Abstract
The hepatitis C virus (HCV) core protein is essential for viral genome encapsidation and plays an important role in steatosis, immune evasion, and hepatocellular carcinoma. It may thus represent a promising therapeutic target to interfere with the HCV life-cycle and related pathogenesis. In this study, we used phage display to generate single-chain variable domain antibody fragments (scFv) to the core protein from bone marrow plasma cells of patients with chronic hepatitis C. An antibody with high-affinity binding (scFv42C) was thus identified, and the binding site was mapped to the PLXG motif (residues 84–87) of the core protein conserved among different genotypes. Whereas scFv42C displayed diffuse cytoplasmic fluorescence when expressed alone in the Huh7 human hepatoma cell line, cotransfection with the core gene shifted its subcellular distribution into that of core protein. The intracellular association of scFv42C with its target core protein was independently demonstrated by the fluorescence resonance energy transfer technique. Interestingly, expression of the single-chain antibody reduced core protein levels intracellularly, particularly in the context of full HCV replication. Moreover, cell proliferation as induced by the core protein could be reversed by scFv4C coexpression. Therefore, scFv42C may represent a novel anti-HCV agent, which acts by sequestering core protein and attenuating core protein–mediated pathogenesis.
The positive-stranded RNA genome of hepatitis C virus (HCV) encodes a single polyprotein of about 3000 amino acids, which is cleaved cotranslationally or posttranslationally by host and viral proteases to yield individual functional viral proteins.1–3 The structural proteins core and E1/E2 are required for the formation of virus particles, while the nonstructural proteins NS2-NS5 are components of a complex required for viral RNA replication.1,4 The core protein, which comprises the N-terminal portion of the HCV polyprotein, is generated by cleavage with signal peptidases at the endoplasmatic reticulum, 2 and forms the nucleocapsid that encompasses the viral genome. Its N-terminal region (residues 1–50) is highly basic and contains RNA and DNA binding domains, as well as putative nuclear localization signals. The C-terminus is hydrophobic and mediates anchorage to the endoplasmic reticulum. 6,7 Two different forms of core protein have been identified. The longer form (p21) is composed of residues 1–191 of the polyprotein and localizes to the cytoplasm in association with the endoplasmic reticulum membrane. 7 The shorter form consists of residues 1–173 (p19) and is derived from p21. It can be translocated to the nucleus depending on the cell line studied. 8
The HCV core protein may possess multiple functions and has been implicated in a diverse array of pathogenetic events including oxidative stress, steatosis, immune suppression, and carcinogenesis. Both in vitro and in vivo experiments have suggested the impact of the core protein on lipid metabolism, cell growth, signaling, apoptosis, and immunity.5,9–13 Hepatic steatosis arises at a high rate in patients with chronic hepatitis C, and a close correlation with intrahepatic core protein expression level has been noted. 14 Consequently, core protein alone was sufficient to induce hepatic steatosis and hepatocellular carcinoma in transgenic mice.11,12 Indeed, transient expression of core protein in human hepatoma cells upregulated transcription of most genes involved in fat/lipid metabolism. 10 The core protein also disregulates growth signaling. It could interact with and maintain an activated form of the RNA-dependent protein kinase (PKR), an enzyme reported to be activated in some cancer cell lines and tissues. 15,16 Moreover, the core protein could promote proliferation of human hepatoma cells by activation of the MAPK/ERK (mitogen-activated protein kinase/extracellular signal-related kinase) pathway17 or the Wnt-1 signaling, 10 the two frequently activated pathways in hepatocellular carcinoma tissues. HCV core protein has also been found to interact with pathways involved in host immune defense, such as the Janus kinase (JAK)–signal transducer and activator of transcription factor (STAT) signaling pathway, a major cascade mediating antiviral response under interferon-α. Core ore protein expression inhibited interferon-α–induced nuclear import of STATs. 18
Due to the lack of a preventive vaccine and effective antiviral therapies, HCV infection remains one of the major health problems worldwide. In this regard, intracellular antibodies, in particular single-chain variable fragments (scFv), have been used successfully to ablate the function of target antigens in different subcellular compartments. The recombinant scFv antibody consists of the variable domain of the heavy chain (VH) connected to the variable domain of the light chain (VL) via a polypeptide linker, thus generating a fully functional antigen-binding unit. 19,20 Tethering the variable domains into a single molecule facilitates expression and stability of antibodies inside cells in comparison to conventional immunoglobulins.21 The scFv antibodies have the potential to alter target protein folding, interaction, modification, or subcellular localization, and have been proposed as therapeutics against cancer and human immunodeficiency virus (HIV). Isolation of high-affinity scFv antibodies specific for the target protein is made possible by phage display technology, which permits the expression of combinatorial libraries of immunoglobulin genes on the surface of filamentous bacteriophages.22,23 Phages displaying antibodies with high affinity against the target molecule can be enriched during several rounds of affinity selection and reamplification (panning), and DNA sequences for such antibodies can be determined directly from the purified phages.
The importance of the HCV core protein in the viral life cycle and pathogenesis makes it an ideal target of the scFv antibodies, because its sequestration may have the potential to abrogate core particle assembly and RNA packaging, as well as its direct effect on cell proliferation and lipid metabolism. Furthermore, core protein is one of the most conserved HCV proteins; a single-chain antibody targeting a conserved epitope in the core protein may have a broad effect on viral isolates belonging to different genotypes. Considering that application of non-human antibodies for gene therapy of human diseases will elicit an immune response, employment of human scFv minimizes such risk, as illustrated by clinical evaluation of two human monoclonal antibodies (mAbs) against hepatitis B virus. 24 Here, we describe the molecular cloning and characterization of a human antibody fragment scFv42C, which has a high affinity to HCV core protein. We also investigated its intracellular distribution, association with the core protein, and its impact on cell proliferation triggered by the core protein.
Patients and Methods
Patients
Bone marrow aspirates were drawn from five patients with chronic hepatitis C who underwent this procedure for medical reasons. All patients had serologically documented HCV infection (HCV antibody by third-generation enzyme-linked immunoassay and serum HCV-RNA by reverse transcription polymerase chain reaction (RT-PCR), genotypes 1 and 3) and had given a written informed consent for the drawing of an additional 5 mL aspirate and 10 mL serum. The procedure was carried out as described.25
Human scFv Library
The cloning procedures of a combinatorial scFv library were described previously. Briefly, human VH and VL fragments were PCR amplified from bone marrow aspirate extracts and cloned into the phagemid vector pAK100.25 This phage display vector allows expression of an scFv as a gene III fusion protein on the surface of filamentous phages including of N-terminal FLAG, C-terminal c-myc, and histidine (His)-tag sequences. The phage library preparation, soluble antibody expression and antibody purification were carried out as described.25
Affinity Selection (Panning) and Phage Enzyme Immunoassay
Recombinant phages bearing human scFv-fragments were selected for their affinity to HCV core protein by panning as described. 25 Phage suspension (approximately 1010 to 1012 diluted in 3% bovine serum albumin [BSA]) was incubated at room temperature for 2 hours in wells coated with 1 μg/well of denatured recombinant HCV core protein fragment covering residues 1–115 (Mikrogen, Munich, Germany). The collected phages (usually 103 to 104) were used to reinfect E. coli XL1-blue growing in the log phase. This panning procedure was repeated four times. The selected phage population after three and four rounds was diluted and plated to obtain individual clones. Enzyme immunoassay (EIA) was used to test their binding to core protein, as described.25 Briefly, plates were coated with the core protein (amino acids [aa] 1–115; Mikrogen) or BSA (as a negative control), incubated with phage clones, and washed. After incubation with horseradish peroxidase (HRP)-conjugated α-M13 antibody diluted 1:5000 and addition of 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonate) substrate, optical density was measured at 450 nm.
