α-Glucuronidase from S. pristinaespiralis was crystallized by the sitting-drop vapour-diffusion method. The crystals belonged to space group R3 and diffracted to a resolution of 1.9 Å.
Keywords: α-glucuronidase, glycosyl hydrolase family 115, Streptomyces pristinaespiralis
Abstract
α-Glucuronidase from Streptomyces pristinaespiralis (SpGlcA115A) is composed of a single-chain peptide containing a catalytic domain belonging to glycosyl hydrolase family 115, a novel family of hemicellulolytic α-glucuronidases. The enzyme catalyzes the hydrolysis of α-linked 4-O-methylglucuronosyl and glucuronosyl residues from both polymeric xylans and oligosaccharides. SpGlcA115A was crystallized at 293 K using the sitting-drop vapour-diffusion method. The crystals belonged to space group R3 and diffracted to a resolution of 1.9 Å.
1. Introduction
Plant cell walls are the most abundant source of biomass in nature and are of increasing importance with the worldwide focus of attention on bioethanol production to combat global warming and to safeguard global energy supplies. Because of competition between food and fuel production, lignocelluloses are expected to be utilized in the future production of ethanol for fuel. Xylan is the second most abundant polysaccharide after cellulose in nature. The efficient hydrolysis of plant xylans using microbial enzymes represents part of our efforts to develop environmentally friendly processes for the conversion of plant biomass. The enzymes that hydrolyse xylan can be divided into two categories: (i) enzymes that degrade the main polysaccharide chain, including endo-β-1,4-xylanases (EC 3.2.1.8) and β-xylosidases (EC 3.2.1.37), and (ii) enzymes that liberate side chains, main-chain substituents and the so-called accessory xylanolytic enzymes, including α-glucuronidases (EC 3.2.1.139), α-l-arabinofuranosidases (EC 3.2.1.55), acetylxylan esterases (EC 3.1.1.72) and feruloyl esterases (EC 3.1.1.73) (Biely, 2003 ▶). Xylan side chains disrupt the attack of xylanases. Therefore, accessory xylanolytic enzymes are important in the hydrolysis of xylan.
Glycosyl hydrolases (GHs) are classified based on the similarity of their amino-acid sequences, which imply both structural and mechanistic relationships (Cantarel et al., 2009 ▶). Among the GH families containing accessory xylanolytic enzymes, only α-glucuronidases (GH67) have been recognized as a family of enzymes that do not attack polymeric xylans (Siika-aho et al., 1994 ▶; Zaide et al., 2001 ▶). The only α-glucuronidase described to date that is capable of liberating 4-O-methyl-d-glucuronic acid (MeGlcA) side chains from hardwood glucuronoxylan is the enzyme present in the cellulolytic system of the wood-rotting fungus Schizophyllum commune (Tenkanen & Siika-aho, 2000 ▶). The N-terminal sequence of this enzyme was reported by Tenkanen & Siika-aho (2000 ▶); however, it did not match any gene sequences in available databases. Recently, we reported the isolation of an extracellular α-glucuronidase from the xylanolytic system of the xylose-fermenting xylanolytic yeast Pichia stipitis CBS 6054, the genome sequence of which recently became available; the results of this study led to the emergence of a new family of α-glucuronidases: GH115 (Ryabova et al., 2009 ▶). Actinomycetes have many kinds of hemicellulases; we are interested in the lignocellulose-degrading enzyme system of actinomycetes (Ichinose et al., 2009 ▶; Fujimoto et al., 2010 ▶). A BLAST search using the deduced amino-acid sequence of P. stipitis α-glucuronidase showed similarity to a hypothetical protein from Streptomyces pristinaespiralis (GenBank accession No. EDY63299; 34% identity and 51% similarity; Ryabova et al., 2009 ▶). EDY63299 appeared to involve a contiguous xylan-degrading gene cluster together with a GH3 β-xylosidase (EDY63298) and a GH10 β-xylanase (EDY63301) in the S. pristinaespiralis genome, suggesting that there is high possibility that the strain possesses an α-glucuronidase. The gene encoding the putative α-glucuronidase was cloned and the recombinant protein was successfully expressed in Escherichia coli. The purified protein showed activity towards birchwood xylan and beechwood xylan. The crystal structures of GH115 α-glucuronidases should provide important information describing their substrate-recognition mechanisms, how they differ from GH67 enzymes and how they recognize the side chains of polymers. For this reason, we analyzed the crystal structure of S. pristinaespiralis α-glucuronidase (SpGlcA115A).
