when depleted by RNAi, reduces centrosome number in the early embryo. Here we focus on this factor, PP2AcLET-92, the sole C. elegans protein phosphatase 2A catalytic subunit.
To confirm the SZY-20-PP2AcLET-92 interaction, we immunoprecipitated SZY-20 from whole worm extracts, and analyzed the immunoprecipitated material by western blotting. In agreement with our mass spectrometry results, we detected PP2AcLET-92 among the material precipitated with α-SZY-20 antibodies (Figure 1A). Given our data indicating a physical association between SZY-20 and PP2AcLET-92 it seemed plausible that SZY-20 is a substrate of PP2A. Consistent with this idea, our mass spectrometry data indicates that SZY-20 is indeed a phosphoprotein (Figure S1A–C), and depletion of PP2AcLET-92 results in a significant and reproducible reduction in mobility of SZY-20 on SDS-PAGE gels (Figure 1B). The change in mobility appears to be due to phosphorylation, as phosphatase treatment of SZY-20 from let-92(RNAi)-treated worms restored its normal mobility (Figure S1D). We conclude that PP2AcLET-92 physically associates with SZY-20 in vivo and regulates the phosphorylation level of SZY-20.
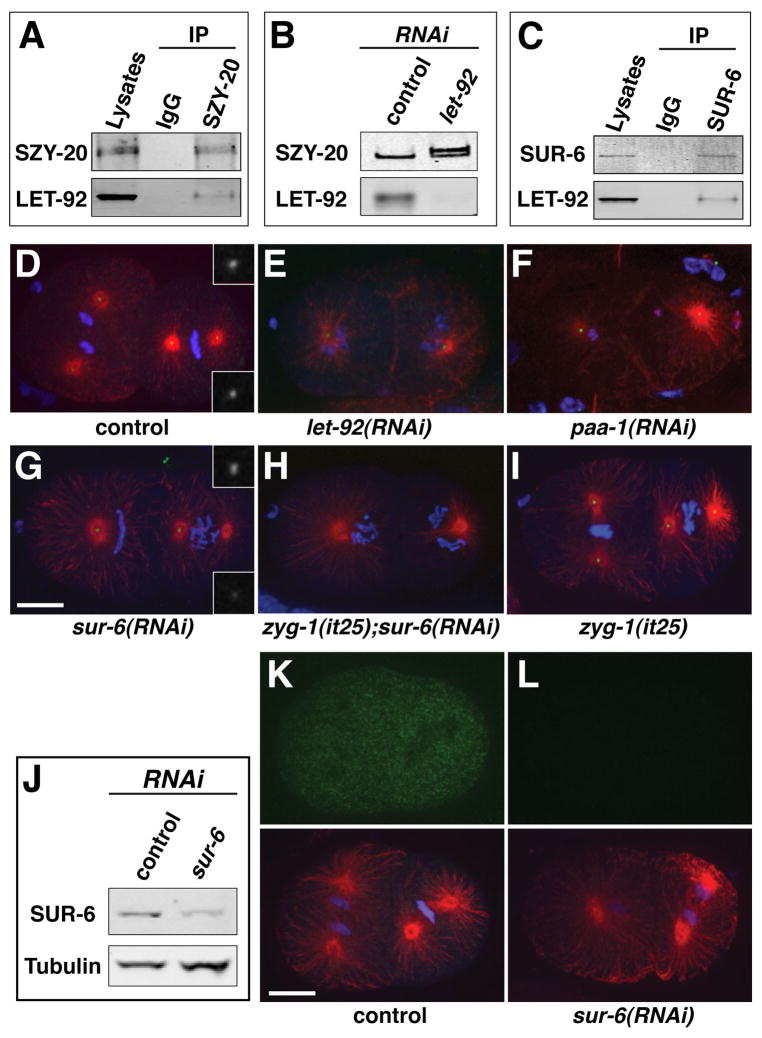
Figure 1. PP2AcLET-92 -SUR-6 regulates centriole duplication.
