Abstract
The evolution of mitochondria from ancestral bacteria required that new protein transport machinery be established. Recent controversy over the evolution of these new molecular machines hinges on the degree to which ancestral bacterial transporters contributed during the establishment of the new protein import pathway. Reclinomonas americana is a unicellular eukaryote with the most gene-rich mitochondrial genome known, and the large collection of membrane proteins encoded on the mitochondrial genome of R. americana includes a bacterial-type SecY protein transporter. Analysis of expressed sequence tags shows R. americana also has components of a mitochondrial protein translocase or “translocase in the inner mitochondrial membrane complex.” Along with several other membrane proteins encoded on the mitochondrial genome Cox11, an assembly factor for cytochrome c oxidase retains sequence features suggesting that it is assembled by the SecY complex in R. americana. Despite this, protein import studies show that the RaCox11 protein is suited for import into mitochondria and functional complementation if the gene is transferred into the nucleus of yeast. Reclinomonas americana provides direct evidence that bacterial protein transport pathways were retained, alongside the evolving mitochondrial protein import machinery, shedding new light on the process of mitochondrial evolution.
Keywords: mitochondria, protein import, Reclinomonas americana, transport pathway, translocon, SecY
Introduction
Mitochondria are derived from bacterial symbionts that were resident in the cytoplasm of what were to become the first eukaryotic cells (Gray 1992; Lang et al. 1999; Gray et al. 2004). The evolution of mitochondria involved a progressive and massive transfer of genetic material from the ancestral endosymbiont to the host cell nucleus (Fox 1983; Andersson and Kurland 1999; Lang et al. 1999; Kurland and Andersson 2000). This dramatic process necessitated evolution of mechanisms to get these “mitochondrial” proteins, made on the host cell ribosomes, back into the proto-mitochondrion. An elaborate protein import pathway emerged and is a conserved feature of all eukaryotes (Dolezal et al. 2006; Lister and Whelan 2006; Burri and Keeling 2007; Neupert and Herrmann 2007; Chacinska et al. 2009; Kutik et al. 2009; Alcock et al. 2010). The protein import machinery enables mitochondrial proteins to be recognized for translocation across the outer membrane via a translocase in the outer mitochondrial membrane (TOM complex) and then sorted to one of the four submitochondrial compartments by the SAM (sorting and assembly machinery) and TIM (translocase in the inner mitochondrial membrane) complexes (Rehling et al. 2003; Koehler 2004; Neupert and Herrmann 2007; Chacinska et al. 2009).
Conflicting models have been proposed to explain the evolution of the TOM and TIM complexes in mitochondria (Cavalier-Smith 2006; Gross and Bhattacharya 2009; Alcock et al. 2010). Although all models agree that the TOM and TIM complexes were installed into the bacterial endosymbiont, there is controversy over 1) the order in which the TOM and TIM complexes were installed, 2) which components of the TOM and TIM complexes were the first to be established, and 3) whether those protein components were derived from the host or the symbiont genome. Recent work on the TIM complex has gone some way to resolving these conflicts. Bioinformatic analysis of extant α-proteobacteria and experimental work on the model α-proteobacterium Caulobacter crescentus demonstrated that four core components of the TIM complex have bacterial orthologs, providing evidence that the TIM complex at least was derived from preexisting proteins coded for by the endosymbiont (Boorstein et al. 1994; Rassow et al. 1999; Clements et al. 2009). However, all models for the evolution of protein import require a considerable time line through which the early TIM (and TOM) complexes would have been inefficient at protein transport (Cavalier-Smith 2006; Dolezal et al. 2006; Clements et al. 2009; Gross and Bhattacharya 2009; Kutik et al. 2009), leaving open the question of how protein transport could suffice for mitochondrial biogenesis in the earliest stages of evolution of the organelle.
Few clues to the early stages of mitochondrial evolution are to be found in humans and model organisms such as yeast, given that their remnant mitochondrial genomes are very small, and thus, the transfer of genetic material from the ancestral endosymbiont to the host cell nucleus appears to have reached a virtual end point. However, large-scale sequencing of mitochondrial genomes has identified a remarkable set of organisms that have retained a large collection of genes in their mitochondrial genome (Lang et al. 1999; Gray et al. 2004). Diverse species have retained around 40 protein-coding genes, including the cryptomonad Rhodomonas salina (40 proteins), the green alga Chara vulgaris (39 proteins), the amoeba Dictyostelium discoideum (38 proteins), and the stramenopile Dictyota dichotoma (37 proteins) (Ogawa et al. 2000; Turmel et al. 2003; Hauth et al. 2005; Oudot-Le Secq et al. 2006). In one group, crucial evidence about protein transport pathways has been retained. The jakobids are a clade of free-living, heterotrophic flagellates whose mitochondrial genomes encode over 60 proteins and the jakobid R. americana has the most bacterial-like mitochondrial genome reported to date (Lang et al. 1997, 1999; Edgcomb et al. 2001; Archibald et al. 2002). Reclinomonas americana was first discovered in the 1990s, and its mitochondrial genome was sequenced soon after (Flavin and Nerad 1993; O'Kelly 1993; Lang et al. 1997). The mitochondrial genome of R. americana carries 98 genes, 67 of which encode proteins (Lang et al. 1999; Gray et al. 2004). Many of these protein-coding genes are no longer present in the mitochondrial genomes of fungi, plants, or animals (Lang et al. 1999). These include a gene encoding SecY, the core subunit of the translocon/SecY complex. In all bacteria, the SecY complex is the major means for transferring proteins across the inner membrane and assists the assembly of hydrophobic proteins into the inner membrane (Rapoport 2007; Driessen and Nouwen 2008; Mandon et al. 2009). The mitochondrial genome of R. americana also encodes TatA and TatC, two subunits of the TAT complex which can export folded proteins across bacterial inner membranes (Palmer et al. 2005; Natale et al. 2008).