2. Materials and methods
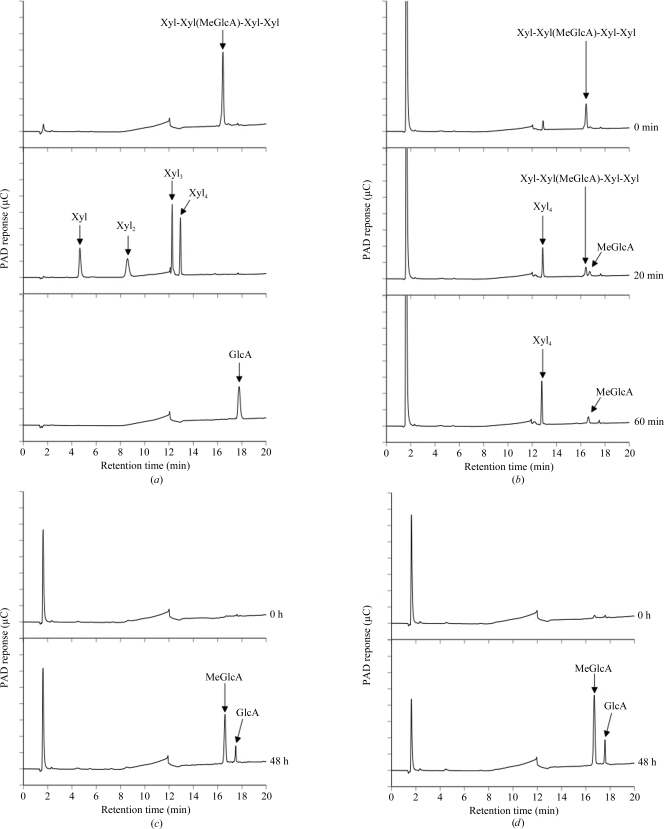
The gene encoding a putative α-glucuronidase (SpriA_010100008072; GenBank accession No. EDY63299) was amplified from S. pristinaespiralis genomic DNA by PCR using Phusion DNA polymerase (Finnzymes, Espoo, Finland) and the following primers: forward, 5′-CAT ATG GCG CCG GGT GGA GGA ACG AGC CGA-3′, and reverse, 5′-AAG CTT ACG CAG CCG CAT CGT CTC TGG AGG-3′. The amplified DNA encoding the mature region of the putative α-glucuronidase was digested with NdeI and HindIII and then cloned into the pET30 expression vector. SpGlcA115A was expressed as a 1032-amino-acid protein in E. coli Tuner (DE3). The transformants were grown in LB medium containing 50 µg ml−1 kanamycin at 310 K until they reached an absorbance of 0.6 at 600 nm. Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside and the culture was continued for 24 h at 298 K. The cells were harvested and resuspended in 50 mM phosphate buffer pH 7.2; they then underwent sonication for 5 min. After centrifugation to remove insoluble material, the supernatant was loaded onto an Ni–NTA agarose (Qiagen GmbH, Hilden, Germany) column (5 × 50 mm). The eluted enzyme was identified by SDS–PAGE as a 111 kDa protein; however, many proteins with different molecular weights were also detected. Therefore, the relevant fractions were pooled, dialyzed against 50 mM phosphate buffer pH 7.0 and applied onto a DEAE-Sepharose (GE Healthcare, USA) column (5 × 50 mm) equilibrated with the same buffer. The bound materials were eluted with a linear gradient of sodium chloride (0–1 M) and identified by SDS–PAGE. The relevant fractions were pooled, concentrated and applied onto a Superose 12 10/300 GL (GE Healthcare; 10 × 300 mm) column equilibrated with 20 mM Tris–HCl buffer pH 7.0 containing 0.2 M sodium chloride. The eluted enzyme was identified by SDS–PAGE and the final