Crystals of the thiaminase type II from S. aureus are orthorhombic, belonging to space group P212121 with unit-cell parameters a = 103.5, b = 104.1, c = 109.6 Å, and diffracted to 2.6 Å resolution.
Keywords: thiaminase type II, TenA, Staphylococcus aureus
Abstract
Thiaminase type II (TenA) catalyzes the deamination of aminopyrimidines, including the cleavage of thiamine to 4-amino-5-hydroxymethyl-2-methylpyrimidine and 5-(2-hydroxyethyl)-4-methylthiazole in the metabolism of thiamine (vitamin B1), in Staphylococcus aureus (Sa). SaTenA was crystallized by the vapour-diffusion method and the resulting crystal diffracted to 2.6 Å resolution usng synchrotron radiation. The crystal is orthorhombic, belonging to space group P212121 with unit-cell parameters a = 103.5, b = 104.1, c = 109.6 Å. With four molecules in the asymmetric unit, the Matthews coefficient is 2.85 Å3 Da−1. Initial attempts to solve the structure by molecular-replacement techniques were successful.
1. Introduction
Staphylococcus aureus can cause a wide spectrum of human and animal diseases ranging from benign skin infections to severe diseases such as pneumonia, meningitis and osteomyelitis (Fey et al., 2003 ▶; Lowy, 1998 ▶). This human pathogen has developed specific strategies for persistence in humans (Kauffman et al., 1993 ▶; Powers et al., 1990 ▶). Although not traditionally considered as an intracellular pathogen, the bacterium can survive in a variety of cells, hiding from the human immune system (Lowy, 1998 ▶). Furthermore, S. aureus has developed resistance against various antibiotics, which makes treatment of this bacterial infection difficult, especially in immune-deficient people (Tumbarello et al., 1998 ▶; Weinke et al., 1992 ▶).
Because of this, investigations to identify novel drug targets for the development of new chemotherapeutics against this pathogen are unceasing in the pharmaceutical industry. In this context, pathogen-specific vitamin syntheses represent an ideal drug target because the corresponding pathways are not present in human metabolism and thus potential cross-reaction with the host is avoided; also, their novelty as targets should minimize latent drug resistance.
Thiamine pyrophosphate (TPP), the active form of vitamin B1, is present in all organisms as an essential cofactor of a variety of key enzymes (Pohl et al., 2004 ▶). In contrast to plants, some protozoa and bacteria, humans and other mammals are unable to synthesize thiamine and are completely dependent on uptake of this essential nutrient from their diet. In humans, a deficiency of exogenous vitamin B1 can result in Wernicke’s disease and beriberi (Ogershok et al., 2002 ▶; Platt & Lu, 1936 ▶).
Bacteria possess an enzyme known as TenA that has been shown to be a type II thiaminase. The biological function of TenA is involved in the salvage of degraded products of thiamine and promotes the production of degradative enzymes in Bacillus subtilis (Jurgenson et al., 2009 ▶).
Crystal structures of TenA enzymes from Helicobacter pylori HP1287 (Barison et al., 2009 ▶), B. subtilis (Toms et al., 2005 ▶), Sulfolobus solfataricus (PDB code 2qzc; Joint Center for Structural Genomics, unpublished work) and Staphylococcus epidermidis (PDB code 3no6; Joint Center for Structural Genomics, unpublished work) have been determined and analyzed. Here, we report the crystallization and data collection to 2.6 Å resolution of TenA from S. aureus (SaTenA). Its highest degree of sequence identity is 57.7% with TenA from S. epidermidis.
2. Materials and methods
2.1. Protein expression and purification
The open reading frame of SaTenA was previously cloned into the E. coli expression plasmid pASK-IBA3 (Müller et al., 2009 ▶), which encodes a C-terminal Strep-tag that allows one-step purification of the recombinant fusion protein using Strep-Tactin Sepharose. The construct was transformed into Escherichia coli BLR (DE3) expression cells (Stratagene, Germany). Briefly, single colonies were picked and grown overnight in Luria–Bertani medium. The bacterial culture was diluted 1:100 and grown at 310 K until the A 600 reached 0.5. Expression was initiated with 200 ng ml−1 anhydrotetracycline and the cells were grown for 4 h at 310 K before being harvested. The cell pellet was resuspended in 100 mM Tris–HCl pH 8.0, 100 mM NaCl, sonicated and centrifuged at 75 000g for 1 h at 277 K. The recombinant Strep-Tag fusion protein was purified according to the manufacturer’s recommendation (Institut für Bioanalytik, Germany). The eluted proteins from the affinity chromatography were analyzed by SDS–PAGE and the protein was visualized by Coomassie staining (Sambrook et al., 1989 ▶).
Eluted samples were concentrated to 10 mg ml−1 in resuspension buffer. The isolated enzyme consists of 229 amino-acid residues and has a molecular weight of 26 kDa. The C-terminal Strep-Tag comprises ten additional residues (SAWSHPQFEK) and was not cleaved off.