DNA Sequencing and Variable Gene Alignment
Custom sequencing was carried out by MWG Biotech, Ebersberg, Germany. Alignment of the nucleotide sequences of rearranged V genes to their closest germline V, D, and J segments was performed using V-base DNA Plot (www.mrc-cpe.cam.ac.uk) and IgBlast softwares (www.ncbi.nlm.nih.gov/igblast/). Determination of immunoglobulin families was performed according to Kabat et al. 29 Multiple sequence alignment was performed with ClustalX program. 26
Expression and Purification of scFv42C Antibody
The scFv42C in pAK100 phagemid vector was transformed into the bacterial nonsuppressor strain JM83 to obtain the soluble form of the scFv antibody fragment devoid of the gIII phage sequence. Expression of scFv was induced with 0.5 mM isopropyl-beta-D-thiogalactopyranoside (IPTG). After an overnight culture, the supernatant was collected and used for binding experiments against HCV core protein by Western blot. The coding region of scFv42C was subcloned into pAK400 vector for epitope mapping, purification and binding assay of the HCV core.27
Phage Display Peptide Library to Map the Epitope Recognized by scFv42C
To identify peptide sequences recognized by the scFv42C antibody, the Ph.D.-7 phage display library peptides kit containing linear peptides (randomly presenting 7–12 amino acids) (New England Biolabs) was used. This library was screened by EIA with the soluble form of scFv42C according to the manufacturer’s protocol. Selected clones were sequenced and the amino acid sequence of the presented peptides was deduced to determine the consensus binding motif.
Mammalian Expression Constructs of scFv42C and HCV Core Protein
The complementary DNA (cDNA) encoding scFv42C and scFvIR (irrelevant scFv) antibody fragments were cloned into pEYFP-N1 (Clontech) with its C-terminus fused to enhanced yellow fluorescence protein (YFP; namely, scFv42C-YFP and scFvIR-YFP). The scFv42C cDNA was also cloned into pEF/myc/cyto vector under control of the EF1a promoter (Invitrogen) (namely pEF/sc42). The cDNA encoding the full-length core protein (aa 1–191 of 1b genotype, accession # AY365213) was cloned into pcDNA3.1/Zeo(+) vector (pZeo+/core) as described.10 Alternatively, its cDNA was subcloned into pRc/CMV and pECFP-C1 vector (Clontech) with N-terminal fusion of the enhanced cyan fluorescence protein (CFP; namely, CFP-C191).
Cell Culture and Transfection Experiments
The hepatoma cell line Huh-7 was cultured in DMEM:F-12 (1:1) or Dulbecco’s modified Eagle medium (DMEM) as described previously.10,25,28 Huh-7 cells (5 × 105) grown on chamber slides (Nunc) were transfected with 10 μg of CFP-C191 DNA (or 10 μg of Core-pRc/CMV for immunfluorescence study), or 10 μg of scFv42C-YFP/scFvIR-YFP DNA, or both using Lipofectamine 2000 (Invitrogen) according to the manufacturer protocols. Alternatively, Huh-7 cells (5 × 105) grown in six-well plates were cotransfected with 0.5 μg of pZeo+/core and pEF/sc42 using polyamine as a carrier10 and harvested 2 days later for Western blot analysis. To determine the effect of scFv42C antibody in the context of HCV replication, Huh-7.5 cells (6 × 106) were electroporated with 5 μg of replicon RNA (JFH1; see Wakita et al. 43 for details) produced from in vitro transcription (Epicentre Biotechnologies, Madison, WI) and 2 μg of pEF/sc42 plasmid or the empty vector using NucleoFactor kit T/Electroporator (Amaxa, Gaithersburg, MD). Cells were harvested 3 days later for Western blot and immunofluorescent staining of the core protein. Alternatively, JFH1 transfected Huh-7.5 cells were cultured at subconfluent status for 76 days followed by electroporation with 2 μg of pEF/sc42 or empty vector. A nonrelevant single-chain antibody pEF/sc75 was included as a control as well. Cell proliferation was measured 2 days later, whereas the HCV replication was detected 3 days later by Northern blot.
Immunofluorescence, Confocal Laser Scanning Microscopy, and Fluorescence Resonance Energy Transfer Technique
Huh-7 cells (5 × 105) were grown on chamber slides (Nunc) and transfected with HCV core (CFP-C191 or Core-pRc/CMV) or scFv42C-YFP/scFvIR-YFP alone or cotransfected with both plasmids for colocalization experiments and fluorescence resonance energy transfer (FRET) analysis. Slides were washed with phosphate-buffered saline (PBS) at 24 hours after transfection and fixed for 15 minutes in 4% paraformaldehyde. After washing, protein expression was examined, using a Zeiss laser-scanning confocal microscope (LSM 510 META; Zeiss, Oberkochen, Germany) in multitracking mode to prevent interference of the dyes. The YFP/CFP coexpression was detected using the META-Scan, to avoid bleed-through of CFP into the YFP channel. CFP was excited at a wavelength of 405 nm, and YFP was excited at 514 nm. To measure FRET, we quantitated the quencing of donor fluorescence dye in the presence of the energy transfer acceptor. The degree of FRET was determined using the LSM-Image-Examiner 3.1 software (Zeiss). Cotransfection of CFP-CD95 and scFv42C-YFP as well as CFP-191 and scFvIR-YFP served as a negative control in FRET analysis. For indirect immunofluorescence, cells were permeabilized with 0.1% Triton X-100 in PBS for 10 minutes and blocked with 1% BSA/PBS for 30 minutes. The immunofluorescence staining was perfomed with the primary antibody anti-HCV core (ABR; 1:200) and a cyanine-3–conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA; 1:500).
