Abstract
Mice contain at least seven nonallelic forms of the H1 histones, including the somatic variants H1a-e and less closely related variants H1 degrees and H1t. The mouse H1 degrees and H1c (H1var.1) genes were isolated and characterized previously. We have now isolated, sequenced and studied the expression properties of two additional mouse H1 genes, termed H1var.2 and H1var.3. Extensive amino acid and nucleotide sequence comparisons were made between the two genes and other mammalian H1 histone genes. A high degree of nucleotide sequence identity was seen between the H1var.2, rat H1d and human H1b genes, even well beyond the coding region, indicating that these genes are likely homologues. Unlike the previously characterized mouse H1var.1 gene which produces both nonpolyadenylated and polyadenylated mRNAs, the H1var.2 and H1var.3 genes produce only typical, replication dependent, nonpolyadenylated mRNAs.
Full text
PDF


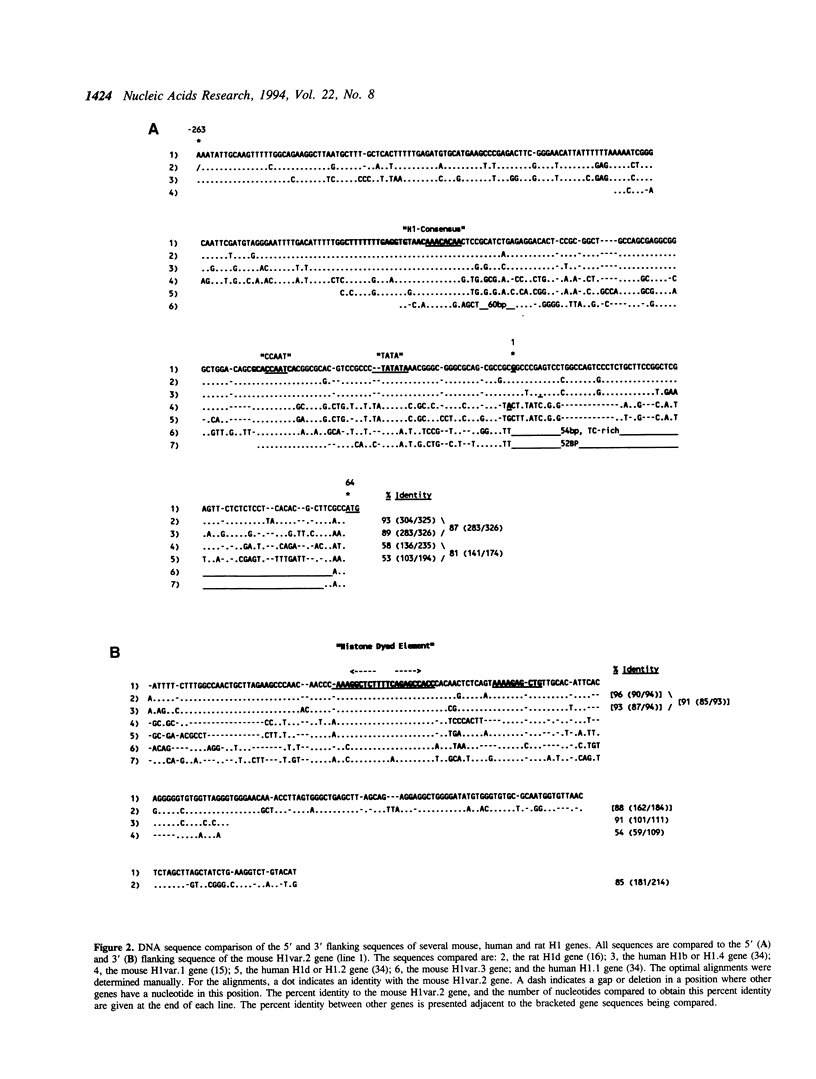




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albig W., Kardalinou E., Drabent B., Zimmer A., Doenecke D. Isolation and characterization of two human H1 histone genes within clusters of core histone genes. Genomics. 1991 Aug;10(4):940–948. doi: 10.1016/0888-7543(91)90183-f. [DOI] [PubMed] [Google Scholar]
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986 Feb 20;187(4):591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Brown D. T., Sittman D. B. Identification through overexpression and tagging of the variant type of the mouse H1e and H1c genes. J Biol Chem. 1993 Jan 5;268(1):713–718. [PubMed] [Google Scholar]
- Cheng G. H., Nandi A., Clerk S., Skoultchi A. I. Different 3'-end processing produces two independently regulated mRNAs from a single H1 histone gene. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7002–7006. doi: 10.1073/pnas.86.18.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G. H., Skoultchi A. I. Rapid induction of polyadenylated H1 histone mRNAs in mouse erythroleukemia cells is regulated by c-myc. Mol Cell Biol. 1989 Jun;9(6):2332–2340. doi: 10.1128/mcb.9.6.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole K. D., Kandala J. C., Kremer E., Kistler W. S. Isolation of a genomic clone encoding the rat histone variant, H1d. Gene. 1990 May 14;89(2):265–269. doi: 10.1016/0378-1119(90)90015-j. [DOI] [PubMed] [Google Scholar]
- Coles L. S., Wells J. R. An H1 histone gene-specific 5' element and evolution of H1 and H5 genes. Nucleic Acids Res. 1985 Jan 25;13(2):585–594. doi: 10.1093/nar/13.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croston G. E., Kerrigan L. A., Lira L. M., Marshak D. R., Kadonaga J. T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991 Feb 8;251(4994):643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- Croston G. E., Laybourn P. J., Paranjape S. M., Kadonaga J. T. Mechanism of transcriptional antirepression by GAL4-VP16. Genes Dev. 1992 Dec;6(12A):2270–2281. doi: 10.1101/gad.6.12a.2270. [DOI] [PubMed] [Google Scholar]
- Doenecke D., Tönjes R. Differential distribution of lysine and arginine residues in the closely related histones H1 and H5. Analysis of a human H1 gene. J Mol Biol. 1986 Feb 5;187(3):461–464. doi: 10.1016/0022-2836(86)90446-8. [DOI] [PubMed] [Google Scholar]
