The seryl-tRNA synthetase from C. albicans was crystallized by the sitting-drop vapour-diffusion method using ammonium sulfate as precipitant. The crystals belonged to the hexagonal space group P6122 and diffraction data were collected to 2.0 Å resolution at a synchrotron source.
Keywords: aminoacyl-tRNA synthetases, SerRS, genetic code ambiguity, Candida albicans
Abstract
The seryl-tRNA synthetase (SerRS) from Candida albicans exists naturally as two isoforms resulting from ambiguity in the natural genetic code. Both enzymes were crystallized by the sitting-drop vapour-diffusion method using 3.2–3.4 M ammonium sulfate as precipitant. The crystals belonged to the hexagonal space group P6122 and contained one monomer per asymmetric unit, despite the synthetase existing as a homodimer (with a molecular weight of ∼116 kDa) in solution. Diffraction data were collected to 2.0 Å resolution at a synchrotron source and the crystal structures of unliganded SerRS and of its complexes with ATP and with a seryl-adenylate analogue were solved by molecular replacement. The structure of C. albicans SerRS represents the first reported structure of a eukaryotic cytoplasmic SerRS.
1. Introduction
Aminoacyl-tRNA synthetases (aaRSs) produce the aminoacyl-tRNA substrates for protein synthesis. In order to translate the genome into proteome with high fidelity, aaRSs exhibit strict substrate specificity and are able to differentiate tRNAs and amino acids efficiently by following conserved genetic code rules (Ibba & Söll, 1999 ▶).
A unique genetic code is used in several species of the fungal CTG clade, where leucine CUG codons are ambiguously decoded as serine and leucine (Santos & Tuite, 1995 ▶; Santos et al., 1996 ▶; Gomes et al., 2007 ▶; Butler et al., 2009 ▶). This genetic code ambiguity is introduced by a mutant serine-tRNA (tRNACAGSer) which is recognized by both the leucyl-tRNA and seryl-tRNA synthetases (LeuRS and SerRS, respectively; Suzuki et al., 1997 ▶). In the cellular context, tRNACAGSer is charged with 95–97% serine and 0.6–5% leucine (Gomes et al., 2007 ▶). Hence, SerRS is the main operating aaRS for the translation of CUG codons. However, the SerRS gene contains a single CUG codon whose mistranslation results in the synthesis of two SerRS isoforms, SerRS_Ser197 (major isoform) and SerRS_Leu197 (minor isoform), which are constitutively expressed. The evolutionary pathway of CUG codons in the CTG-clade is poorly understood and led us to hypothesize that structural characterization of both SerRS variants should unveil novel features of the fungal tRNA-serylation system and may provide new clues about this novel codon-reprogramming event with implications for understanding the evolution of the genetic code of Candida albicans and, eventually, new features of its biology and pathogenesis.
2. Material and methods
2.1. Cloning of two C. albicans SerRS variants
The C. albicans SerRS coding sequence (GenBank accession No. AF290915) was amplified from strain CAI4 genomic DNA by PCR using the primers SerRS1 (5′-GGA ATT CCA TAT GTT AGA CAT TAA TGC ATT TCT CG-3′) and SerRS2 (5′-GGA TCC CGC TTT TCT TAC CTT TAG CTT TTT TAA C-3′). The PCR fragment was digested with NdeI and BamHI endonucleases and ligated to NdeI/SalI-digested pT7-7 expression plasmid (USB). The resulting expression vector encodes the full-length C. albicans SerRS followed by a 17-residue linker and a C-terminal hexahistidine (His6) tag (the linker and tag sequence is PKNTTSVKKAKGKNGSRHHHHHH).
The two natural C. albicans SerRS isoforms were generated by site-directed mutagenesis using primers 5′-GCT TTA ATC AAC TAC GGT TTA TCG TTT TTG AGT AGC AAA GGA TAC G-3′ and 5′-CGT ATC CTT TGC TAC TCA AAA ACG ATA AAC CGT AGT TGA TTA AAG C-3′ for the SerRS_Ser197 variant and primers 5′-CTT TAA TCA ACT ACG GTT TAT TGT TTT TGA GTA GCA AAG GAT ACG-3′ and 5′-CGT ATC CTT TGC TAC TCA AAA ACA ATA AAC CGT AGT TGA TTA AAG-3′ for the SerRS_Leu197 isoform.