In the context of the current debate over mitochondrial evolution, R. americana provides a means to address two important questions 1) Has some specific block to gene transfer occurred in R. americana, perhaps due to the high hydrophobicity of the proteins encoded on the remnant genes, which might be linked to a need to maintain the bacterial protein transport pathways? 2) Does R. americana have both bacterial (e.g., SecY) and mitochondrial (e.g., TIM) protein transport machinery for mitochondrial protein assembly?
Here, we show that the mitochondrial SecY protein (RaSecY) has structural features conserved with the recently characterized bacterial SecY structures (Zimmer et al. 2008) and find that many of the membrane proteins encoded in the mitochondrial genome of R. americana have topologies and signal sequences that would make them substrates of the SecY complex. We used one of these mitochondrially encoded proteins, RaCox11, to test whether these proteins are incapable of being “imported” back into mitochondria if the gene is transferred into the nucleus. We show that RaCox11 can be imported into yeast mitochondria using the mitochondrial protein import pathway that depends on TOM and TIM complexes. When transferred into the nucleus of yeast cells, the gene encoding RaCox11 functionally complements a Δcox11 yeast mutant, demonstrating that there is no impediment to the transfer of this gene to the nucleus. Furthermore, analysis of expressed sequence tags (ESTs) from R. americana shows that the organism has a mitochondrial TIM machinery. This evidence demonstrates a general evolutionary scenario for an overlap between the installation of the TIM complex and the loss of the bacterial protein transport pathways mediated by the SecY complex. Taken together, these data further suggest that R. americana stands as an exciting example of evolutionary transition in which the ancestral and derived protein transport pathways coexist and could assist one another in the assembly of proteins into the mitochondrial inner membrane.
Materials and Methods
Reclinomonas americana Culture and Cryo-Electron Microscopy
Reclinomonas americana (ATCC 50394) was obtained from the American Type Culture Collection (ATCC; Manassas, Virginia) and cultured in wheat grass extract medium supplemented with Klebsiella pneumoniae at 25 °C. The bacterium was grown on a yeast extract plate with glucose as a carbohydrate source at 30 °C for 24 h prior to inoculation into the wheat grass extract medium. Cells were collected from 20-ml cultures by centrifugation (5,000 × g, 1 min). Droplets (5 μl) of the cell suspension were sandwiched between type A brass freezer hats (ProSciTech, Thuringowa, Qld), and the enclosed cell suspensions were frozen using a Leica EM High Pressure Freezer (Wien, Austria). Frozen cell pellets were subjected to freeze substitution in 0.1% (w/v) uranyl acetate in acetone at −90 °C for 48 h and the temperature raised to −50 °C at 6 °C/h in a Leica EM automated freeze-substitution unit. Samples were washed 3 times with changes of acetone and infiltrated with a graded series of Lowicryl HM20 low temperature resin (Polysciences, Warrington, PA) in acetone consisting of 25% resin (8 h), 50% resin (16 h), 75% (8 h), and 100% (16 h). The infiltrated samples were placed in a fresh change of 100% resin in gelatine capsules, polymerized under UV light for 48 h at −50 °C, and brought to room temperature at 6 °C/h. The soft sample blocks were then hardened under UV light for a further 24 h. Embedded cells in blocks were sectioned with a diamond knife on a Leica Ultracut R microtome and ultra-thin sections (90 nm) were collected onto pioloform-coated 200 mesh hexagonal copper grids. The grids are viewed in a Phillips CM120 Biotwin transmission electron microscope at 120 kV. Images were captured with a Gatan Multiscan 600CW digital camera.
Sequence Analysis
Hydropathy calculations were carried out according to Claros et al. (1995). The sequence alignment and analysis were performed using ClustalW (Larkin et al. 2007) and JalView (Waterhouse et al. 2009). The color-coded cartoon representation of the AaSecY structure was created using PyMol (DeLano 2002). Hidden Markov model analysis was carried out as previously described (Likic et al. 2010).
Cloning and Yeast Expression
The open-reading frame of RaCox11 and ScCox11 was amplified from genomic DNA prepared from R. americana and Saccharomyces cerevisiae (strain W303α), respectively. The DNA fragments were cloned into the yeast expression plasmid p425-MET25. Overlap polymerase chain reaction was used to construct the fusion sequence MTS-RaCox11, corresponding to residues 1–86 of ScCox11. A haploid Δcox11 knockout yeast strain (cox11:KANR, BY4741 background, MATα, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) was purchased from Open Biosystems. Because Δcox11 yeast mutants tend to lose mitochondrial DNA, haploid cells were crossed with BY4742 (MATa) strain and the resultant, respiratory-competent, diploid cells were transformed with p425-MET25 plasmids to express ScCox11, RaCox11, or MTS-RaCox11. The transformed diploid cells were sporulated and dissected as described previously (Johnson 1994). The kanamycin-resistant, haploid Δcox11 transformants were serially diluted and spotted on solid media containing the fermentable carbon source glucose or the nonfermentable carbon source glycerol and grown on plates incubated at 30 °C.