Immunoblotting
HCV core protein (200 ng, aa 1-115; Mikrogen) was electrophoresed in 0.1% sodium dodecyl sulfate (SDS)/12% polyacrylamide gel (PAGE) and blotted onto a nitrocellulose membrane (Protran; Schleicher und Schuell, Dassel, Germany). As a positive control, one strip of the blot was incubated with anti-core mAb C7-50 (ABR, 1:2000). Another strip was incubated with soluble scFv42C followed by anti-Flag mAb M1 (Sigma, 1:420). After incubation with HRP-conjugated rabbit anti-mouse antibody (Bio-Rad Laboratories; 1:10000), signals were revealed by chemiluminescence using the ECL system (Amersham Pharmacia).
The pellets of Huh-7 cells cotransfected with pZeo+/core and pEF/sc42 were resuspended in 80 μL of H20 and subjected to four cycles of freeze-thaw in dry ice/methanol and 37°C water bath, followed by addition of 20 μL of 5×PBS. After centrifugation at 3000 g for 5 minutes, the cleared lysate was used for detection of core and scFv42C antibody expression. A total of 20 μg of proteins was separated in 0.1% SDS/12% PAGE and transferred to polyvinylidene fluoride membranes. Expression of HCV core protein was detected as described previously using a mAb against core protein, 10 whereas expression of scFv42C antibody was revealed by HisProbe (Pierce Biotechnology) following protocols recommended by the manufacturer. Blots were reprobed with anti–β-actin as protein loading controls (Sigma Aldrich).
Cell Proliferation Assay
Transfected Huh-7 cells were grown in 24-well or 96-well plates (50%–60% confluence), and cell proliferation was performed at day 1 or 2 after transfection by addition of 10 μl of tetrazolium to medium followed by 1–4 hours incubation at 37°C (Cell Counting Kit-8; Dojindo Molecular Technologies, Gaithersburg, MD). The absorbence at 450 nm was measured with a reference wavelength at 620 nm.
Results
Identification of High-Affinity Single-Chain Antibodies Against HCV Core ore Protein
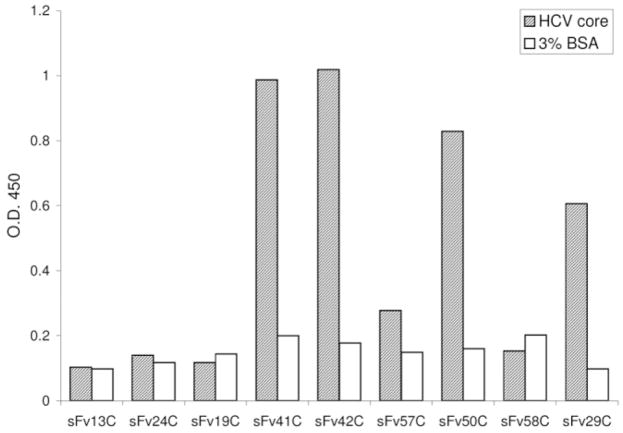
The recombinant scFv displayed on the surface of filamentous phages were screened in a panning reaction with recombinant HCV core protein (aa 1-115) bound to a solid phase. The panning procedure was repeated four times to enrich high-affinity binding phages. Afterward, individual phages were tested again for their affinity with the core protein using phage EIA. Clones with positive EIA results (signal-to-noise ≥ 2.4) were selected for further experiments. As shown in Fig. 1, clones scFv41C, scFv42C, scFv50C, and scFv29C possessed high binding affinity to HCV core. DNA sequencing revealed that two clones (scFv50C and scFv29C) contained incomplete hypervariable domains while the remaining two clones were identical (scFv41C and scFv42C) (data not shown). Thus, clone scFv42C, with the highest binding affinity to the core protein, was chosen for further characterization.
Fig. 1.
Binding of individual phage clones harboring scFv fragments to HCV core protein. The phage population after three or four rounds of panning was diluted, and single phages were amplified. Their reactivity with core protein (aa 1-115) relative to BSA was determined by phage EIA. Clones scFv41C, scFv42C, scFv50C, and scFv29C were thus identified as core protein binders.
Deduced Amino Acid Sequence of the scFv42C Clone
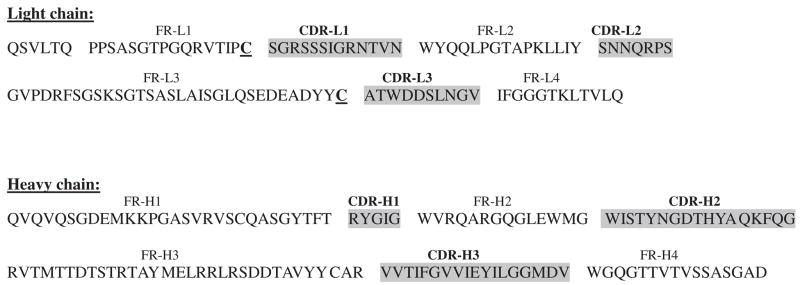
The amino acid sequence coding for the single-chain antibody scFv42C was deduced from the DNA sequence (Fig. 2). The positions of framework regions (FRs) and complementarity determining regions (CDRs) were located according to the Kabat database. 29 The single-chain antibody contains four VL (FR-L1 to FR-L4) and four VH (FR-H1 to FR-H4) domains, respectively, separated by the linker. Based on sequence homology search in the European Molecular Biology Laboratory and GenBank databases, the VH domains belong to the Kabat human heavy chain subgroup VH1 (V-segment: 1–18; D-segment: D3-3/D3–9; J-segment: JH6/JH3), while the VL domains belong to the human lambda light chain subgroup VL1 (V-segment: VL1-16; J-segment: JL2/JL3). The CDRs were mapped to the following aa sequences: CDR-L1: 23–35; CDR-L2: 51–57; CDR-L3: 90–99; CDR-H1: 142–146; CDR-H2: 161–177; and CDR-H3: 210–227.
Fig. 2.
Deduced variable domain sequence of scFv42C. Both DNA strands were sequenced and amino acid sequences of VL and VH were deduced. Indicated are framework regions (FR) and complementarity determining regions (CDR) according to the nomenclature by Kabat et al. 29 Two cysteine residues in the variable regions of VL, which are involved in structurally conserved intrachain disulfide bonds, are shown in bold and underlined.