2.2. Overexpression and purification of recombinant SerRS
Large-scale production of C. albicans SerRS was achieved in Escherichia coli BL21 (DE3) CodonPlus RIL (Stratagene) with culture inoculation by the plating method (Suter-Crazzolara & Unsicker, 1995 ▶). Expression cultures [LB medium with 100 µg ml−1 ampicillin, 34 µg ml−1 chloramphenicol and 1%(w/v) glucose] were incubated at 310 K until the OD600 reached ∼0.3. At this point, the incubation temperature was lowered to 303 K and the bacterial cells were allowed to grow until mid-exponential phase (0.5 < OD600 < 0.7). SerRS expression was induced by the addition of 0.5 mM IPTG (Biosynth) and continued for 3 h. Cells were harvested by centrifugation (8100g, 20 min, 277 K), resuspended in lysis buffer [50 mM Na HEPES pH 7.6, 100 mM KCl, 10 mM MgCl2, 20 mM imidazole and 10%(v/v) glycerol] supplemented with 1 mg ml−1 lysozyme (Sigma) and stored at 253 K until further use.
For SerRS purification, the cell suspensions were thawed and supplemented with protease inhibitors (Complete EDTA-free, Roche) and DNase I (AppliChem) to 5 µg ml−1 final concentration and the cells were disrupted by sonication. The homogenate was centrifuged (13 800g, 25 min, 277 K) and the supernatant was filtered (0.22 µm low-protein-binding PVDF membrane) and loaded onto a 5 ml HisTrap HP column (GE Healthcare) pre-equilibrated in lysis buffer. The bound protein was eluted with a 20–500 mM linear imidazole gradient in lysis buffer. EDTA (500 µM final concentration) was added to the SerRS-containing fractions (which eluted at ∼250 mM imidazole).
The SerRS-containing fractions were pooled, diluted 20-fold in buffer A [20 mM Na HEPES pH 7.6, 10 mM KCl, 5 mM MgCl2, 0.1 mM Na2EDTA and 5%(v/v) glycerol] and loaded onto a 5 ml Econo-Pac High Q cartridge (Bio-Rad) pre-equilibrated in buffer A. The bound SerRS was eluted with a linear 10–500 mM NaCl gradient (in buffer A) over 15 column volumes. The fractions corresponding to the SerRS peak were pooled, dialyzed against buffer B [50 mM Na HEPES pH 7.6, 150 mM KCl, 10 mM MgCl2 and 8%(v/v) glycerol] and concentrated (to ∼20 mg ml−1) using a 10 kDa molecular-weight cutoff ultracentrifugal concentration device (Millipore).
For analysis of the recombinant proteins by gel-filtration chromatography, 50 µl concentrated SerRS sample (4.0 mg ml−1) was applied onto a Superose 12 10/300 column (GE Healthcare) pre-equilibrated in buffer B at room temperature.
2.3. Crystallization
Initial crystallization conditions were obtained in sitting drops using a commercial ammonium-sulfate-based sparse-matrix screen (JBScreen Classic 6, Jena Bioscience). After refinement, reproducible growth of crystals of native recombinant SerRS (both isoforms) was obtained in drops composed of identical volumes of protein solution (8–14 mg ml−1) and reservoir solution [100 mM Na MES pH 5.6–5.8, 3.2–3.4 M ammonium sulfate and 0–2%(v/v) glycerol] equilibrated against a 500 µl reservoir. For preparation of the binary complexes SerRS–SerSA and SerRS–ATP, SerRS_Ser197 was pre-incubated on ice for 2–3 h with 15 mM 5′-O-[N-(l-seryl)sulfamoyl]adenosine (RNA-TEC) or 10 mM ATP (freshly prepared in buffer B; Sigma), respectively, and crystallized using 100 mM Na MES pH 5.8–6.2, 3.3–3.4 M ammonium sulfate and 0–5%(v/v) glycerol as precipitant solution. The crystals were then transferred sequentially to 2.9 and 3.4 M sodium malonate solution (for 5–10 s) and flash-cooled in liquid nitrogen.