Protein Import and Electrophoresis
Isolation of mitochondria was performed according to published protocols (Daum et al. 1982). Mitochondria were purified from S. cerevisiae strains W303α or tom40–97 (Gabriel et al. 2003) strains and incubated with nuclease-free rabbit reticulocyte lysate (Promega) translated 35S-labeled precursors over time-courses at 25 °C in import buffer (0.6 M sorbitol, 50 mM HEPES pH 7.4, 2 mM potassium phosphate pH 7.4, 25 mM KCl, 10 mM MgCl2, 0.5 mM ethylenediaminetetraacetic acid, 1 mM dithiothreitol) in the presence or absence of an energy source (NADH and ATP) as stated. Import was terminated by dissipation of membrane potential (using a cocktail of antimycin, valinomycin, and oligomycin) and placing the reaction on ice (Hulett et al. 2008; Bursać and Lithgow 2009). Postimport samples were treated with trypsin (Sigma–Aldrich) unless stated, centrifuged at 13,000 × g for ten min and mitochondrial extracts analyzed by SDS–PAGE (Chan and Lithgow 2008). Osmotically ruptured mitochondria (mitoplasts) were prepared as previously described (Glick et al. 1992). Where indicated, blue native polyacrylamide gel electrophoresis (BN–PAGE) was used as described (Beilharz et al. 1998; Schägger and Pfeiffer 2000; Chan and Lithgow 2008). The cytochrome c oxidase complex migrates as a doublet of bands representing the supercomplexes formed between the cytochrome c oxidase (complex IV) and cytochrome bc1 reductase (complex III) as previously described (Schägger and Pfeiffer 2000). Where necessary, the dried sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels were analyzed by phosphorimaging, with signals quantified using ImageQuant software.
Alkaline Extraction of Membrane Proteins
After import assays, the mitochondrial pellet was washed in import buffer and then resuspended in 100 mM sodium carbonate solution (pH 10.8) to extract the soluble and peripheral membrane proteins. The sample was incubated on ice for 30 min and centrifuged at 100,000 × g in a TLA120.2 rotor (45,000 rpm) for 1 h. The supernatant fraction (containing soluble and peripheral membrane proteins) and pellet fraction (containing the membranes including the integral membrane proteins) were separated and analyzed by SDS–PAGE (Burri et al. 2006).
Results
Mitochondrial Morphology in R. americana
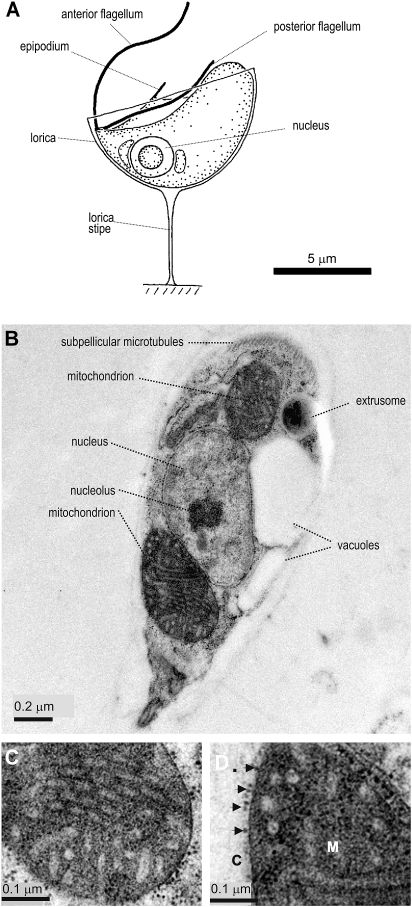
In order to visualize the mitochondrial morphology of R. americana, we developed a procedure for cryo-fixation of the organism and sectioned the cells for transmission electron microscopy. The cells are well preserved by this method with the cell shape indicative of the living cells observed by light microscopy (fig. 1A). Well-developed tubular cristae are evident in the mitochondrial profiles (fig. 1B,C). Reclinomonas americana lives in aerobic environments, and the highly developed cristae is consistent with a dependence on mitochondria for cellular ATP-production. Particles of the size of ribosomes, ∼ 20 nm, are seen on the surface of the outer mitochondrial membrane (fig. 1D). Ribosomes have previously been identified on the mitochondrial surface of yeast cells and represent polysomes translating mitochondrial proteins for import into the organelle (Kellems et al. 1974, 1975; Lithgow 2000).
FIG. 1.
Ultrastructure of R. americana. (A) Drawing of Reclinomonas americana from light micrographs and (B) longitudinal section through cryopreserved samples of R. americana, visualized by TEM. (C) Longitudinal section through posterior mitochondrion to show details of cristae membranes. (D) Higher magnification view of particles (arrows) closely apposed to patches of the mitochondrial outer membrane (M, mitochondrial matrix; C, cytosol).