Purification of scFv42C and Western Blot Analysis of its Binding to HCV Core Protein
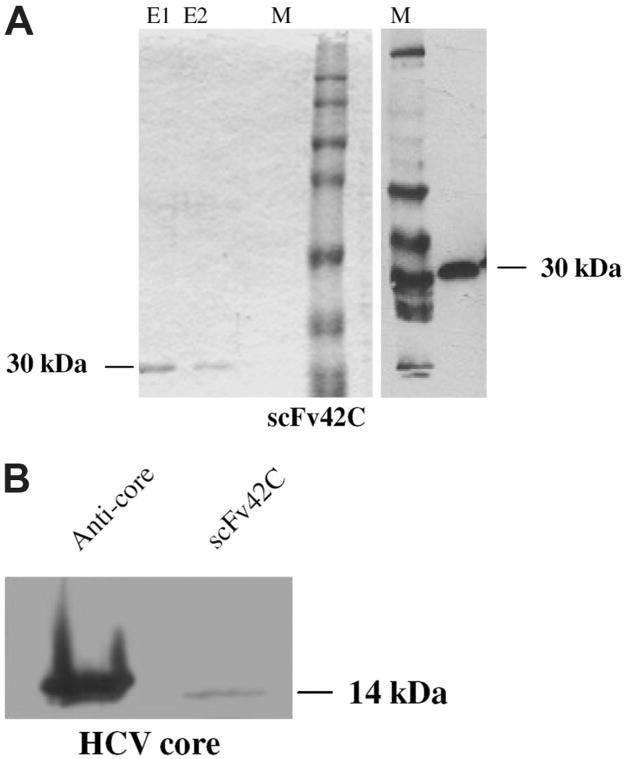
For high-level expression of soluble scFv antibody in E. coli, the scFv42C coding sequence was subcloned into pAK400 vector. A six-histidine (His(6)) tag added to the C-terminus of the protein facilitated purification. After induction with IPTG, soluble scFv42C antibody was harvested from periplasmic space and purified using a nickel-nitrilotriacetic acid column. According to Coomassie Blue staining of the protein gel, the first two elutions contained a pure 30-kDa band expected for the scFv42C antibody (Fig. 3A, left). As expected, this protein reacted with an anti-His antibody in Western blot (Fig. 3A, right). Moreover, the purified antibody could recognize the core protein (aa 1-115) in Western blot, albeit the signal was much weaker than that of mouse anti-core antibody (Fig. 3B).
Fig. 3.
Purification of functional scFv42C and its binding to HCV core protein. (A) Detection of purified scFv42C. Coomassie blue staining of purified scFv42C separated in SDS-PAGE (left). The scFv42C antibody was purified using Ni-NTA agarose. E1: 1st elution; E2: 2nd elution. The predicted size of scFv is 30 kDa. Immunoblotting of purified scFv42C with α-his mAb (right). M: full-range rainbow molecular mass protein standard. (B) Binding of bacterially expressed and purified scFv42C fragment to HCV core protein. The 14 kDa of core protein (aa 1-115, Mikrogen) was separated in 12% SDS-PAGE and blotted onto membrane. Lane 1: detection with α-HCV-core antibody (C7–50, ABR) followed by α-mouse-HRP. Lane 2: detection with FLAG-tagged scFv42C, followed by α-FLAG mAb and α-mouse-HRP. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Mapping of the scFv42C Binding Motif on the Core Protein
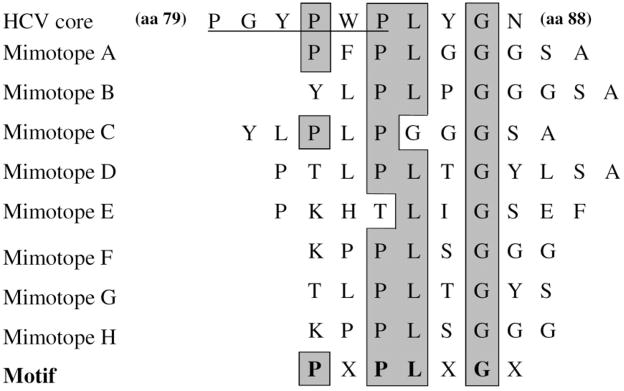
In order to identify the recognition site of scFv42C on the HCV core protein, the Ph.D.-7 phage display library, which expresses random 7-mer to 12-mer peptides on phage surface, was screened. The library was incubated with microtiter plates coated with the purified scFv42C. After extensive washing, the bound phages were eluted, amplified, and sequenced. Sequence alignment revealed the consensus binding motif PLXG on the majority of phages, where X can be any amino acid (Fig. 4). Of the three conserved residues, proline and leucine are absent in only one of the eight mimotopes, respectively, whereas glycine is preserved in all mimotopes. Indeed, residues 84–87 of the HCV core protein (PLYG) conform to this motif, and is highly conserved among different genotypes. Assignment of the binding motif to residues 84–87 was further supported by the presence of a proline at the second position upstream of the PLXG sequence in two of the eight mimotopes: PFPLGG in mimotope A and PLPGGG in mimotope C, which matches the PWPLYG PLYG sequence (residues 82–87) in the core protein.
Fig. 4.
Identification of scFv42C binding motif in HCV core protein by multiple sequence alignment. Core protein sequence (residues 79–88) is shown on the top, followed by deduced amino acid sequences of eight phage clones (mimotopes) from the Ph.D.-7 library with high-affinity binding to scFv42C. The bottom summarizes the consensus binding motif. Shown in gray and boxed are residues conserved between the mimotopes and core protein. In the top line, the putative Jak binding site on the core protein is underlined.