2.4. SerRS thermal stability characterization
To characterize protein stability, the melting temperature (T m) of SerRS was determined by following its behaviour in solution by DLS (Zetasizer Nano, Malvern). SerRS (50 µl 1.0 mg ml−1) was heated from 298 to 326 K in a 45 µl quartz cuvette (Hellma Inc.). In order to reduce evaporation, the protein solution was overlaid with paraffin oil (20 µl). Protein aggregation was monitored by measuring Z-average diameter (nm) and scattering intensity (kilocounts per second) as a function of temperature, using 1 K temperature increments and 3 min equilibration before each measurement. The resulting inflection point of the particle-size curve was defined as T m.
2.5. Data collection and processing
All diffraction data were collected at 100 K from single crystals at the European Synchrotron Radiation Facility (ESRF, Grenoble, France). All data sets were collected using an ADSC Q4R detector on beamline ID14-EH1. The SerRS_Ser197 and SerRS–SerSA data sets were collected to 2.0 Å resolution in 0.5° oscillation steps (270 frames). Data for the SerRS–ATP complex were collected to 2.55 Å resolution in 1° oscillation steps over a range of 120°. The SerRS_Leu197 diffraction images were collected at 2.3 Å resolution in 0.6° oscillation steps (200 frames). Diffraction images were integrated with MOSFLM (Leslie, 1999 ▶) and scaled with SCALA (Collaborative Computational Project, Number 4, 1994 ▶).
2.6. Structure solution
The three-dimensional structure of C. albicans SerRS was solved by molecular replacement (MR) with Phaser (McCoy, 2007 ▶) using the catalytic domain of Pyrococcus horikoshii SerRS as the search model (PDB entry 2dq0; the model contained residues 107–447 of chain A with all non-identical non-glycine residues truncated to Ala; Itoh et al., 2008 ▶). The SerRS model was rebuilt with ARP/wARP (Perrakis et al., 2001 ▶) on electron-density maps calculated using the phases from the MR solution and the C. albicans SerRS sequence information. Refinement (energy-gradient minimization, simulated-annealing and restrained individual B-factor refinement) with CNS (Brünger et al., 1998 ▶) was alternated with manual model building using Coot (Emsley & Cowtan, 2004 ▶). The coordinates of SerRS_Ser197 were used as an MR search model to solve the three-dimensional structures of SerRS_Leu197, SerRS–SerSA and SerRS–ATP.
3. Results and discussion
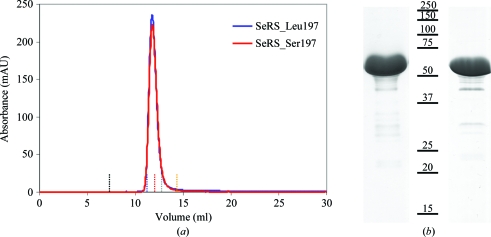
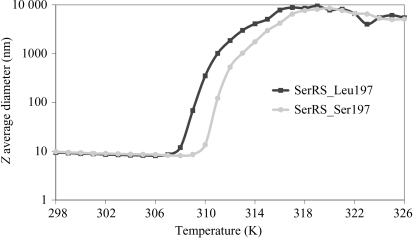
Recombinant C. albicans SerRS was purified in two chromatographic steps, yielding essentially pure material as judged by size-exclusion chromatography and SDS–PAGE analysis (Fig. 1 ▶). The protein yields were 10 and 4 mg of purified SerRS_Ser197 and SerRS_Leu197, respectively, per litre of culture. The apparent molecular weights of the proteins, as determined by gel-filtration chromatography (Fig. 1 ▶ a, Table 1 ▶) and dynamic light scattering (data not shown), suggest that they exist as homodimers in solution, as observed for other class II synthetases (First, 2005 ▶). In order to further understand the impact of Ser to Leu replacement on the dimer structure, the thermal stability of each C. albicans SerRS isoform was evaluated by dynamic light scattering (DLS; Fig. 2 ▶). The melting curve demonstrated that the presence of Leu at position 197 slightly decreases protein stability, presumably owing to the loss of polar interactions in SerRS_Leu197.
Figure 1.