Mitochondrial Biogenesis from the Inside Out
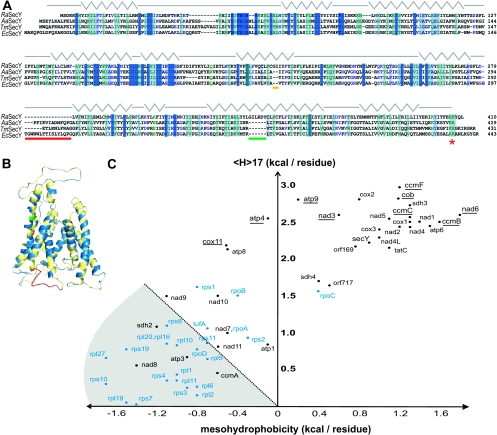
Of the 67 proteins encoded in the mitochondrial genome of R. americana (Lang et al. 1997), 26 have at least one transmembrane segment predicted by the dense alignment surface method DAS (Cserzö et al. 1997; supplementary table 1, Supplementary Material online). SignalP predictions suggest at least five of these membrane proteins have a signal peptide that would further enhance their ability to be recognized by bacterial-type protein translocation machinery (Bendtsen et al. 2004; supplementary table 1, Supplementary Material online). The mitochondrial genome of R. americana encodes a protein with sequence similarity to bacterial SecY proteins (Lang et al. 1999), and the protist sequence (RaSecY) aligns faithfully with bacterial SecY sequences, including those from Aquifex aeolicus and Thermotoga maritima (fig. 2A) whose crystal structures have recently been reported (Zimmer et al. 2008). Using the sequence alignment in figure 2A, we mapped all residues from RaSecY having a conservation score of 7.5 or better onto the AaSecY structure (PDB code 3DL8 chain G). Figure 2B shows that the sequence conservation is spread across the protein’s length and includes 1) the hydrophobic characteristics of the transmembrane segments, 2) the overall predicted conformity to the protein fold, and 3) a dibasic motif in the C-terminal domain. In Escherichia coli, the C-terminus of SecY, and particularly K434 of the dibasic motif, is important for translocation coupling with SecA (Mori and Ito 2006; Karamanou et al. 2008; Zimmer et al. 2008) and the dibasic motif is the most highly conserved feature in this “C6” domain of SecY, in both γ-proteobacteria-like E. coli and in α-proteobacteria from which mitochondria were derived (Chiba et al. 2002). Attempts to express RaSecY in recombinant form, for structural analysis and with a view to raising an antiserum for immunogold labeling of the electron micrographs, were unsuccessful. Antiserum recognizing SecY from E. coli (Boy and Koch 2009) does not cross-react with extracts made from R. americana, nor from the α-proteobacterium C. crescentus (data not shown).
FIG. 2.
The mitochondrially-encoded proteome of Reclinomonas americana. (A) Sequence alignment of the mitochondrial RaSecY with SecY from the bacteria Aquifex aeolicus (AaSecY; 21% identity), Thermotoga maritima (TmSecY; 12% identity), and Escherichia coli (EcSecY; 17% identity) highlighting those residues having a conservation score of 7.5 or greater (max 11) with the most conserved residues show in darker colors. The secondary structural elements corresponding to the AaSecY crystal structure (PDB code 3DL8 chain G) are shown with gray lines indicating loops and turns and zig-zags indicating helices. Two insertions are indicated by horizontal orange and green lines and a deletion of an interhelix loop indicated by a red line. The conserved bacterial SecY dibasic motif (KK/RR) is indicated by a red star. (B) AaSecY crystal structure (PDB code 3DL8 chain G; Zimmer et al. 2008) is shown in gold. Highlighted in blue are those conserved residues from RaSecY (conservation score of 7.5 or better) from the sequence alignment. The locations of the two insertions in RaSecY are shown in orange and green and the large loop between transmembrane segments 6 and 7, absent in RaSecY, is shown in red. (C) The mitochondrial genome was conceptually translated and proteome subject to hydropathy analysis as previously described (Chan et al. 2006). An arbitrary line is drawn to distinguish the more hydrophilic proteins (gray-shaded sector) from the more hydrophobic. The hydrophobicity values are provided in supplementary table S1 (Supplementary Material online) (membrane proteins) and supplementary table S2 (Supplementary Material online) (nonmembrane proteins). The individual sequences are color coded according to their predicted location in the inner membrane (black) or matrix (blue). Underlined are those sequences for which a bacterial-type signal sequence or signal anchor was detected (see supplementary table S1, Supplementary Material online). Seven ribosomal proteins (rpS12, S13, S14, L14, L18, L32, L34) have 17 scores below 0 and, being so extremely hydrophilic, are not displayed on this set of axes. Similarly, TatA is not displayed having a mesohydrophobicity score of −4.4 (see supplementary table S1, Supplementary Material online).
Beyond SecY itself, approximately half of the mitochondrially encoded proteins are subunits of the gene expression machinery (supplementary table 2, Supplementary Material online) that would be localized in the matrix, functioning to produce the membrane proteins encoded on the mitochondrial genome.