(A) PP2AcLET-92 coprecipitates with SZY-20. (B) Depletion of PP2AcLET-92 results in a decrease in SZY-20 mobility consistent with hyperphosphorylation. (C) PP2AcLET-92 coprecipitates with SUR-6. (D–I) Embryos of indicated genotype stained for microtubules (red), centrioles (SAS-4, green), and DNA (blue). Depletion of SUR-6 in wild-type embryos (G) results in centriole duplication defects such as monopolar spindles (left blastomere) or asymmetric spindles (right blastomere). Insets show symmetrical staining of wild-type spindle poles (D) and asymmetrical staining of sur-6(RNAi) spindle poles (G). (H, I) RNAi of sur-6 enhances the centriole duplication defect of zyg-1(it25) embryos raised at permissive temperature. (J) A SUR-6 immunoblot demonstrating the specificity of the α-SUR-6 antibody. RNAi of sur-6 results in a reduction in the intensity of the SUR-6 band. (K, L) Immunostaining reveals a broad distribution of SUR-6 (green) in the control embryo (K) and a loss of this staining in the sur-6(RNAi) embryo (L). Lower panels show the same embryos stained for microtubules (red) and DNA (blue). Bars, 10μm. For supporting data see Figure S1, Table S1, and movies S1 and S2.
Disruption of the PP2A holoenzyme blocks centriole duplication
In C. elegans embryos, a maternal block to centrosome duplication is manifest by a distinct phenotype: a bipolar first division followed by formation of monopolar spindles at the two-cell stage (O’Connell et al., 2001). This phenotype arises because the two sperm centrioles can separate and set up the poles of the first bipolar spindle, but can only organize monopolar spindles when partitioned to the daughter cells. Strikingly, we found that depletion of PP2AcLET-92, although pleiotropic in phenotype, resulted in a nearly identical centrosome defect: a bipolar spindle during first division followed by monopolar spindles during second division (Figure 1E and Movie S1). In addition, PP2AcLET-92-depleted embryos fail to properly align their chromosomes on a metaphase plate, exhibit a delay in cell cycle progression, and display defects in mitotic exit such as a failure to reform nuclei and to decondense chromatin. A defect in mitotic exit results from depletion of PP2A-B55α in human cells (Schmitz et al., 2010), indicating that this role is conserved between nematodes and humans. To quantify the effect of PP2AcLET-92 depletion on centrosome duplication, we performed live imaging of let-92(RNAi) embryos expressing GFP-SPD-2 to mark centrosomes and mCherry-histone to mark chromatin (Movie S2). Reducing PP2AcLET-92 completely blocks centrosome duplication (0/34 successful events, n=18 embryos). We conclude that PP2AcLET-92 is required for centrosome duplication in C. elegans.
The B55 regulatory subunit SUR-6 directs the role of PP2A in centriole assembly
PP2A functions as a heterotrimeric complex consisting of a catalytic subunit, a scaffolding subunit, and a regulatory subunit. In C. elegans, the scaffolding subunit is encoded by a single gene paa-1. Depletion of PP2AaPAA-1 produces defects in chromosome alignment, cell cycle progression, and mitotic exit that are similar to those induced by PP2AcLET-92 depletion (data not shown). In addition, depletion of PAA-1 results in the formation of monopolar spindles during the second embryonic cell division (Figure 1F), confirming the involvement of the PP2A holoenzyme in centrosome duplication. The specificity of the PP2A holoenzyme is determined by association with a specific regulatory subunit. By homology, we identified eight genes in C. elegans that encode PP2A regulatory subunits. We used RNAi to knockdown each of these genes and screened for a centrosome duplication defect. Only RNAi of sur-6, encoding a B55 family member involved in Ras/Raf/MEK/ERK signaling (Kao et al., 2004), produced an obvious duplication defect (Figure 1G). In these embryos we observed both monopolar spindles and asymmetric spindles in which one spindle pole was smaller than the other (Figure 1G). Asymmetric spindles arise when centriole duplication is partially inhibited (Delattre et al., 2004; Kirkham et al., 2003). In contrast to let-92(RNAi), the effect of sur-6(RNAi) was relatively mild; embryos did not possess obvious defects in cell cycle progression or mitotic exit but did occasionally exhibit anaphase bridging of chromatin. Also unlike let-92(RNAi), sur-6(RNAi) did not completely block centrosome duplication: 24% of spindles were monopolar and 47.5% were asymmetric (Table S1). The lack of a completely penetrant centriole duplication defect could reflect incomplete depletion of SUR-6 protein by RNAi (Figure 1J), or alternatively functional redundancy between SUR-6 and other PP2A regulatory subunits. To address this, we re-screened these genes using a sensitized genetic background. At the semipermissive temperature of 20°C, centrosomes in the temperature-sensitive zyg-1(it25) mutant fail to duplicate just 2.9% of the time. (Figure 1I and Table S1). When subjected to RNAi of each regulatory gene, only sur-6(RNAi) had a significant effect, increasing the rate of monopolar spindle formation 22-fold to 64% (Figure 1H and Table S1). These results suggest that the role of PP2A in centrosome duplication is predominantly, if not entirely, directed by SUR-6, and that the lack of a more robust centriole duplication defect upon sur-6(RNAi) is due to incomplete depletion.