Intracellular Colocalization of scFv42C with HCV Core Protein
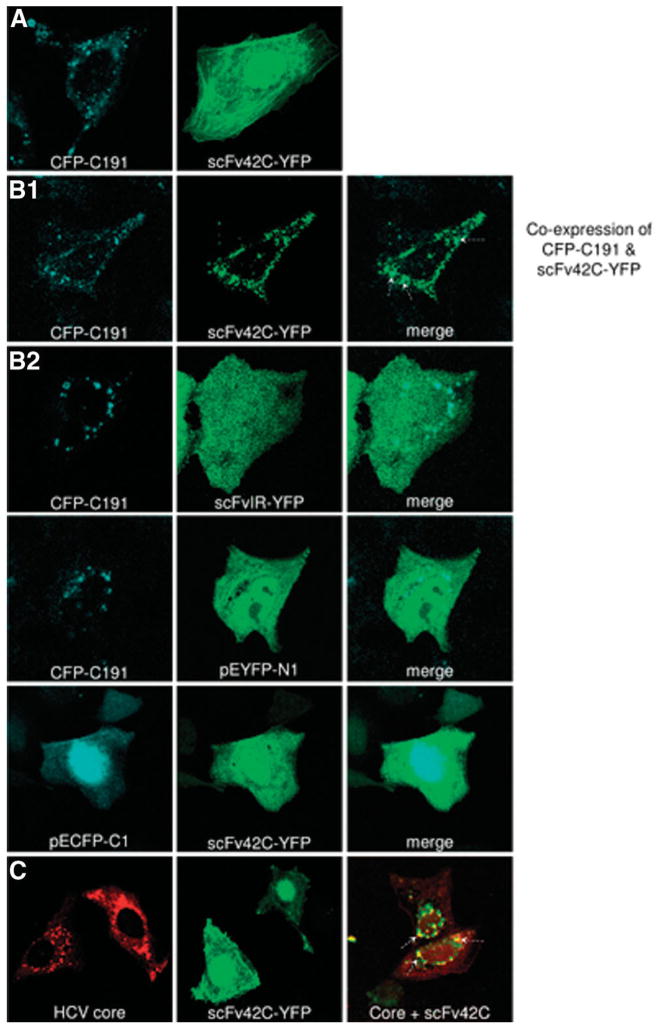
Confocal laser scanning microscopy was used to investigate intracellular interaction between scFv42C and core protein. To facilitate their detection, CFP was as attached to the N-terminus of core protein (CFP-191), whereas YFP was tagged to the C-terminus of scFv42C (scFv42C-YFP) or scFvIR (scFvIR-YFP). Huh-7 cells were transfected with HCV core or scFv42C/scFvIR construct alone, or cotransfected with both constructs. Cells were fixed 24 hours later to detect core protein by the CFP tag (405 nm), and scFv42C/scFvIR by the YFP tag (514 nm). As expected, HCV core protein was found in the cytoplasmic and perinuclear region, with a globular-like or vesicular-like pattern (Fig. 5A, left; Fig. 5C, left). Coexpression of scFv42C did not alter core protein localization (Fig. 5B1, left; Fig. 5C). In contrast, the expression of scFv42C-YFP produced a diffuse fluorescence pattern throughout the cytoplasm when transfected alone (Fig. 5A, right; Fig. 5C, middle). However, coexpression of scFv42C with the core protein shifted the scFv42C expression pattern to globular-like, which colocalized with HCV core (Fig. 5B1, middle and right panels). An irrelevant antibody fragment scFvIR-YFP, which does not recognize HCV core protein, was used as a negative control. As expected, this antibody fragment did not show any colocalization with the core protein (Fig. 5B2). Similar negative results were observed when using empty vectors (pECFP-C1 and pEYFP-N1) (Fig. 5B2). In addition perinuclear patterns of the scFv42C expression were also observed by indirect immunofluorescence in cotransfected cells, which again overlapped with core protein (Fig. 5C). Thus, the three-dimensional analysis confirmed the colocalization of these two proteins, indicative of their physical interaction.
Fig. 5.
Colocalization of HCV core protein with scFv42C. (A) Huh-7 cells were transfected with CFP-tagged core protein (CFP-C191) (left) or YFP-tagged scFv42C (scFv42C-YFP) (right). (B) Huh-7 cells were cotransfected with CFP-C191 and scFv42C-YFP/scFvIR-YFP or control vectors and analyzed with a confocal laser scanning microscope scope using a planar 63 × objective 24 hours later. CFP was excited at wavelength of 405 nm, whereas YFP was excited at 488 nm. (C) Huh-7 cells were transfected with Core-pRc/CMV (left), or scFv42C-YFP (middle), or both constructs (right). Colocalization was confirmed by merging core and scFv42C images [(B) right panel and (C) right panel, white arrows].
Intracellular Interaction of scFv42C with Core Protein as Evidenced by FRET Analysis
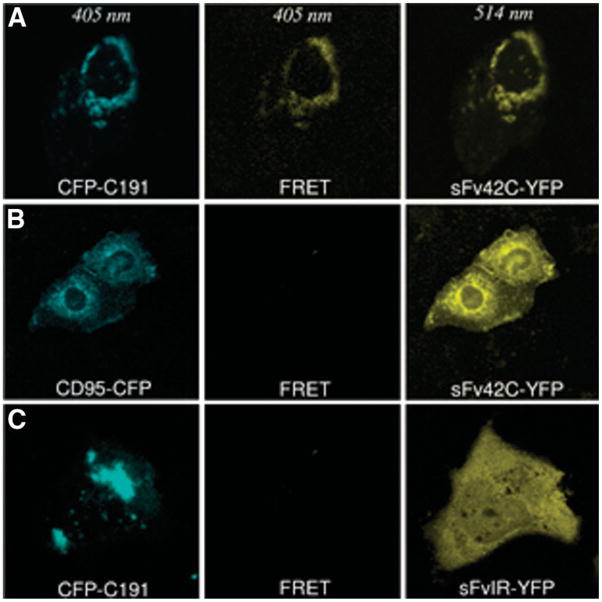
FRET technology represents a novel tool to measure protein-protein interactions. We used the FRET technique to further demonstrate intracellular interaction between CFP-tagged core protein (CFP-C191) and YFP-tagged single-chain antibody (scFv42C-YFP). If the two proteins interact with each other (close proximity <10 nm), the donor chromophore (that is, CFP) in its excited state can transfer energy by a nonradiative long-range dipole-dipole coupling mechanism to an acceptor chromophore (that is, YFP). Indeed, scFv42C-YFP could be detected at excitation of 405 nm wavelength in cells cotransfected with CFP-tagged core protein but not with CFP-tagged CD95, a control protein (Fig. 6A,B, middle panels). In cells cotransfected with CFP-191 and the irrelevant antibody scFvIR-YFP, the FRET signal was negative at excitation of 405 nm (Fig. 6C). These data strongly suggest that HCV core protein and scFv42C are closely associated with each other in the perinuclear region of Huh-7 cells.
Fig. 6.
FRET analysis. (A) Specific interaction between scFv42C and HCV core protein. Huh-7 cells were cotransfected with CFP-C191 and scFv42C-YFP constructs. CFP was detected at a wavelength of 405 nm and YFP at 514 nm. The observed YFP-specific spectrum in perinuclear region using 405 nm excitation clearly reveals an assignment of the resonance energy from CFP-C191 to sFv42C-YFP (FRET, middle panel). (B, C) Negative controls of FRET. Huh-7 cells were cotransfected with CD95-CFP and sFv42C-YFP or CFP-C191 and scFvIRYFP. No YFP-specific spectrum was detected using 405 nm excitation (middle panel).