The Hydrophobicity Hypothesis and Mitochondrial Gene Transfer
The hydrophobicity hypothesis is an explanation of why some membrane proteins remain trapped on mitochondrial genomes (von Heijne 1986; Popot and de Wry 1990; Claros et al. 1995; Daley and Whelan 2005; de Grey 2005). The relative hydrophobicity of a group of proteins can be determined by calculating the average regional hydrophobicity (“mesohydrophobicity” <H>60–80) and the maximal local hydrophobicity (<H>17). Claros et al. (1995) previously showed that a scatter plot of these two values for the yeast mitochondrial genome leaves all proteins in the upper right quadrant of the graph. Similar studies on plants and a range of protists report similar findings (Daley et al. 2002; Waller et al. 2003; Cardol et al. 2006). We calculated <H>60–80 and <H>17 for the 67 proteins encoded on the mitochondrial genome of R. americana (supplementary tables S1 and S2, Supplementary Material online). We find that more than half of the proteins encoded on the mitochondrial genome are relatively hydrophilic (fig. 2C), with 23 shown in the gray-shaded sector and a further 10 having such hydrophilic values for <H>60–80 and <H>17 that they sit outside the axes as shown in figure 2C. A group of hydrophobic membrane proteins have values of <H>17 greater than 2.0, and many of these have signal sequences predicting for their interaction with a bacterial translocon (these genes are underlined in fig. 2C). Hydrophobicity alone does not explain the set of proteins that remain encoded on the mitochondrial genome of R. americana.
The Gene Encoding RaCox11 Can Be Transferred to the Nucleus
Cox11 is an integral membrane protein, anchored by a transmembrane segment to the mitochondrial inner membrane, with a copper-binding chaperone domain exposed to the intermembrane space (Carr et al. 2005; Khalimonchuk et al. 2005). It thereby assists assembly of cytochrome c oxidase (Cobine et al. 2006). In jakobids like R. americana, the Cox11 protein (RaCox11) is perhaps unique, being encoded in the mitochondrial genome, as genome sequencing projects invariably find the cox11 gene transferred to the nucleus. In yeast, for example, the gene encoding Cox11 is found on chromosome XVI in the nucleus and the Cox11 protein from S. cerevisiae (ScCox11) is imported before assembly into the mitochondrial inner membrane (Carr et al. 2005; Khalimonchuk et al. 2005). Multiple sequence alignment of ScCox11 and RaCox11 with Cox11 homologs from various species of bacteria shows that the yeast protein is made with an N-terminal extension of 85 residues (supplementary fig. S1, Supplementary Material online) that includes a predicted targeting sequence: residues 1–30 of ScCox11 show the characteristic features of a cleavable mitochondrial targeting sequence, as predicted by MitoP (Claros and Vincens 1996). Given this unique situation, RaCox11 was chosen for further study.
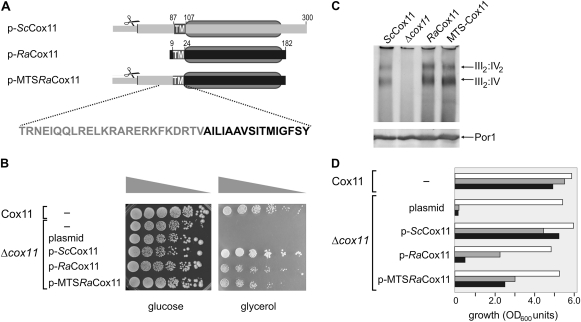
In order to construct a model system to test whether the RaCox11 gene can retain function if transferred to the nucleus, we transformed yeast lacking their own COX11 gene (Δcox11 yeast) with plasmids encoding ScCox11, RaCox11, or MTS-RaCox11 (fig. 3A). MTS-RaCox11 has the 86 residues extension of ScCox11 fused to the N-terminus of RaCox11. Mutant Δcox11 yeast grows on fermentable carbon sources like glucose but fail to grow on nonfermentable carbon sources, such as glycerol because the cytochrome c oxidase complex cannot be assembled in the absence of Cox11 (Banting and Glerum 2006). The growth rate, as judged by colony size, of Δcox11 yeast on the fermentable carbon source glucose is unaffected by any of the plasmids (fig. 3B). The plasmid encoding the yeast ScCox11 complements the growth of Δcox11 yeast on the nonfermentable carbon source glycerol. The MTS-RaCox11 construct also complements growth on glycerol, demonstrating that RaCox11 is assembled into a form that can deliver copper into the cytochrome c oxidase complex to sustain electron transport.
FIG. 3.