To determine where in the cell SUR-6 acts, we produced affinity-purified anti-SUR-6 polyclonal sera. The sera identified a band of the correct size by immunoblotting, and the intensity of this band was significantly reduced by sur-6(RNAi)(Figure 1J). Further, the SUR-6 antibody also coprecipitated PP2AcLET-92 from worm extracts (Figure 1C). Immunostaining of embryos produced diffuse cytoplasmic staining that was significantly reduced by sur-6(RNAi) (Figure 1K, L). This result suggests that the function of PP2AcLET-92-SUR-6 is not limited to the centrosome and may function broadly in the cell to regulate centriole duplication.
The SZY-20-PP2AcLET-92 interaction plays a minor role in regulating centriole duplication
We next wanted to determine where in the centriole duplication pathway PP2A functions. SZY-20 negatively regulates centriole duplication (Song et al., 2008) and appears to be a substrate of PP2AcLET-92-SUR-6 (Figures 1B, S1D and S1E). Thus, PP2A might promote duplication by repressing SZY-20. To address this, we depleted PP2AcLET-92 in strain OC470, a szy-20(bs52) mutant that expresses GFP-SPD-2 and mCherry-histone. Strong knockdown of PP2AcLET-92 in this strain still produced a complete block to duplication (n=38 events), indicating that the centriole duplication defect of let-92(RNAi) embryos is not due to an overactive SZY-20 protein. However, the szy-20(bs52) mutation could partially suppress a weaker knockdown of PP2AcLET-92 (7 of 26 centrosomes duplicated in szy-20(bs52) mutants versus 0/18 in controls), suggesting that one minor role for PP2A is to repress SZY-20. Overall, failure of the szy-20(bs52) mutation to strongly affect the let-92(RNAi) duplication defect indicates that the critical role of PP2A in centriole duplication lies elsewhere in the pathway.
PP2A functions closely with ZYG-1 and SAS-5
We have shown that genes encoding core centriole assembly factors exhibit robust genetic interactions with each other (Kemp et al., 2004). Mutations in two different centriole assembly genes often fail to complement one another, with the double heterozygotes exhibiting high levels of embryonic lethality and centriole duplication errors. This interaction, termed non-allelic non-complementation, occurs when two genes function closely together in a common process; in a double heterozygote, the process is rendered inefficient enough to yield an observable phenotype. To determine if let-92 also behaves in this manner, we asked if the let-92(s504) mutation would fail to complement the zyg-1(b1) allele. Animals heterozygous for either zyg-1(b1) or let-92(s504) had low levels of embryonic lethality, yet strikingly the zyg-1(b1)/+; let-92(s504)/+ double heterozygotes exhibited 100% embryonic lethality (Figure 2A). We also tested the zyg-1(b1) mutation against the let-92(s677) allele and again detected a very strong interaction: 61% (n=1197) of the embryos produced by zyg-1(b1)/+; let-92(s677)/+ hermaphrodites were inviable compared to 2.8% (n=611) for let-92(s677)/+ progeny. We then examined embryos from zyg-1(b1)/+; let-92(s504)/+ hermaphrodites and observed a prominent centrosome duplication defect (Figure 2B–D). Live DIC imaging of these embryos revealed that the block to centriole duplication was complete (0 of 48 centrioles duplicated; movie S3). These results show that PP2AcLET-92 behaves genetically in the same manner as other core centriole duplication factors and indicate that PP2AcLET-92 functions closely with ZYG-1.
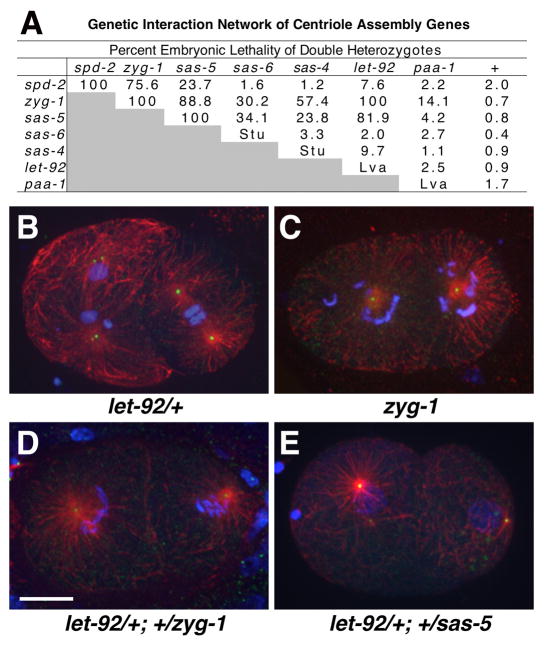
Figure 2. PP2AcLET-92 genetically interacts with other centriole duplication factors.