The scFv42C Antibody Reduced Core Protein Level but not Viral Replication per se in Transient Transfection
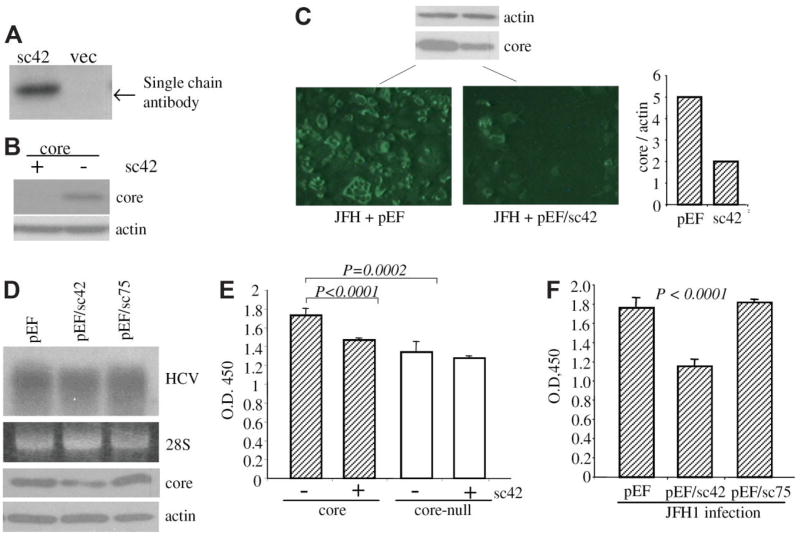
Intracellular interaction with single-chain antibody may affect the target protein function and/or stability. To investigate the effect of scFv42C antibody on HCV core protein expression, Huh-7 cells were cotransfected with scFv42C and the core expression construct (1b genotype). Expression of scFv42C antibody was readily detectable 2 days after transfection (Fig. 7A). Interestingly, core protein level was reduced or undetectable in the presence of the scFv42C antibody (Fig. 7B and data not shown). To further examine whether the scFv42C antibody has any effect on core protein processed from HCV polyprotein during viral replication, Huh-7.5 cells were electroporated with scFv42C and HCV replicon of 2a genotype, a highly replicating HCV strain. Indeed, expression of scFv42C antibody greatly reduced core protein detection by immunofluorescent staining (Fig. 7C). In this regard, the C7–50 mAb employed for the staining recognizes an epitope (aa 21–40) distant from the scFv42C-binding site (aa 84–87). The reduction of core protein level in replicon-transfected cells was validated by Western blot and quantified by densitometry (Fig. 7C, upper and lower right). Interestingly, reduction of core protein as revealed by immunofluorescent staining was more dramatic than that detected by Western blot, indicating both sequestration and depletion by the scFv42 antibody. These data also prove that the scFv42C antibody recognizes core protein of different genotypes (1b and 2a in this study). To evaluate the effect of the antibody on HCV replication, Huh-7.5 cells chronically infected with the JFH strain were electroporated with the scFv42C or control constructs, and viral replication was examined by Northern blot hybridization 3 days later. Results showed expression of the scFv42C had no effect on viral RNA replication in transient transfection experiments as compared to the control cells (Fig. 7D).
Fig. 7.
Effect of scFv42C on core protein expression, replication, and cell proliferation. (A) Intracellular expression of scFv42C. Huh-7 cells were transfected with pEF/sc42 or empty vector (vec) and harvested 2 days later. Expression of scFv42C was detected by Western blot using HisProbe. (B) Effect of scFv42C antibody on core protein level. Huh-7 cells were co-transfected with 0.5 μg of pZeo+/core and pEF/sc42 or empty vector. Core protein was analyzed by Western blot at day 2 after transfection. Actin was detected in parallel as loading controls. (C) Immune staining of core protein expressed from JFH1 replicon. Huh-7.5 cells were coelectroporated with 5 μg of replicon RNA and 2 μg pEF/sc42 plasmid or empty vector, and seeded in 24-well plates with cover slips. Cells were fixed 3 days later and stained with anti-core antibody followed by ultraviolet microscopy as described. 10 Cells seeded in six-well plates were subject to Western blot of core (10 μL lysate) and β-actin (2 μL lysate). The relative ratio of the core versus actin in Western blot was measured by densitometry (right panel). (D) Effect of scFv42C on HCV replication. Huh-7.5 cells were electroporated with 5 μg of JFH replicon RNA and continuously cultured under subconfluent condition for 76 days. Cells were then electroporated with 2 μg plasmid DNA encoding scFv42C (pEF/sc42), or nonrelevant scFv (pEF/sc75), or empty vector (pEF). Viral replication was examined by Northern hybridization. The 28S RNA served as loading controls. Core and β-actin expression was also shown. (E) Effect of scFv42C on cell proliferation. Huh-7 cells were cotransfected with pZeo +/core (or a core-null mutant as a control) (0.19 μg/six wells of a 96-well plate) and same amount of pEF/sc42 (or empty vector). Cell proliferation was measured by a modified MTT assay (cholecystokinin-8 assay) at day 1 after transfection. Core-null: core cDNA with a stop codon immediately downstream of the ATG codon. (F) Effect of single-chain antibody on cell proliferation in the context of HCV replication. HCV-infected Huh-7.5 cells as described in (D) were seeded in 96-well plates after electroporation with the plasmids indicated. Cell proliferation was measured 2 days later as described above. Data are presented as the mean ± standard deviation (n = 6). Statistical analysis was done by Student t test.
The scFv42C Antibody Blunted Core Protein-Mediated Cell Proliferation
We previously demonstrated that expression of core protein in Huh-7 cells was associated with increased cell proliferation. 10 To explore whether scFv42C interaction with core protein can abrogate the growth-promoting effect of the core protein, we compared the cell proliferation rate in the presence or absence of scFv42C. Consistent with the previous observation, expression of core protein significantly increased cell proliferation (Fig. 7D; compare core and core-null transfected cells in the absence of scFv42C, P < 0.0002). Interestingly, expression of scFv42C antibody did not change growth rate of cells cotransfected with a core null-mutant, but significantly reduced cell proliferation in cells expressing the core protein (Fig. 7D; P < 0.0001). To further validate the role of the scFv42C antibody in cell proliferation in the context of HCV replication, Huh-7.5 cells with chronic HCV infection were established. The cells were continuously cultured for 76 days after electroporation with the JFH1 replicon RNA. Under subconfluent culture conditions, viral replication and protein translation persisted at a low level compared to those in transient transfection (data not shown). Interestingly, cell proliferation was reduced to a great extent in chronically infected cells after transfection with the scFv42C, but not the nonrelevant antibody (Fig. 7F; P < 0.0001).
Discussion
Recombinant antibodies have become important tools for prevention, diagnosis, and treatment of a broad range of diseases including infectious diseases, cancer, neurological disorders, and autoimmune diseases. 30 Recombinant single-chain antibodies (scFvs) have some advantages over intact antibodies because of their smaller size, ease of production, and improved tissue penetration. In the study of HIV, intracellular expression of scFvs targeting the tat or VIF protein inhibited HIV replication, 31,32 whereas scFvs against the integrase conferred cellular resistance to HIV infection. 33 As for cancer therapy, single-chain antibodies targeting endothelial cell receptor-tyrosine kinases, vascular endothelial growth factor receptor 2 and Tie-2 and activating transcription factor-1 reduced the malignant potential of melanoma cells. 34,39 Growth of lung cancer cells could be reduced by intracellular immunization with a single-chain antibody recognizing lung cancer–associated common antigens. 35 Moreover, recombinant adenovirus expressing scFvs against erbB-2 oncogene led to reversal of the malignant phenotype of ovarian cancer and promoted initiation of a clinical trial by using adenoviral-mediated gene therapy. 36
HCV infection causes a series of liver diseases including steatosis, fibrosis, and cirrhosis, which eventually may lead to hepatocellular carcinoma. Single-chain antibodies may prove useful in the therapy of chronic HCV infection. Indeed, scFvs against the viral nonstructural proteins NS3 (helicase) and NS5B (RNA-dependent RNA polymerase) have been isolated, and have proved effective in inhibiting their enzymatic activities both in vitro and in transfected cells. 25,28,37,38 Although these antibodies have not been tested for their effect in the context of the entire HCV polyprotein and HCV replication, they represent potential candidates for intracellular immunization against HCV infection. The core protein has been proposed to be a major viral factor contributing to HCV pathogenesis. Thus, targeting the viral core protein may alleviate HCV-related pathological changes.