2.2. Crystallization of SaTenA
The purified SaTenA was dialyzed against 100 mM Tris–HCl buffer pH 8.0; it was then concentrated to 10 mg ml−1 using an Amicon Ultra centrifugal filter device (Millipore; 3 kDa molecular-weight cutoff) and used for initial screening of crystallization conditions. Preliminary crystallization experiments were carried out at 293 K in 96-well crystallization plates (NeXtal QIA1 µplates, Qiagen) using the following commercially available crystallization screening kits: JCSG+ Suite, ComPAS, Classics and Cryo (Qiagen). A total of 384 crystallization conditions were screened by the sitting-drop vapour-diffusion method using a Honeybee 961 robot (Genomic Solutions Ltd). Droplets of 600 nl in volume (300 nl protein solution plus 300 nl reservoir solution) were equilibrated against 35 µl reservoir solution. Initial crystals appeared after 5 d using a solution consisting of 0.2 M sodium acetate, 0.1 M Tris–HCl pH 8.5 and 26%(w/v) polyethylene glycol (PEG) 4000. The condition that yielded crystals was further optimized in order to obtain crystals that were suitable for X-ray analysis. A drop containing 2 µl protein solution was mixed with 2 µl precipitant and equilibrated against 1 ml reservoir solution consisting of 0.2 M sodium acetate, 0.1 M Tris–HCl pH 8.5 and 28%(w/v) PEG 3350 using the hanging-drop technique in 24-well Linbro plates. Crystals grew to dimensions of 0.1 × 0.1 × 0.4 mm after 7 d (Fig. 1 ▶).
Figure 1.
A crystal of SaTenA (0.1 × 0.1 × 0.4 mm; the scale bar is 0.1 mm in length) obtained using 0.2 M sodium acetate, 0.1 M Tris–HCl pH 8.5 and 28%(w/v) PEG 3350.
2.3. Diffraction experiment
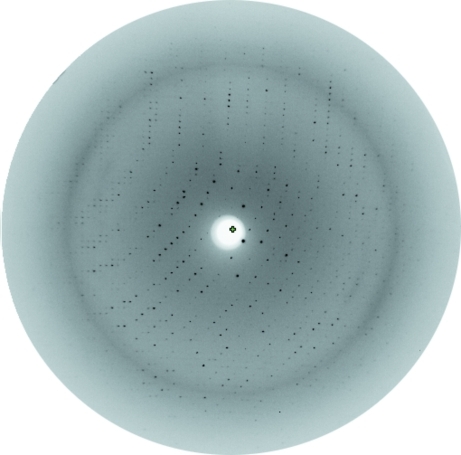
Diffraction data were collected to a resolution of 2.60 Å using synchrotron radiation of wavelength 0.8123 Å on the consortium’s fixed-wavelength beamline X13 at HASYLAB/DESY, Hamburg, Germany (Fig. 2 ▶). For cryocooling, crystals were transferred to reservoir solution containing 10%(v/v) glycerol before flash-freezing them in a nitrogen stream at 100 K prior to X-ray diffraction analysis. Data were collected at a crystal-to-detector distance of 256 mm and with 0.8° oscillation per image. Initial crystal characterization and space-group assignment were performed using the HKL-2000 software package (Otwinowski & Minor, 1997 ▶) and scaling was performed using SCALEPACK (Otwinowski & Minor, 1997 ▶).
Figure 2.
X-ray diffraction pattern of an SaTenA crystal recorded using a MAR CCD detector.
3. Results and discussion
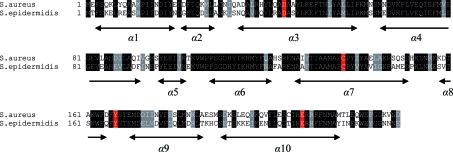
SaTenA was cloned with an affinity tag and expressed in E. coli BLR (DE3) cells. The protein consists of 229 amino acids with a molecular weight of 26 840 Da as calculated from the primary sequence. Sequence comparison of SaTenA within the PDB revealed high homology to the related protein from S. epidermidis (PDB code 3no6; Joint Center for Structural Genomics, unpublished work). The sequence identity between the two models is 57.7%. The sequence alignment and the putative active-site residues Asp44, Cys137, Tyr167 and Glu208 are shown in Fig. 3 ▶ (Jurgenson et al., 2009 ▶). The crystallization procedure is described above. A complete diffraction data set was collected to 2.60 Å resolution from a single crystal. Data-collection statistics are reported in Table 1 ▶. A total of 424 031 measured reflections in the resolution range 30–2.6 Å merged to 31 741 unique reflections with an overall R merge of 4.2%. Analysis of the diffraction intensities confirmed the space group to be the orthorhombic space group P212121, with unit-cell parameters a = 103.5, b = 104.1, c = 109.6 Å. With four molecules in the asymmetric unit, the calculated Matthews coefficient is 2.85 Å3 Da−1 (Matthews, 1968 ▶), corresponding to a solvent content of 56%. A preliminary solution of the structure was obtained by molecular replacement with the program MOLREP (Vagin & Teplyakov, 2010 ▶) using TenA from S. epidermidis as the search model. The best results gave a correlation coefficient of 51.8% and an R factor of 45.8% at 15–3.5 Å resolution for four molecules in the asymmetric unit. Examination of the best solution revealed good crystal packing and no clashes between symmetry-related molecules. Model building with Coot (Emsley & Cowtan, 2004 ▶), sequence adaptation and further refinement of the model continue.