(A) A genetic interaction network of centriole duplication genes. Shown is the percent embryonic lethality of indicated double heterozygotes. Single heterozygotes are listed in last column (+). Some single homozygotes exhibit a sterile uncoordinated (Stu) phenotype and others a larval lethal (Lva) phenotype. Between 345 and 1,521 embryos were counted for each strain. (B–E) Embryos of indicated genotype stained for microtubules (red), centrioles (SAS-4, green), and DNA (blue). Note that let-92 (B) and zyg-1 heterozygotes (not shown) assemble bipolar spindles, but zyg-1 homozygotes (C), and let-92/+; zyg-1/+ (D) and let-92/+; +/sas-5 double heterozygotes assemble monopolar spindles. Bar, 10 μm. For supporting data see Movie S3.
We next asked if we could use this type of genetic interaction to map the role of PP2A, as well as other factors, in the centriole assembly pathway. We constructed and quantified embryonic lethality in all possible double heterozygotes from a set of spd-2, zyg-1, sas-5, sas-6, sas-4, let-92 and paa-1 mutations (Figure 2A). Most pertinent to the current study is the behavior of let-92, which shows strong interactions with zyg-1 and sas-5 but weak or no interactions with spd-2, sas-6, and sas-4. Again, these double heterozygotes exhibit centriole duplication defects (Figure 2E). These results suggest that the role of PP2AcLET-92 is most closely tied to zyg-1- and sas-5-dependent steps. Also of interest, spd-2 only shows a strong interaction with zyg-1, consistent with the only known role of SPD-2 in localizing ZYG-1 to sites of centriole assembly (Delattre et al., 2006; Pelletier et al., 2006). In contrast, zyg-1 shows strong genetic interactions with multiple components including spd-2, sas-5, sas-4, and let-92, suggesting that ZYG-1 functions at multiple steps in this pathway.
PP2A is required for recruitment of SAS-5 and SAS-6
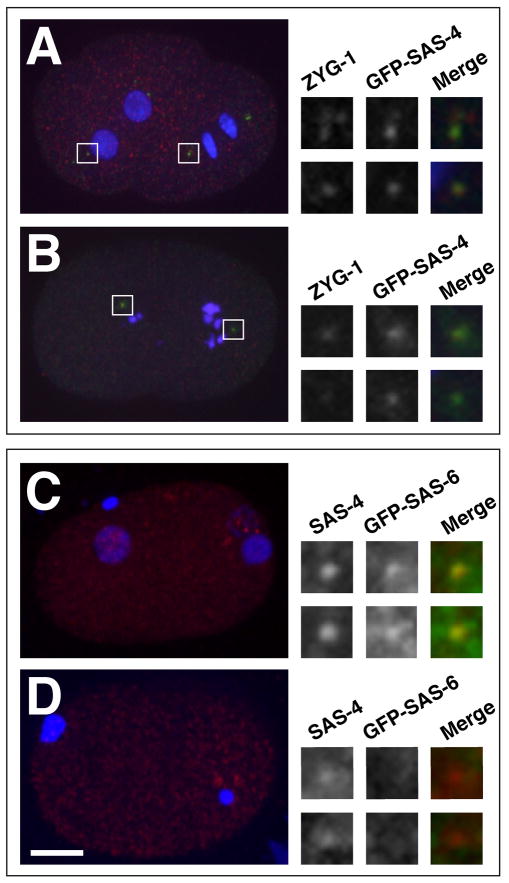
A molecular hierarchy describing the order and dependencies with which duplication factors localize to assembling centrioles has been described (Delattre et al., 2006; Pelletier et al., 2006). To determine where in this pathway PP2AcLET-92-SUR-6 functions, we analyzed the effect of PP2AcLET-92 depletion on the recruitment of duplication factors. We found that let-92(RNAi) does not block localization of ZYG-1 to nascent centrioles (Figure 3A and B). We then examined SAS-5 and SAS-6, two downstream effectors of ZYG-1 that form a complex in vivo and are mutually dependent for localization to assembling centrioles (Dammermann et al., 2004; Delattre et al., 2004; Leidel et al., 2005). To examine the localization of the SAS-5/6 complex, we followed GFP-SAS-6 utilizing a previously established recruitment assay (Pelletier et al., 2006). In this assay, matings between wild-type males and transgenic females carrying a germ line-expressed gfp-sas-6 gene allows one to examine the accumulation of GFP-SAS-6 protein around the unlabeled sperm-derived centrioles. Consistent with previous observations, we found that in control embryos, GFP-SAS-6 was initially detected at centrioles during the first S phase (Figure 3C). In contrast, we often failed to detect centriole-associated GFP-SAS-6 in let-92(RNAi) embryos (Figure 3D). Of ten one-cell let-92(RNAi) embryos that ranged from S phase to mid prophase, only two showed even a faint GFP-SAS-6 signal around one of the two centrioles. We conclude that PP2AcLET-92-SUR-6 is required for proper localization of the SAS-5/6 complex to sites of centriole assembly.