preparation was used as the purified enzyme (Fig. 1 ▶, lane 2). α-Glucuronidase activity was assessed by thin-layer chromatography analysis as described by Ryabova et al. (2009 ▶) and by high-performance anion-exchange chromatography with a pulsed amperometric detection system (HPAEC-PAD; Dionex, Sunnyvale, California, USA). Birchwood xylan and beechwood xylan purchased from Sigma (St Louis, Missouri, USA) were also used as substrates. Glucuronic acid (GlcA) and xylose were from Nacalai Tesque (Kyoto, Japan). Xylooligosaccharides were from Megazyme (Wicklow, Ireland). The reaction mixture consisted of 2%(w/v) substrate, 50 mM sodium acetate buffer pH 5.0 and 15 µg ml−1 enzyme. The mixtures were incubated at 313 K for 48 h and the reaction was then stopped by boiling for 10 min at 373 K. For the hydrolysis of aldopentauonic acid [2′′-O-α-(4-O-methyl-α-d-glucuronosyl)-xylotetraose; Xyl-Xyl(MeGlcA)-Xyl-Xyl; Biely et al., 1997 ▶], the enzyme (1.5 µg ml−1) was incubated with 0.001%(w/v) substrate in 50 mM sodium acetate buffer pH 5.0 at 313 K for 1 h. The samples were analyzed using a CarboPac PA1 column (Dionex) and elution with 0.1 M NaOH (0–5 min) followed by a linear gradient (5–35 min) of sodium acetate (0–0.4 M) at a flow rate of 1 ml min−1. SpGlcA115A liberated MeGlcA from Xyl-Xyl(MeGlcA)-Xyl-Xyl (Fig. 2 ▶ b) and liberated both MeGlcA and glucuronic acid from birchwood or beechwood xylan (Figs. 2 ▶ c and 2 ▶ d).
Figure 1.
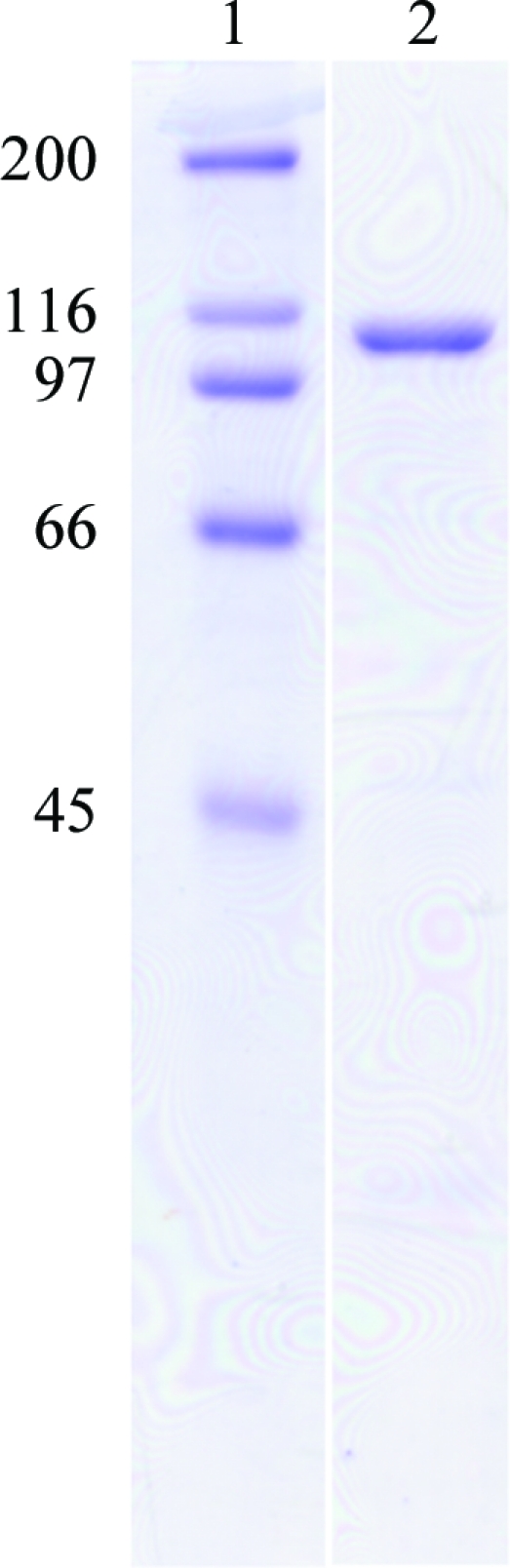
SDS–PAGE analysis of recombinant SpGlcA115A. The protein was subjected to SDS–PAGE on a 10% polyacrylamide gel and the bands were visualized by Coomassie Brilliant Blue R-250 staining. Lane 1, molecular-mass markers (kDa); lane 2, purified recombinant SpGlcA115A. The molecular mass of SpGlcA115A as estimated from SDS–PAGE was found to be 111 kDa. This was in agreement with the predicted molecular mass of 114 kDa.