In this study, a human mAb directed against HCV core protein was isolated using phage display technology. Further characterization revealed binding of the soluble form of scFv42C to HCV core, whether immobilized on membranes or inside cells. The sequence critical for scFv42C binding was mapped to a linear PLXG sequence by phage display peptide library, with possible contribution by another proline upstream (the extended motif being PXPLXG). This his motif corresponds to aa 82–87 of the HCV core protein (PWPLYG) which is found to be conserved among different HCV genotypes. Consistent with the conservation of the scFv42C binding motif, scFv42C can recognize the viral core protein derived from 1b and 2a genotypes as shown in this study (Fig. 7). Interestingly, PWPLYG partially overlaps with the putative binding site of JAK signaling proteins in the core protein (PXXPXP, aa 79–84), which has been implicated in modulation of the JAK-STAT signaling pathway. 40,41,42
Intracellular interaction of scFv42C with HCV core protein resulted in core protein depletion and/or sequestration thus reducing core protein–related pathogenesis such as induction of cell proliferation. On the other hand, intracellular expression of the core antibody may also interrupt viral life cycle. In this study, we found that the scFv42C antibody did not directly affect HCV replication in transient transfection. As expected, the core protein is not required for viral genome replication, but is necessary for virus particle assembly. We assume that binding of the scFv42C antibody to viral core protein may interfere with nucleocapsid formation or RNA encapsidation. Indeed, our preliminary data indicated that the viral particle formation was reduced in the presence of the scFv42C antibody (J. Li et al., unpublished). Thus, scFv42C may represent a promising antiviral agent to abrogate viral life cycle or mitigate pathogenesis associated with HCV core protein.
Acknowledgments
We are grateful to Dr. Charles Rice, Rockefeller University, New York, for providing Huh-7.5 cells and Dr. Takaji Wakita, National Institute of Infectious Diseases, Tokyo, for providing the JFH1 replicon construct.
Supported by Deutsche Forschungsgemeinschaft, grant HE 2655/3-3, 3-2, 3-1, by the Forschungskommission of the Heinrich-Heine University Düsseldorf, grant 701010324, the van-Meeteren Foundation; and by grants DK066950, CA109733 from the U.S. National Institutes of Health. J. Li was a Liver Scholar of the American Liver Foundation.
Abbreviations
- HCV
hepatitis C virus
- FRET
fluorescence resonance energy transfer technique
- JAK-STAT
Janus kinase-signal transducer and activator transcription factor
- scFv
single-chain variable fragment
Footnotes
This work is dedicated to Renate Heimann.
Potential conflict of interest: Nothing to report.
References
- 1.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 3.Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hope RG, Murphy DJ, McLauchlan J. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J Biol Chem. 2002;277:4261–4270. doi: 10.1074/jbc.M108798200. [DOI] [PubMed] [Google Scholar]
- 5.Jin DY, Wang HL, Zhou Y, Chun AC, Kibler KV, Hou YD, et al. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 2000;19:729–740. doi: 10.1093/emboj/19.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwer B, Ren S, Pietschmann T, Kartenbeck J, Kaehlcke K, Bartenschlager R, et al. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J Virol. 2004;78:7958–7968. doi: 10.1128/JVI.78.15.7958-7968.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki R, Sakamoto S, Tsutsumi T, Rikimaru A, Tanaka K, Shimoike T, et al. Molecular determinants for subcellular localization of hepatitis C virus core protein. J Virol. 2005;79:1271–1281. doi: 10.1128/JVI.79.2.1271-1281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi SI, Ichikawa M, Kajita T, et al. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048–6055. doi: 10.1128/jvi.72.7.6048-6055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otsuka M, Kato N, Taniguchi H, Yoshida H, Goto T, Shiratori Y, et al. Hepatitis C virus core protein inhibits apoptosis via enhanced Bcl-xL expression. Virology. 2002;296:84–93. doi: 10.1006/viro.2002.1371. [DOI] [PubMed] [Google Scholar]
- 10.Fukutomi T, Zhou Y, Kawai S, Eguchi H, Wands JR, Li J. Hepatitis C virus core protein stimulates hepatocyte growth: correlation with upregulation of wnt-1 expression. Hepatology. 2005;41:1096–1105. doi: 10.1002/hep.20668. [DOI] [PubMed] [Google Scholar]
- 11.Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, et al. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol. 1997;78(Pt 7):1527–1531. doi: 10.1099/0022-1317-78-7-1527. [DOI] [PubMed] [Google Scholar]
- 12.Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, et al. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 13.Hahn CS, Cho YG, Kang BS, Lester IM, Hahn YS. The HCV core protein acts as a positive regulator of fas-mediated apoptosis in a human lymphoblastoid T cell line. Virology. 2000;276:127–137. doi: 10.1006/viro.2000.0541. [DOI] [PubMed] [Google Scholar]
- 14.Fujie H, Yotsuyanagi H, Moriya K, Shintani Y, Tsutsumi T, Takayama T, et al. Steatosis and intrahepatic hepatitis C virus in chronic hepatitis. J Med Virol. 1999;59:141–145. doi: 10.1002/(sici)1096-9071(199910)59:2<141::aid-jmv3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Spaziani A, Alisi A, Sanna D, Balsano C. Role of p38 MAPK and RNA-dependent protein kinase (PKR) in hepatitis C virus core-dependent nuclear delocalization of cyclin B1. J Biol Chem. 2006;281:10983–10989. doi: 10.1074/jbc.M512536200. [DOI] [PubMed] [Google Scholar]
- 16.Yan XB, Battaglia S, Boucreux D, Chen Z, Brechot C, Pavio N. Mapping of the interacting domains of hepatitis C virus core protein and the double-stranded RNA-activated protein kinase PKR. Virus Res. 2007;125:79–87. doi: 10.1016/j.virusres.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Sato Y, Kato J, Takimoto R, Takada K, Kawano Y, Miyanishi K, et al. Hepatitis C virus core protein promotes proliferation of human hepatoma cells through enhancement of transforming growth factor alpha expression via activation of nuclear factor-kappaB. Gut. 2006;55:1801–1808. doi: 10.1136/gut.2005.070417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melen K, Fagerlund R, Nyqvist M, Keskinen P, Julkunen I. Expression of hepatitis C virus core protein inhibits interferon-induced nuclear import of STATs. J Med Virol. 2004;73:536–547. doi: 10.1002/jmv.20123. [DOI] [PubMed] [Google Scholar]