Figure 3. PP2AcLET-92 is required for recruitment of the SAS-5/6 complex.
(A) Control and (B) let-92(RNAi) embryos stained for ZYG-1 (red), GFP-SAS-4 (green) and DNA (blue). ZYG-1 localizes to centrosomes in PP2AcLET-92 -depleted embryos (insets). (C) Control and (D) let-92(RNAi) embryos stained for endogenous SAS-4 (red) GFP-SAS-6 (green-insets only) and DNA (blue). (C) In control embryos, GFP-SAS-6 is recruited to centrioles as evidence by the overlap between the GFP-SAS-6 and SAS-4 signals (insets). (D) In let-92(RNAi) embryos, GFP-SAS-6 does not accumulate around the centrioles. Note that control and let-92(RNAi) embryos are approximately the same age based on position of DNA and separation of centrioles. The DNA of let-92(RNAi) embryos however stays highly condensed.
PP2AcLET-92-SUR-6 regulates ZYG-1 and SAS-5 levels
We next tested for direct effects of PP2AcLET-SUR-6 depletion on centriole assembly factors. Using quantitative immunoblotting, we measured the levels of ZYG-1, SAS-5, and SAS-6 proteins in control and PP2AcLET-92-depleted embryos. Depleting PP2AcLET-92, SUR-6, or PAA-1 led to reproducible declines in the levels of ZYG-1 and SAS-5 (Figure 4A and B). We also noted that SAS-5 mobility decreased slightly upon depletion of PP2A subunits (Figure 4B). Quantitatively, PP2AcLET-92-depleted embryos possess 48% ± 25 (n=6) as much ZYG-1, and 40% ± 15 (n=7) as much SAS-5 as control embryos. As evident in Figure 3B, the reduction in ZYG-1 protein level does not completely block the localization of ZYG-1 to centrosomes. However, we are currently unsure if the centrosome levels of ZYG-1 are slightly reduced upon PP2AcLET-92 depletion. In contrast to ZYG-1 and SAS-5, the level of SAS-6 appears unaffected by depletion of PP2AcLET-92 or SUR-6 (Figure 4C). Thus, a striking parallel exists between our genetic and biochemical analyses; genetically let-92 interacts with zyg-1 and sas-5 but not with sas-6, and biochemically, loss of PP2AcLET-92 affects the levels of ZYG-1 and SAS-5 but not of SAS-6.
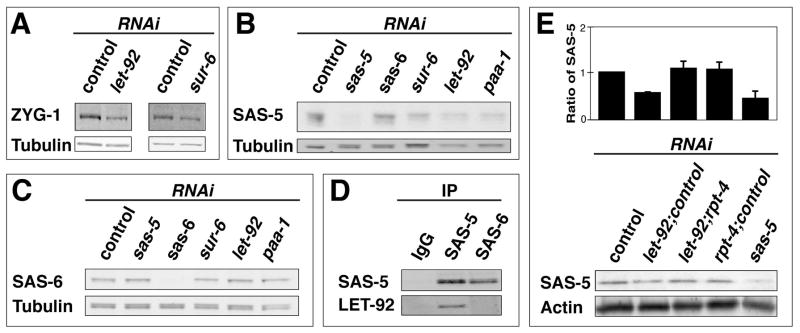
Figure 4. The PP2AcLET-92 -SUR-6 complex regulates the levels of ZYG-1 and SAS-5.
(A–C) Quantitative immunoblots of embryonic extracts showing that RNAi of let-92, sur-6, or paa-1 reduces the levels of ZYG1 and SAS-5 but not the levels of SAS-6. (D) PP2AcLET-92 coprecipitates with SAS-5, but not with SAS-6. (E) Weak rpt-4(RNAi) restores SAS-5 levels in PP2AcLET-92-depleted embryos. Shown is a graph depicting the average relative levels of SAS-5 with standard deviations from three experiments, and a representative blot. The reduction of SAS-5 levels upon let-92(RNAi) is less pronounced in these experiments due to dilution of let-92 bacteria with control bacteria. Ratios of RNAi bacteria used: let-92:control, 1:1; let-92:rpt-4:control: 4:1:3; rpt-4:control, 1:7. For supporting data see movie S4.