Purification of C. albicans SerRS isoforms. (a) Analytical gel-filtration chromatography profile of the recombinant proteins showing that they both behave as dimers in solution with an apparent molecular weight of 116 kDa; vertical dotted lines highlight the elution volumes of the molecular-weight markers shown in Table 1 ▶ (black, blue dextran; blue, aldolase; red, albumin; grey, ovalbumin; yellow, chymotrypsin). (b) Coomassie Blue-stained SDS–PAGE (12.5%) analysis of recombinant SerRS isoforms (left, SerRS_Leu197; right, SerRS_Ser197). Both samples contained identical minor impurities that did not interfere with the crystallization process.
Table 1. Sample elution volume on a Superose 12 10/300 column (GE Healthcare).
| Elution volume (ml) | Molecular weight (kDa) | |
|---|---|---|
| Blue dextran | 7.43 | |
| Aldolase | 11.33 | 158 |
| Albumin | 12.21 | 67 |
| Ovalbumin | 13.00 | 43 |
| Chymotrypsin | 14.74 | 25 |
| SerRS_Ser197 | 11.79 | 116 |
| SerRS_Leu197 | 11.73 | 118 |
Figure 2.
Differential incorporation of serine or leucine at the CUG position 197 of SerRS alters the thermal stability. Comparison of the thermal denaturation profiles of SerRS_Ser197 and SerRS_Leu197 shows that the incorporation of Leu into both monomers of SerRS induces a decrease in thermal stability.
For both SerRS isoforms crystals appeared after 2 or 3 d (Fig. 3 ▶). The SerRS_Ser197 crystals and cocrystals were successfully cryoprotected in sodium malonate pH 6.0 solution. However, better diffraction data were obtained for the SerRS_Leu197 crystals when the pH of the cryoprotecting solution was lowered to 5.0. All SerRS crystals analyzed belonged to the hexagonal space group P6122, with unit-cell parameters a = b = 90.1, c = 276.8 Å (crystallographic statistics are reported in Table 2 ▶). Assuming the presence of one C. albicans SerRS molecule in the asymmetric unit, the calculated Matthews coefficient is 3.22 Å3 Da−1, which corresponds to a solvent content of 61.8% (Matthews, 1968 ▶).
Figure 3.

Recombinant C. albicans SerRS_Ser197 crystal belonging to space group P6122.
Table 2. Statistics of data collection and processing.
Values in parentheses are for the highest resolution shell.
| SerRS_Ser197 | SerRS_Leu197 | SerRS–SerSA† | SerRS–ATP† | |
|---|---|---|---|---|
| X-ray source | ESRF ID14-EH1 | ESRF ID14-EH1 | ESRF ID14-EH1 | ESRF ID14-EH1 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Wavelength (Å) | 0.934 | 0.934 | 0.934 | 0.934 |
| Space group | P6122 | P6122 | P6122 | P6122 |
| Unit-cell parameters (Å) | a = b = 90.1, c = 276.8 | a = b = 92.5, c = 273.6 | a = b = 91.0, c = 275.8 | a = b = 91.2, c = 275.8 |
| Solvent content (%) | 58.9 | 60.6 | 59.7 | 59.8 |
| Resolution (Å) | 78.09–2.00 (2.11–2.00) | 80.06–2.30 (2.42–2.30) | 78.81–2.00 (2.11–2.00) | 79.06–2.55 (2.69–2.55) |
| No. of reflections (total/unique) | 955175/43773 | 624090/31871 | 1144615/46397 | 314641/23155 |
| Multiplicity | 21.8 (15.0) | 19.6 (12.4) | 24.7 (14.9) | 13.6 (13.3) |
| Completeness (%) | 95.7 (78.0) | 100.0 (100.0) | 99.5 (98.9) | 100.0 (100.0) |
| Rmerge‡ (%) | 6.9 (23.0) | 9.8 (44.2) | 9.1 (56.2) | 8.5 (57.3) |
| Rp.i.m.§ (%) | 1.4 (6.0) | 2.1 (12.8) | 1.8 (14.9) | 2.4 (16.1) |
| 〈I/σ(I)〉 | 7.8 (3.1) | 6.0 (1.7) | 6.9 (1.4) | 8.0 (1.3) |
The complexes with SerSA and ATP were prepared using the Ser_Ser197 isoform.