Functional complementation of Δcox11 yeast mutants by RaCox11 expressed on a gene in the nucleus. (A) The domain structure of the ScCox11, RaCox11, and MTS-RaCox11 constructs borne by yeast expression plasmids. The MTS-RaCox11 fusion is constructed from residues 1 to 85 of ScCox11 and residues 9–182 from RaCox11. Designated with scissors, an MPP cleavage site is predicted after residue 30 ScCox11. TM denotes the predicted transmembrane segment. The gray oval defines the boundaries of the copper-binding CtaG_Cox11 domain (Pfam 04442) in ScCox11 (residues 105–253) and RaCox11 (residues 25–178). (B) Yeast cells were transformed to express the indicated construct and their growth tested in serial dilution experiments. Equal cell numbers were serially diluted onto medium containing glucose or glycerol as a carbon source and incubated at 25 °C. (C) The transformed yeast strains were grown on medium containing glucose and mitochondria isolated from mid-log phase cultures. Samples of mitochondria (100 μg protein) were analyzed by BN–PAGE, the mitochondrial samples were probed with antisera recognizing the subunit Cox4p to detect cytochrome c oxidase, which migrates as a doublet of bands at ∼1,000 kDa and ∼750 kDa, representing the III2:IV2 and III2:IV supercomplexes formed between cytochrome c oxidase (IV) and cytochrome bc1 reductase (III) as previously described (Schägger and Pfeiffer 2000). Duplicate samples of mitochondria were analyzed by SDS–PAGE and immunoblotting with an antiserum recognizing the outer membrane protein Por1, as a control for the amount of mitochondrial membrane proteins in each of the samples. (D) Transformed yeast cells were grown in liquid media until mid-log phase. The cell numbers measured at 20 h of culture are shown from cultures (white bars) containing glucose as a carbon source, (gray bars) containing glycerol as a carbon source, or (black bars) containing glycerol as a carbon source and supplemented with 50 μM BCDS. The data are representative of five independent experiments.
RaCox11 also complements the defects of Δcox11 yeast for growth on glycerol (fig. 3B) and is sufficient to restore assembly of the cytochrome c oxidase complex as judged by BN–PAGE. Mitochondria were purified from Δcox11 yeast transformed with the ScCox11, RaCox11, MTS-RaCox11, or a control plasmid and analyzed by BN–PAGE and immunoblotting, confirming that both MTS-RaCox11 and RaCox11 restore the assembly of cytochrome c oxidase (fig. 3C).
The experiments in figure 3 demonstrate that, in the context of evolution, transfer of the RaCox11-encoding gene to the nucleus would not necessarily interrupt respiratory function because RaCox11 can be imported into mitochondria even without appendage of a mitochondrial targeting sequence. This is consistent with previous work predicting that many bacterial proteins have intrinsic mitochondrial targeting sequence characteristics (Lucattini et al. 2004). The N-terminus of RaCox11 could form a short basic, amphipathic helix (with positively charged residues K3, R5, and K6), and is followed by a helical transmembrane segment of amphipathic character, which could explain this “minimalist importability.” But is MTS-RaCox11 delivered to mitochondria even more efficiently in vivo? To test this, we monitored the growth of yeast in liquid media with glycerol as a carbon source in the presence or absence of disodium bathocuproine disulphonate (BCDS). BCDS decreases the availability of copper in the growth medium, accentuating copper-dependent growth defects. Figure 3D shows that cells expressing MTS-RaCox11 are reproducibly better (∼50% of wild-type growth) able to grow on glycerol in the presence of BCDS than yeast expressing RaCox11 (∼10% of wild-type growth), suggesting that the appendage of a mitochondrial targeting sequence improves the efficiency of RaCox11 import.
Addition of a Targeting Sequence Promotes Efficient Import of RaCox11 into Mitochondria
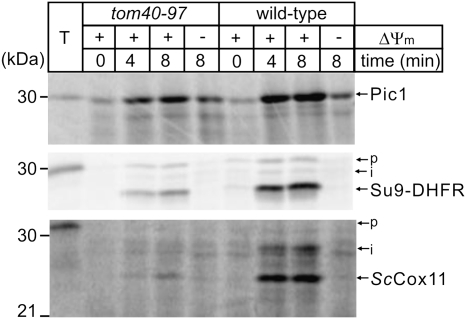
The import of ScCox11 has not previously been analyzed. Like other mitochondrial inner membrane proteins, we found ScCox11 is imported into mitochondria isolated from wild-type yeast in a time-dependent manner, with import dependent on the presence of a membrane potential (fig. 4). The import of proteins in vitro is often relatively inefficient, relying on in vitro-transcribed RNA being translated in vitro into protein, which is then presented posttranslationally to isolated mitochondria resuspended in buffer. However, like Su9-DHFR and Pic1, the import of ScCox11 is relatively efficient in these assays—within 8 min, more than 5% of the added precursor protein (T) was processed to a faster migrating species that represents the mature form of Cox11.
FIG. 4.
ScCox11 is imported and assembled via the TIM23 pathway. Mitochondria were isolated from either wild-type yeast or from the tom40–97 yeast strain (Gabriel et al. 2003). Mitochondria (50 μg protein) were incubated with [35S]-labeled Su9-DHFR, PiC, or ScCox11 for the indicated time and analyzed by SDS–PAGE. Precursors (p) were processed to intermediate (i) and mature forms as indicated by arrows. (−ΔΨ) refers to reactions pretreated with 0.1 μM valinomycin. A control lane (T) shows 5% of the total [35S]-labeled precursor proteins.