To test if PP2AcLET-92-SUR-6 controls the levels of SAS-5 directly, we immunoprecipitated SAS-5 from wild-type embryonic extracts and probed for PP2AcLET-92. Consistent with a direct interaction between SAS-5 and PP2AcLET-92, we detected PP2AcLET-92 in the precipitated material (Figure 4D). We also immunoprecipitated SAS-6 from wild-type embryonic extracts but in contrast to the SAS-5 pull-down experiment, we did not detect PP2AcLET-92 among the coprecipitating proteins (Figure 4D). It should be noted that a significant fraction of SAS-5 exists in a cytoplasmic complex with SAS-6 (Leidel et al., 2005). Thus, it was unexpected that we failed to detect an interaction between SAS-6 and PP2AcLET-92. This may indicate that PP2AcLET-92 interacts predominantly with the pool of SAS-5 not bound to SAS-6.
Our findings that PP2AcLET-92 physically interacts with SAS-5 and controls SAS-5 levels suggests that PP2A promotes the stability of SAS-5, and that in the absence of PP2A, the reduced amount of SAS-5 (as well as ZYG-1) is unable to support centriole duplication. To investigate this, we determined what effect blocking SAS-5 degradation would have on PP2AcLET-92-depleted embryos. We knocked down both PP2AcLET-92 and RPT-4, an essential component of the 26S proteosome, and assayed centriole behavior. Remarkably, we found 58% of centrioles (n=26) duplicated in such embryos while centrioles never duplicated (n=24) in paired let-92(RNAi) controls (Movie S4). Further, knockdown of RPT-4 could also suppress the genetic interaction between zyg-1 and let-92. Specifically, we found (using slightly less stringent conditions than used for the genetic interaction experiments) that 70% of centrosomes (n=10) duplicated in zyg-1(b1)/+; let-92(s504)/+ double heterozygotes subject to RPT-4 knockdown compared to 10% (n=10) in controls. Further, despite the fact that RPT-4 knockdown reduces the viability wild-type embryos (27% viability), we found that it actually increases the viability of embryos produced by zyg-1(b1)/+; let-92(s504)/+ double heterozygotes 10-fold from 0.3% (n=292) in controls to 3.0% (n=448) in rpt-4-treated animals. Finally, we investigated the effect of RPT-4 depletion on SAS-5 levels and found that partial knockdown of RPT-4 could suppress the reduction of SAS-5 observed in PP2AcLET-92-depleted embryos (Figure 4E). Therefore in PP2AcLET-92-depleted embryos, the level of SAS-5 positively correlates with the ability of centrioles to duplicate.
Discussion
While the involvement of kinases in centriole duplication has been established, the potential involvement of phosphatases has not been well explored. Our identification of PP2AcLET-92-SUR-6 as a positive regulator of duplication indicates that centriole duplication is governed by both activating and inhibitory phosphorylation events. A recent proteomic study in Drosophila found the fly PP2A catalytic and scaffolding subunits associated with embryonic centrosomes (Muller et al., 2010). Consistent with our results, depletion of either PP2A subunit in fly or human tissue culture cells reduced centrosome number suggesting a defect in centriole duplication or segregation (Muller et al., 2010). Thus it seems likely that the role of PP2A in centriole duplication is conserved.
We have identified ZYG-1 and SAS-5 as factors that are targeted by PP2AcLET-92-SUR-6: the levels of these proteins are diminished upon PP2AcLET-92 or SUR-6 depletion. ZYG-1 is still detectable at centrosomes in PP2AcLET-92-depleted embryos (Figure 3A, B), although it remains unclear if ZYG-1 levels at the centrosome are reduced relative to the wild type. However, since ZYG-1 and SAS-5 are both required for SAS-6 localization (Delattre et al., 2006; Delattre et al., 2004; Leidel et al., 2005; Pelletier et al., 2006), the reduced levels of ZYG-1 and especially SAS-5 likely contribute to the failure of SAS-6 to localize to nascent centrioles in the absence of PP2AcLET-92-SUR-6. This is supported by our finding that inhibiting proteolysis in PP2AcLET-92-depleted embryos restores both the level of SAS-5 and centriole duplication. Despite this, it still remains possible that PP2AcLET-92-SUR-6 also functions directly (and independent of its role in regulating ZYG-1 and SAS-5 levels) to promote the recruitment of the SAS-5/6 complex.