R
merge = 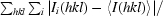
 , where Ii(hkl) and 〈I(hkl)〉 are the observed intensity and the average intensity of multiple observations of symmetry-related reflections, respectively.
, where Ii(hkl) and 〈I(hkl)〉 are the observed intensity and the average intensity of multiple observations of symmetry-related reflections, respectively.
R
p.i.m. = 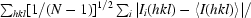
 , where Ii(hkl) is the observed intensity, 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections and N is the multiplicity.
, where Ii(hkl) is the observed intensity, 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections and N is the multiplicity.
The molecular coordinates of the catalytic domain of P. horikoshii SerRS (PDB entry 2dq0; Itoh et al., 2008 ▶) were used to solve the structure of the C. albicans enzyme by the molecular-replacement method. The program Phaser (McCoy, 2007 ▶) located one monomer of C. albicans SerRS_Ser197 in the asymmetric unit (rotation-function Z score of 6.8, translation-function Z score of 25.1). The electron-density map obtained was of good quality and a significant portion of the model (403 out of 471 residues) could be traced automatically with ARP/wARP (Perrakis et al., 2001 ▶). The difference electron-density maps obtained for the SerRS–SerSA and SerRS–ATP complexes showed unambiguous density for the ligands at the SerRS active site. The structures of both C. albicans SerRS isoforms are currently in the final stages of refinement and will be compared in order to evaluate the structural impact of serine/leucine exchange at the CUG codon position.
Acknowledgments
We acknowledge the ESRF (Grenoble, France) for the provision of synchrotron-radiation facilities, the ESRF staff for assistance in using beamline ID14EH1 and Frederico Silva (UP3 – Protein Production and Purification Unit, IBMC, Porto) for providing technical support during protein purification. This work was funded by Fundação para a Ciência e a Tecnologia (FCT), Portugal through grants PTDC/SAU-MII/70634/2006, PTDC/BIA-BCM/64745/2006, PTDC/SAU-GMG/098850/2008, PTDC/BIA-MIC/099826/2008 and REEQ/564/BIO/2005 (EU-FEDER and POCI 2010). RR is an FCT (BD/15233/2004) and Doctoral Programme on Experimental Biology and Biomedicine (University of Coimbra) PhD fellow.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Butler, G. et al. (2009). Nature (London), 459, 657–662.
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- First, E. A. (2005). The Aminoacyl-tRNA Synthetases, edited by M. Ibba, C. Francklyn & S. Cusack, pp. 328–352. Georgetown: Landes Bioscience.
- Gomes, A. C., Miranda, I., Silva, R. M., Moura, G. R., Thomas, B., Akoulitchev, A. & Santos, M. A. (2007). Genome Biol. 8, R206. [DOI] [PMC free article] [PubMed]
- Ibba, M. & Söll, D. (1999). Science, 286, 1893–1897. [DOI] [PubMed]
- Itoh, Y., Sekine, S., Kuroishi, C., Terada, T., Shirouzu, M., Kuramitsu, S. & Yokoyama, S. (2008). RNA Biol. 5, 169–177. [DOI] [PubMed]
- Leslie, A. G. W. (1999). Acta Cryst. D55, 1696–1702. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J. (2007). Acta Cryst. D63, 32–41. [DOI] [PMC free article] [PubMed]
- Perrakis, A., Harkiolaki, M., Wilson, K. S. & Lamzin, V. S. (2001). Acta Cryst. D57, 1445–1450. [DOI] [PubMed]
- Santos, M. A., Perreau, V. M. & Tuite, M. F. (1996). EMBO J. 15, 5060–5068. [PMC free article] [PubMed]
- Santos, M. A. & Tuite, M. F. (1995). Nucleic Acids Res. 23, 1481–1486. [DOI] [PMC free article] [PubMed]
- Suter-Crazzolara, C. & Unsicker, K. (1995). Biotechniques, 19, 202–204. [PubMed]
- Suzuki, T., Ueda, T. & Watanabe, K. (1997). EMBO J. 16, 1122–1134. [DOI] [PMC free article] [PubMed]