Figure 2.
HPAEC-PAD analysis of hydrolysis products from aldopentauronic acid and xylans generated by recombinant SpGlcA115A. (a) Standards, (b) hydrolysis products from Xyl-Xyl(MeGlcA)-Xyl-Xyl, (c) hydrolysis products from birchwood xylan, (d) hydrolysis products from beechwood xylan. Abbreviations: Xyl, xylose; Xyl2, xylobiose; Xyl3, xylotriose; Xyl4, xylotetraose; MeGlcA, 4-O-methyl-d-glucuronic acid; GlcA, glucuronic acid.
The protein solution was concentrated to 3 mg ml−1 [A 280(1 mm) = 1.35 U] by ultrafiltration using a YM-30 membrane (Millipore, Massachusetts, USA) and filtered through a 0.1 µm membrane (Millipore, Massachusetts, USA). The protein was pooled in 20 mM Tris–HCl buffer pH 7.0 containing 0.2 M NaCl for crystallization trials. Sparse-matrix crystal screening was performed using several crystallization kits from Emerald BioStructures, Washington, USA (Wizard Screens I and II), Hampton Research, California, USA (Index HT) and Qiagen, Hilden, Germany (JCSG+ Suite). Sitting-drop vapour-diffusion trials were set up in 96-well Intelli-Plates (Art Robbins Instruments, California, USA) at 293 K using 50 µl reservoir solution, with each drop consisting of 0.3 µl protein solution and 0.3 µl reservoir solution. Within a few days, several crystals were observed using polyethylene glycol 2000 monomethyl ester (PEG 2000 MME), PEG 3000, PEG 3350 or PEG 10 000 as the precipitating agent at pH 5.5–7.0. Crystallization conditions were optimized manually by refinement of the pH and the concentration of the precipitant solution using CrystalClear Strips 96-well sitting-drop plates (Douglas Instruments, Berkshire, England). The quality of the obtained crystals was checked from diffraction images obtained using an R-AXIS VII X-ray diffractometer and a MicroMax-007 X-ray generator (Rigaku, Tokyo, Japan). Because of the good reproducibility of the crystals, the condition using 16%(w/v) PEG 2000 MME was adopted.
Native diffraction data were collected on beamline BL-NE3 of the Photon Factory-Advanced Ring (PF-AR), High Energy Accelerator Research Organization, Tsukuba, Japan. Crystals were soaked in reservoir solution containing 15% glycerol, immediately scooped into a 0.2 mm nylon loop (Hampton Research, California, USA) and flash-frozen in a nitrogen stream at 95 K. Diffraction data were collected with a 3 s exposure time for 1° oscillations over a total of 300° at a wavelength of 1.00 Å with a Quantum 270 CCD detector (ADSC, California, USA). Data were integrated and scaled using DENZO and SCALEPACK from the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
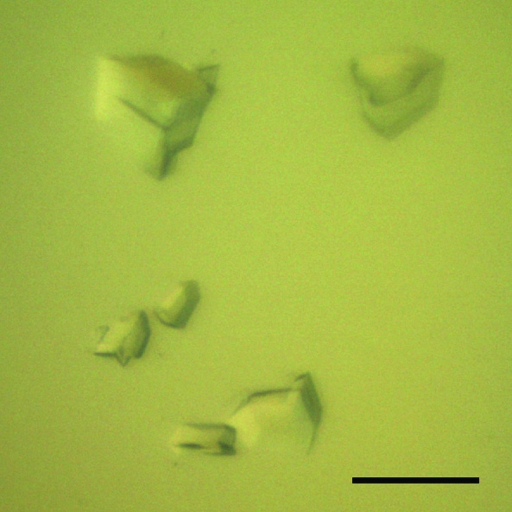
The reservoir solution of the optimized crystallization condition for SpGlcA115A was composed of 16%(w/v) PEG 2000 MME and 0.1 M Tris pH 7.0. Crystals with maximum dimensions of 100 × 100 × 50 µm were consistently obtained using 50 µl reservoir solution and drops consisting of 2 µl protein solution and 2 µl reservoir solution (Fig. 3 ▶). Using synchrotron radiation, the crystals diffracted to a maximum resolution of 1.9 Å and belonged to the rhombohedral space group R3, with unit-cell parameters a = b = 188.4, c = 97.8 Å, α = β = 90, γ = 120° in the hexagonal setting. The processing statistics of the collected data are summarized in Table 1 ▶. Assuming the presence of one molecule in the asymmetric unit, the Matthews coefficient was calculated to be 2.81 Å3 Da−1 (Matthews, 1968 ▶), corresponding to a solvent content of 56.3%. We are currently preparing a selenomethionine-substituted SpGlcA115A crystal (15 methionines per molecule) for phase determination by the MAD technique.