In order to determine whether ScCox11 is inserted into the inner mitochondrial membrane by the canonical “TIM23 pathway,” we used yeast strain tom40–97 (Gabriel et al. 2003). This strain carries a single point mutation in Tom40 that debilitates the transfer of imported proteins to the TIM23 complex but has little effect on the transfer of proteins to other aspects of the protein import machinery such as the carrier pathway (Rehling et al. 2003; Koehler 2004; Neupert and Herrmann 2007; Chacinska et al. 2009). Figure 4 shows that transfer of the carrier protein Pic1 to the TIM22 complex in the inner membrane yields more than 50% the efficiency of wild-type import rates in tom40–97 mitochondria. However, the transfer of the control protein Su9-DHFR to the TIM23 complex is reduced by more than 75%. ScCox11 too is strongly inhibited from import into tom40–97 mitochondria, suggesting that it is assembled in the inner membrane via stop-transfer mediated by the TIM23 pathway. This is consistent with the sequence features in the N-terminal targeting domain of ScCox11 (supplementary fig. S1, Supplementary Material online).
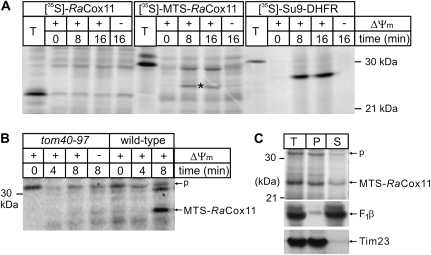
Translation of RaCox11 in vitro yields numerous products that appear to represent protein aggregates: they are resistant to trypsin and migrate as species larger than the predicted size of RaCox11 (∼18 kDa, see fig. 5A). There is no evidence in these assays that RaCox11 is imported in a time- and ΔΨm-dependent manner. However, the addition of the mitochondrial targeting sequence from ScCox11, that is, MTS-RaCox11, improves the import efficiency with a fraction of the protein becoming protected from trypsin over time (fig. 5A) and some of this being processed to a mature form (fig. 5A, *). MTS-RaCox11 is imported into mitochondria using the TOM–TIM23 pathway because the protein’s import is blocked in tom40–97 mutants (fig. 5B). When mitochondria are extracted with 0.1 M sodium carbonate, the majority of the [35S]-labeled MTS-RaCox11 remains integrated in the membranes, like the integral membrane protein Tim23, whereas peripherally associated membrane proteins like the β-subunit of the F1F0-ATPase are extracted into the supernatant (fig. 5C). Although the heterologous MTS may have assisted in this import, the minimal conclusion is that the RaCox11 is able to be imported into mitochondria for assembly into the mitochondrial inner membrane.
FIG. 5.
MTS-RaCox11 is imported more efficiently than RaCox11. (A) Mitochondria were isolated from wild-type yeast and aliquots (50-μg mitochondrial protein) were incubated with [35S]-labeled RaCox11, [35S]-labeled MTS-RaCox11, or [35S]-labeled Su9-DHFR. (−ΔΨ) refers to reactions pretreated with 0.1 μM valinomycin. A control lane (T) shows 5% of the total [35S]-labeled proteins. The import reactions were then treated with trypsin and analyzed by SDS–PAGE and phosphorimage analysis. The mature, processed form of MTS-RaCox11, is indicated by an asterisk (*). (B) Mitochondria were isolated from either wild-type yeast or from the tom40–97 yeast strain and incubated with [35S]-labeled MTS-RaCox11 for the indicated time. The precursor (p) and processed forms are indicated with arrows and (−ΔΨ) refers to reactions pretreated with a cocktail of antimycin, valinomycin, and oligomycin. (C) Mitochondria (100-μg mitochondrial protein) were incubated with [35S]-labeled MTS-RaCox11 for eight min and then reisolated by centrifugation. A sample was prepared for SDS–PAGE (T, total) and a second sample resuspended in 0.1 M sodium carbonate (as described in Materials and Methods) and the membrane pellet (P) and supernatant (S) prepared for SDS–PAGE. The SDS–PAGE gels were analyzed by fluorography and by immunoblots analysis using antisera recognizing the β-subunit of the F1F0-ATP synthase (F1β) or Tim23.
Taken together with the complementation data in figure 3, we conclude that 1) the import of RaCox11 is inefficient, but that 2) in vivo the import pathway can handle RaCox11 sufficiently well to fully complement the assembly of cytochrome c oxidase and enable Δcox11 yeast to grow on a nonfermentable carbon source.
The Mitochondrial Protein Import Machinery in Reclinomonas
The Taxonomically Broad (EST) Database (TBestDB; http://tbestdb.bcm.umontreal.ca/) is a unique resource of EST sequences from R. americana and many other protists (O'Brien et al. 2007). Although it does not provide complete coverage of genes expressed from the nuclear genome, analysis of the R. americana data in TBestDB (supplementary table S3, Supplementary Material online) suggests the presence of at least one member of the Tim23/Tim17/Tim22 family of translocase proteins, at least two small TIM chaperones, and the processing peptidases (MPPα, MPPβ, Imp2), that is, major components of the TIM pathways for biogenesis of inner membrane proteins (fig. 6). In addition, R. americana has the characteristic cytosolic chaperones Hsp70 and Hsp90 (Simpson et al. 2006) that assist hydrophobic proteins in the initial stages of protein import (Chacinska et al. 2002; Young et al. 2003; Chan et al. 2006; Zara et al. 2009). Without a complete genome to analyze, we cannot make a comprehensive statement on the further components of the protein import machinery used by R. americana to handle the import and assembly of membrane proteins encoded by nuclear genes.