- Drabent B., Kardalinou E., Doenecke D. Structure and expression of the human gene encoding testicular H1 histone (H1t). Gene. 1991 Jul 22;103(2):263–268. doi: 10.1016/0378-1119(91)90284-i. [DOI] [PubMed] [Google Scholar]
- Drabent B., Kunz C., Doenecke D. A rat histone H2B pseudogene is closely associated with the histone H1d gene. Biochim Biophys Acta. 1993 Feb 20;1172(1-2):193–196. doi: 10.1016/0167-4781(93)90293-m. [DOI] [PubMed] [Google Scholar]
- Harris M. E., Böhni R., Schneiderman M. H., Ramamurthy L., Schümperli D., Marzluff W. F. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991 May;11(5):2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hurt M. M., Bowman T. L., Marzluff W. F. A common transcriptional activator is located in the coding region of two replication-dependent mouse histone genes. Mol Cell Biol. 1991 Jun;11(6):2929–2936. doi: 10.1128/mcb.11.6.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- La Bella F., Zhong R., Heintz N. Cell type-specific expression of a human histone H1 gene. J Biol Chem. 1988 Feb 15;263(5):2115–2118. [PubMed] [Google Scholar]
- Lachman H. M., Cheng G. H., Skoultchi A. I. Transfection of mouse erythroleukemia cells with myc sequences changes the rate of induced commitment to differentiate. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6480–6484. doi: 10.1073/pnas.83.17.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Threshold phenomena and long-distance activation of transcription by RNA polymerase II. Science. 1992 Sep 18;257(5077):1682–1685. doi: 10.1126/science.1388287. [DOI] [PubMed] [Google Scholar]
- Lennox R. W., Oshima R. G., Cohen L. H. The H1 histones and their interphase phosphorylated states in differentiated and undifferentiated cell lines derived from murine teratocarcinomas. J Biol Chem. 1982 May 10;257(9):5183–5189. [PubMed] [Google Scholar]
- Marzluff W. F., Pandey N. B. Multiple regulatory steps control histone mRNA concentrations. Trends Biochem Sci. 1988 Feb;13(2):49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McKnight S., Tjian R. Transcriptional selectivity of viral genes in mammalian cells. Cell. 1986 Sep 12;46(6):795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Ohe Y., Hayashi H., Iwai K. Human spleen histone H1. Isolation and amino acid sequence of a main variant, H1b. J Biochem. 1986 Aug;100(2):359–368. doi: 10.1093/oxfordjournals.jbchem.a121722. [DOI] [PubMed] [Google Scholar]
- Ohe Y., Hayashi H., Iwai K. Human spleen histone H1. Isolation and amino acid sequences of three minor variants, H1a, H1c, and H1d. J Biochem. 1989 Nov;106(5):844–857. doi: 10.1093/oxfordjournals.jbchem.a122941. [DOI] [PubMed] [Google Scholar]
- Shimamura A., Sapp M., Rodriguez-Campos A., Worcel A. Histone H1 represses transcription from minichromosomes assembled in vitro. Mol Cell Biol. 1989 Dec;9(12):5573–5584. doi: 10.1128/mcb.9.12.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Wellman S. E., Marzluff W. F. Sequences of four mouse histone H3 genes: implications for evolution of mouse histone genes. J Mol Evol. 1986;23(3):242–249. doi: 10.1007/BF02115580. [DOI] [PubMed] [Google Scholar]
- Yang Y. S., Brown D. T., Wellman S. E., Sittman D. B. Isolation and characterization of a mouse fully replication-dependent H1 gene within a genomic cluster of core histone genes. J Biol Chem. 1987 Dec 15;262(35):17118–17125. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]