How might the PP2AcLET-92-SUR-6 complex regulate ZYG-1 and SAS-5 levels? Our finding that SAS-5 physically associates with PP2AcLET-92 in vivo suggests that PP2AcLET-92-SUR-6 acts to stabilize SAS-5 by dephosphorylation. Regulating protein stability via phosphorylation is common; in vertebrates, Plk4 negatively regulates its own levels by targeting itself for destruction via autophosphorylation (Guderian et al., 2010; Holland et al., 2010; Sillibourne et al., 2010). Autophosphorylation promotes association of Plk4 with βTr-CP, an adapter for the SCF E3 ubiquitin ligase. In flies, inhibition of SCFβTr-CP leads to elevated levels of Plk4 and centriole amplification (Cunha-Ferreira et al., 2009; Rogers et al., 2009), suggesting that autophosphorylation may function to maintain proper levels of Plk4.
In summary, PP2AcLET-92-SUR-6 is a core centriole duplication factor and regulates the levels of ZYG-1 and SAS-5. Unlike the other core duplication factors, PP2AcLET-92-SUR-6 does not appear to function exclusively at centrosomes, as we find that depletion of RSA-1, a regulatory chain that targets PP2AcLET-92 to centrosomes (Schlaitz et al., 2007), does not inhibit centrosome duplication (Figure 2A) and SUR-6 is not enriched at centrosomes (Figure 1K). Thus, PP2AcLET-92-SUR-6 likely acts within the cytoplasm. Future studies aimed at identifying the specific residues within SAS-5, and possibly ZYG-1, targeted by PP2AcLET-92-SUR-6 will reveal how inhibitory phosphorylation events contribute to the fidelity of centriole duplication.
Experimental Procedures
Immunoprecipitation and Quantitative Immunoblotting
For whole worm lysates, N2 worms were grown in liquid culture until gravid, harvested by centrifugation, washed several times in lysis buffer (50 mM HEPES, pH7.4, 1 mM EDTA, 1 mM MgCl2, 200 mM KCl, 10% glycerol: (Cheeseman et al., 2004), and gently pelleted. Worm slurries (7ml) were frozen on dry ice and lysed using a One Shot Cell Disrupter (Constant Systems Limited, Northants, UK) set at 40 psi. Crude lysates were centrifuged three times at 382K rpm for 45 min; the resulting high-speed supernatants were used for immunoprecipitation.
For pull-down assays, antibodies to SUR-6, SAS-5, SAS-6 and SZY-20 (Song et al., 2008) were bound to Dynabeads coupled to protein A (Invitrogen) at a ratio of 1μg antibodies to1μl beads. Anti-rabbit-IgG (Invitrogen) served as a negative control. Antibodies were incubated with beads in PBST (1×PBS, 0.1% Triton X-100), rotated overnight at 4°C, and cross-linked to beads according to manufacturer’s instructions. The beads were then mixed with 1 ml of the whole worm extract and incubated with rotation overnight at 4°C. The lysate was removed and the beads washed with PBST ten times at room temperature.
For quantitative immunoblotting, embryonic proteins were fractionated on a NuPAGE Bis-Tris Gel (Invitrogen) and transferred to nitrocellulose. α-SZY-20 (Song et al., 2008), α-SPD-2 (Kemp et al., 2004), α-ZYG-1 (Kemp et al., 2007), α-PP2Ac (Clone 1D6, Millipore), α-actin (Thermo Scientific), and DM1A (Sigma) were used at a 1:500–1500 dilution. IRDye secondary antibodies (LI-COR Biosciences) were used at 1:15,000. Blots were imaged using the Odyssey Infrared Imaging System (LI-COR Biosciences) and analyzed using the same software or ImageJ v1.40G. For quantifying ZYG-1 and SAS-5 levels, values were normalized against tubulin or actin.
RNAi
To feed dsRNA, L4 larvae were placed on a lawn of bacteria that carried a plasmid for inducible expression of let-92, paa-1, sur-6, sas-5, or rpt-4 dsRNA (Source Bioscience plc, Nottingham, UK), and allowed to feed for one day at 25°C. Bacteria carrying the dsRNA-feeding vector L4440 served as a negative control. For dual RNAi, plates were seeded with mixtures of bacteria in specified ratios based on the optical densities of the cultures; the total amount of bacteria added to the plate was kept constant by adding the appropriate amount of L4440 bacteria. To test for suppression of let-92 by rpt-4, we compared the effect of a 1:1 mixture of L4440 and let-92 bacteria (0% duplication, n=24 events) to a 3:4:1 ratio of L4440, let-92, and rpt-4 bacteria (58% duplication, n=26 events) as they both contained the same fraction of let-92 bacteria. To test for suppression of let-92(RNAi) by szy-20(bs52), we compared N2 and szy-20(bs52) animals grown on a 1:1 mixture of L4440 and let-92 bacteria (strong PP2AcLET-92 knockdown) or on a 3:1 mixture of L4440 and let-92 bacteria (weak knockdown). To test zyg-1(b1)/+; let-92(s504)/+ double heterozygotes for suppression by rpt-4, we compared double heterozygotes grown on L4440 to those grown on a 7:1 mixture of L4440 and rpt-4 bacteria.