Figure 3.
Sequence alignment of S. aureus and S. epidermidis TenA. The secondary-structure regions are indicated and the active-site residues are shown in red.
Table 1. Summary of data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Data-collection parameters | |
| Wavelength (Å) | 0.8123 |
| Temperature (K) | 100 |
| Oscillation range (°) | 0.8 |
| Crystal-to-detector distance (mm) | 256 |
| Data-integration statistics | |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 103.5, b = 104.1, c = 109.6 |
| Resolution limits (Å) | 30–2.60 |
| Total No. of reflections | 424031 |
| No. of unique reflections | 31741 |
| Multiplicity | 5.1 (4.9) |
| Completeness (%) | 95.3 (95.0) |
| Rmerge† | 0.042 (0.48) |
| Mean I/σ(I) | 23.0 (3.3) |
| Molecules per asymmetric unit | 4 |
| VM‡ (Å3 Da−1) | 2.85 |
| Solvent content (%) | 56 |
R
merge = 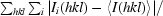
 , where 〈I(hkl)〉 is the mean intensity of the observations Ii(hkl) of reflection hkl.
, where 〈I(hkl)〉 is the mean intensity of the observations Ii(hkl) of reflection hkl.
For four molecules in the asymmetric unit.
Acknowledgments
This project was supported by a joint grant from the Deutscher Akademischer Austauschdienst (DAAD), Germany and by a grant from Deutsche Forschungsgemeinschaft (WR 124/2). JD, MP, CW and CB are members of the Hamburg School for Structure and Dynamics in Infection (SDI) which is supported by the Hamburg Ministry of Science and Research and Joachim Herz Stiftung as part of the Hamburg Initiative for Excellence in Research (LEXI).
References
- Barison, N., Cendron, L., Trento, A., Angelini, A. & Zanotti, G. (2009). FEBS J. 276, 6227–6235. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fey, P. D., Salid-Salim, B., Rupp, M. E., Hinrichs, S. H., Boxrud, D. J., Davis, C. C., Kreiswirth, B. N. & Schlievert, P. M. (2003). Antimicrob. Agents Chemother. 47, 196–203. [DOI] [PMC free article] [PubMed]
- Jurgenson, C. T., Begley, T. P. & Ealick, S. E. (2009). Annu. Rev. Biochem. 78, 569–603. [DOI] [PMC free article] [PubMed]
- Kauffman, C. A., Terpenning, M. S., He, X., Zarins, L. T. & Ramsey, M. A., Jorgensen, K. A., Sottile, W. S. & Bradley, S. F. (1993). Am. J. Med. 94, 371–378. [DOI] [PubMed]
- Lowy, F. D. (1998). N. Engl. J. Med. 339, 520–532. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Müller, I. B., Bergmann, B., Groves, M. R, Couto, I., Amaral, L., Begley, T. P., Walter, R. D. & Wrenger, C. (2009). PLoS ONE, 4, e7656. [DOI] [PMC free article] [PubMed]
- Ogershok, P. R., Rahman, A., Nestor, S. & Brick, J. (2002). Am. J. Med. Sci. 323, 107–111. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Platt, B. S. & Lu, G. D. (1936). Q. J. Med. 5, 355–374.
- Pohl, M., Sprenger, G. A. & Müller, M. (2004). Curr. Opin. Biotechnol. 4, 335–342. [DOI] [PubMed]
- Powers, K. A., Terpenning, M. S., Voice, R. A. & Kauffman, C. A. (1990). Am. J. Med. 88, 9–13. [PubMed]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press.
- Toms, A. V., Haas, A. L., Park, J.-H., Begley, T. P. & Ealick, S. E. (2005). Biochemistry, 44, 2319–2329. [DOI] [PubMed]
- Tumbarello, M., Tacconelli, E., de Gaetano, K., Ardito, F., Pirronti, T., Cauda, R. & Ortona, L. (1998). J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 18, 39–45. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Weinke, T., Schiller, R., Fehrenbach, F. J. & Pohle, H. D. (1992). Eur. J. Clin. Microbiol. Infect. Dis. 11, 985–989. [DOI] [PubMed]