Figure 3.
Typical crystals of SpGlcA115A obtained using 20%(w/v) PEG 2000 MME and 0.1 M Tris pH 7.0. The scale bar is 100 µm in length.
Table 1. Data-collection statistics for the SpGlcA115A crystal.
Values in parentheses are for the highest resolution shell.
| Beamline | PF-AR BL-NE3 |
| Wavelength (Å) | 1.00 |
| Detector | ADSC Quantum 270 |
| No. of crystals for data collection | 1 |
| Crystal-to-detector distance (mm) | 229.1 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 300 |
| Exposure time per image (s) | 3 |
| Space group | R3 |
| Unit-cell parameters (Å, °) | a = b = 188.4, c = 97.8, α = β = 90, γ = 120 |
| Resolution range (Å) | 50.0–1.90 (1.97–1.90) |
| Rmerge† | 0.093 (0.391) |
| Completeness (%) | 99.7 (97.0) |
| Multiplicity | 9.4 (8.1) |
| Average I/σ(I) | 23.6 (4.4) |
| Unique reflections | 101979 (9902) |
| Observed reflections | 960656 |
| Mosaicity range (°) | 0.51–0.88 |
| Overall B factor from Wilson plot (Å2) | 20.1 |
R
merge = 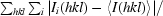
 , where I
i(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where I
i(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
Acknowledgments
We would like to thank the scientists and staff at the Photon Factory for the use of synchrotron radiation. This work was financially supported in part by a Grant-in-Aid (Development of Biomass Utilization Technologies for Revitalizing Rural Areas) from the Ministry of Agriculture, Forestry and Fisheries of Japan and grant VEGA 2/0001/10 from the Slovak Grant Agency.
References
- Biely, P. (2003). Handbook of Food Enzymology, edited by J. R. Whitaker, A. Voragen & D. Wong, pp. 879–916. New York: Marcel Dekker.
- Biely, P., Vršanská, M., Tenkanen, M. & Kluepfel, D. (1997). J. Biotechnol. 57, 151–166. [DOI] [PubMed]
- Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V. & Henrissat, B. (2009). Nucleic Acids Res. 37, D233–D238. [DOI] [PMC free article] [PubMed]
- Fujimoto, Z., Ichinose, H., Maehara, T., Honda, M., Kitaoka, M. & Kaneko, S. (2010). J. Biol. Chem. 285, 34134–34143. [DOI] [PMC free article] [PubMed]
- Ichinose, H., Fujimoto, Z., Honda, M., Harazono, K., Nishimoto, Y., Uzura, A. & Kaneko, S. (2009). J. Biol. Chem. 284, 25097–25106. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ryabova, O., Vršanská, M., Kaneko, S., van Zyl, W. H. & Biely, P. (2009). FEBS Lett. 583, 1457–1462. [DOI] [PubMed]
- Siika-aho, M., Tenkanen, M., Buchert, J., Puls, J. & Viikari, L. (1994). Enzyme Microb. Technol. 16, 813–819.
- Tenkanen, M. & Siika-aho, M. (2000). J. Biotechnol. 78, 149–161. [DOI] [PubMed]
- Zaide, G., Shallom, D., Shulami, S., Zolotinsky, G., Golan, G., Baasov, T., Shoham, G. & Shoham, Y. (2001). Eur. J. Biochem. 268, 3006–3016. [DOI] [PubMed]