Cytology
The following antibodies were used at a 1:500–2000 dilution: DM1A (Sigma), α-GFP (Roche), α-SPD-2 (Kemp et al., 2004), α-SPD-5 (Hamill et al., 2002), α-ZYG-1 (O’Connell et al., 2001), α-SAS-4 (Song et al., 2008), and Alexa Fluor 488 and 568 secondary antibodies (Invitrogen). Methods and imaging systems for indirect immunofluorescence and confocal and 4D-DIC microscopy have been described (Peters et al.; Song et al., 2008). Image processing was performed with ImageJ v1.40G and Adobe Photoshop CS5.
The SAS-6 recruitment assay was performed essentially as described (Pelletier et al., 2006). Briefly, wild-type males were mated overnight to OD103 females on RNAi plates at 25°C. The next day embryos were immunostained using the α-SAS-4 antibody to mark centrioles and the anti-GFP antibody to detect GFP-SAS-6.
Supplementary Material
Acknowledgments
We thank members of the O’Connell and Song labs, David Weisblat, Alexander Dammerman, Tony Hyman, Markus Decker, Karen Oegema, Martin Srayko, and Orna Cohen-Fix, for sharing strains and reagents and/or providing advice. Some strains were provided by The Caenorhabditis Genetics Center and The National Bioresource Project- Japan. This work was supported by the Intramural Research Program of the National Institutes Health (NIH) and by the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos E, Dominguez P, Gonzalez C. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr Biol. 2008;18:1209–1214. doi: 10.1016/j.cub.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli M, Zhang W, Laue E, Callaini G, Glover DM, Bettencourt-Dias M. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gonczy P. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol. 2004;6:656–664. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J Cell Sci. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao G, Tuck S, Baillie D, Sundaram MV. C. elegans SUR-6/PR55 cooperates with LET-92/protein phosphatase 2A and promotes Raf activity independently of inhibitory Akt phosphorylation sites. Development. 2004;131:755–765. doi: 10.1242/dev.00987. [DOI] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome Maturation and Duplication in C. elegans Require the Coiled-Coil Protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- Kemp CA, Song MH, Addepalli MK, Hunter G, O’Connell K. Suppressors of zyg-1 Define Regulators of Centrosome Duplication and Nuclear Association in Caenorhabditis elegans. Genetics. 2007;176:95–113. doi: 10.1534/genetics.107.071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Busso C, Fluckiger I, Gonczy P. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev Cell. 2009;17:900–907. doi: 10.1016/j.devcel.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Leidel S, Gonczy P. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Marshall WF. Centriole evolution. Curr Opin Cell Biol. 2009;21:14–19. doi: 10.1016/j.ceb.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Schmidt D, Steinbrink S, Mirgorodskaya E, Lehmann V, Habermann K, Dreher F, Gustavsson N, Kessler T, Lehrach H, et al. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010;29:3344–3357. doi: 10.1038/emboj.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Peters N, Perez DE, Song MH, Liu Y, Muller-Reichert T, Caron C, Kemphues KJ, O’Connell KF. Control of mitotic and meiotic centriole duplication by the Plk4-related kinase ZYG-1. J Cell Sci. 2010;123:795–805. doi: 10.1242/jcs.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz AL, Srayko M, Dammermann A, Quintin S, Wielsch N, MacLeod I, de Robillard Q, Zinke A, Yates JR, 3rd, Muller-Reichert T, et al. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 2007;128:115–127. doi: 10.1016/j.cell.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, Goris J, Trinkle-Mulcahy L, Lamond AI, Poser I, et al. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol. 2010;12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne JE, Tack F, Vloemans N, Boeckx A, Thambirajah S, Bonnet P, Ramaekers FC, Bornens M, Grand-Perret T. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol Biol Cell. 2010;21:547–561. doi: 10.1091/mbc.E09-06-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MH, Aravind L, Muller-Reichert T, O’Connell KF. The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev Cell. 2008;15:901–912. doi: 10.1016/j.devcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Gonczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.