- 19.Chen SY, Khouri Y, Bagley J, Marasco WA. Combined intra- and extracellular immunization against human immunodeficiency virus type 1 infection with a human anti-gp120 antibody. Proc Natl Acad Sci U S A. 1994;91:5932–5936. doi: 10.1073/pnas.91.13.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SJ, Park Y, Hong HJ. Antibody engineering for the development of therapeutic antibodies. Mol Cell. 2005;20:17–29. [PubMed] [Google Scholar]
- 21.Schouten A, Roosien J, Bakker J, Schots A. Formation of disulfide bridges by a single-chain Fv antibody in the reducing ectopic environment of the plant cytosol. J Biol Chem. 2002;277:19339–19345. doi: 10.1074/jbc.M201245200. [DOI] [PubMed] [Google Scholar]
- 22.Gram H, Marconi LA, Barbas CF, 3rd, Collet TA, Lerner RA, Kang AS. In vitro selection and affinity maturation of antibodies from a naive combinatorial immunoglobulin library. Proc Natl Acad Sci U S A. 1992;89:3576–3580. doi: 10.1073/pnas.89.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 24.Galun E, Eren R, Safadi R, Ashour Y, Terrault N, Keeffe EB, et al. Clinical evaluation (phase I) of a combination of two human monoclonal antibodies to HBV: safety and antiviral properties. Hepatology. 2002;35:673–679. doi: 10.1053/jhep.2002.31867. [DOI] [PubMed] [Google Scholar]
- 25.Tessmann K, Erhardt A, Haussinger D, Heintges T. Cloning and molecular characterization of human high affinity antibody fragments against Hepatitis C virus NS3 helicase. J Virol Methods. 2002;103:75–88. doi: 10.1016/s0166-0934(02)00017-4. [DOI] [PubMed] [Google Scholar]
- 26.Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 27.Krebber A, Bornhauser S, Burmester J, Honegger A, Willuda J, Bosshard HR, et al. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J Immunol Methods. 1997;201:35–55. doi: 10.1016/s0022-1759(96)00208-6. [DOI] [PubMed] [Google Scholar]
- 28.Artsaenko O, Tessmann K, Sack M, Haussinger D, Heintges T. Abrogation of hepatitis C virus NS3 helicase enzymatic activity by recombinant human antibodies. J Gen Virol. 2003;84:2323–2332. doi: 10.1099/vir.0.19299-0. [DOI] [PubMed] [Google Scholar]
- 29.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. 5. Bethesda, MD: National Institutes of Health; 1991. NIH Publication No. 91–3242. [Google Scholar]
- 30.Souriau C, Hudson PJ. Recombinant antibodies for cancer diagnosis and therapy. Expert Opin Biol Ther. 2003;3:305–318. doi: 10.1517/14712598.3.2.305. [DOI] [PubMed] [Google Scholar]
- 31.Marasco WA, LaVecchio J, Winkler A. Human anti-HIV-1 tat sFv intrabodies for gene therapy of advanced HIV-1-infection and AIDS. J Immunol Methods. 1999;231:223–238. doi: 10.1016/s0022-1759(99)00159-3. [DOI] [PubMed] [Google Scholar]
- 32.Goncalves J, Silva F, Freitas-Vieira A, Santa-Marta M, Malho R, Yang X, et al. Functional neutralization of HIV-1 Vif protein by intracellular immunization inhibits reverse transcription and viral replication. J Biol Chem. 2002;277:32036–32045. doi: 10.1074/jbc.M201906200. [DOI] [PubMed] [Google Scholar]
- 33.Kitamura Y, Ishikawa T, Okui N, Kobayashi N, Kanda T, Shimada T, et al. Inhibition of replication of HIV-1 at both early and late stages of the viral life cycle by single-chain antibody against viral integrase. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:105–114. doi: 10.1097/00042560-199902010-00001. [DOI] [PubMed] [Google Scholar]
- 34.Jean D, Tellez C, Huang S, Davis DW, Bruns CJ, McConkey DJ, et al. Inhibition of tumor growth and metastasis of human melanoma by intracellular anti-ATF-1 single-chain Fv fragment. Oncogene. 2000;19:2721–2730. doi: 10.1038/sj.onc.1203569. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Li G, Tang L, Wang J, Ge XR. The inhibition of lung cancer cell growth by intracellular immunization with LC-1 ScFv. Cell Res. 2002;12:47–54. doi: 10.1038/sj.cr.7290109. [DOI] [PubMed] [Google Scholar]
- 36.Alvarez RD, Barnes MN, Gomez-Navarro J, Wang M, Strong TV, Arafat W, et al. A cancer gene therapy approach utilizing an anti-erbB-2 single-chain antibody-encoding adenovirus (AD21): a phase I trial. Clin Cancer Res. 2000;6:3081–3087. [PubMed] [Google Scholar]
- 37.Heintges T, zu Putlitz J, Wands JR. Characterization and binding of intracellular antibody fragments to the hepatitis C virus core protein. Biochem Biophys Res Commun. 1999;263:410–418. doi: 10.1006/bbrc.1999.1350. [DOI] [PubMed] [Google Scholar]
- 38.Moradpour D, Bieck E, Hugle T, Wels W, Wu JZ, Hong Z, et al. Functional properties of a monoclonal antibody inhibiting the hepatitis C virus RNA-dependent RNA polymerase. J Biol Chem. 2002;277:593–601. doi: 10.1074/jbc.M108748200. [DOI] [PubMed] [Google Scholar]
- 39.Jendreyko N, Popkov M, Rader C, Barbas CF., 3rd Phenotypic knockout of VEGF-R2 and Tie-2 with an intradiabody reduces tumor growth and angiogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102:8293–8298. doi: 10.1073/pnas.0503168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bode JG, Ludwig S, Ehrhardt C, Albrecht U, Erhardt A, Schaper F, et al. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 2003;17:488–490. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida T, Hanada T, Tokuhisa T, Kosai K, Sata M, Kohara M, et al. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J Exp Med. 2002;196:641–653. doi: 10.1084/jem.20012127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosui A, Ohkawa K, Ishida H, Sato A, Nakanishi F, Ueda K, et al. Hepatitis C virus core protein differently regulates the JAK-STAT signaling pathway under interleukin-6 and interferon-gamma stimuli. J Biol Chem. 2003;278:28562–28571. doi: 10.1074/jbc.M210485200. [DOI] [PubMed] [Google Scholar]
- 43.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]