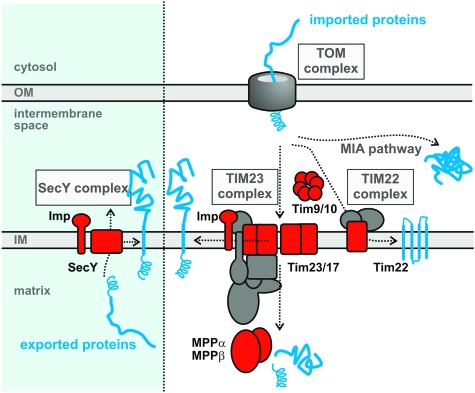
FIG. 6.
A model for mitochondrial protein sorting in Reclinomonas americana. Proteins like RaCox11, encoded on the mitochondrial genome would be “exported” by the SecY complex; the blue panel denotes that this ancestral protein export pathway would have been present in the bacterium that gave rise to mitochondria but has been lost by most eukaryotes. Genes transferred to the nucleus result in proteins, translated in the cytosol, which must be imported via a TOM complex in the outer membrane. For membrane proteins destined for the inner membrane, insertion and assembly can be via either the TIM23 complex or TIM22 complex. Each of these translocase complexes is composed from members of the Tim23/17/22 family of proteins: in some organisms, there is a single TIM complex (Gentle et al. 2007; Schneider et al. 2008). Reclinomonas americana has at least one member of the Tim23/17/22 protein family (indicated in red, the TBestDB accessions are RAL00006644, RAL00001769, and RAL00003686). Polytopic membrane proteins are ferried to the TIM complex by small TIM chaperones, such as Tim9 and Tim10; R. americana has examples of both proteins (RAL00003758 and RAL00003938, respectively). Imported proteins can be processed by MPPα/β and/or Imp2 and, R. americana has proteases of each type: RAL00003642, RAL00006561, RAL00004675, and RAL00001943.
Discussion
Gene Transfer and Mitochondrial Evolution
In R. americana, Cox11 is encoded in the mitochondrial genome. We showed that this RaCox11 contains intrinsic sequences for import into mitochondria. Previously, Lucattini et al. (2004) found that 5–10% of proteins encoded in bacterial genomes have intrinsic targeting information and predicted this was true in the bacterial ancestor of mitochondria, serving as a “preadaptation” (i.e., exaptation) that would have facilitated gene transfers during mitochondrial evolution. The finding that RaCox11 contains intrinsic sequences for import into mitochondria provides experimental evidence in support of this hypothesis; genes from the ancestral endosymbiont could have been transferred to the nucleus and supported at least basic cellular functions even before additional mitochondrial targeting sequences were constructed by exon-shuffling and other mechanisms that improve targeting efficiency (Brennicke et al. 1993; Kadowaki et al. 1996; Kurland and Andersson 2000; Gray et al. 2001).
Comparative sequence analysis of mitochondrial genomes shows that both “the tempo and pattern” of mitochondrial gene loss is episodic (Adams and Palmer 2003), and extant organisms have from 0 to 67 protein-coding genes on their mitochondrial genomes. Typically, the retained genes encode highly hydrophobic proteins, often involved in redox reactions such as those employed in oxidative phosphorylation (von Heijne 1986; Popot and de Wry 1990; Claros et al. 1995; Daley et al. 2002; Daley and Whelan 2005; de Grey 2005). However, hydropathy analysis of the very many proteins encoded on the mitochondrial genome of R. americana shows the majority are relatively hydrophilic and have homologs in yeast and other organisms whose genes have already been successfully transferred to the nucleus. We experimentally verified this observation, showing that RaCox11 can be expressed in the nucleus and the protein imported into mitochondria sufficiently well to complement the defects of Δcox11 yeast.
Bacterial Protein Transport Pathways in Mitochondria
In bacteria, the SecY complex (Rapoport 2007; Driessen and Nouwen 2008; Mandon et al. 2009) is needed to export integral membrane proteins with large periplasmic domains, such as the bacterial homolog of Cox11, and it is reasonable to predict that in RaSecY assists in the assembly of RaCox11. But the experiments reported here also demonstrate that the mitochondrial TOM and TIM machinery would be capable of importing RaCox11 if the gene had been transferred to the nucleus and the protein made in the cytosol. Is there some other biological impediment preventing the transfer of the gene encoding RaCox11 from the mitochondrial genome of R. americana? We cannot exclude the possibility that the continued presence of a SecY complex for the export and assembly of hydrophobic membrane proteins might simply minimize selective pressure for gene transfer to the nucleus.
Sequencing the complete genome of R. americana is required for a full understanding of the protein transport pathways in its unique mitochondrion. Bacterial translocons are composed of the ten transmembrane segments donated by the SecY subunit as well as two very small subunits SecE and SecG (Rapoport 2007; Driessen and Nouwen 2008; Natale et al. 2008; Mandon et al. 2009). We find no evidence of small open-reading frames related to SecE or SecG on the mitochondrial genome of R. americana, and we therefore suggest that these proteins might be encoded in the nuclear genome and the SecE and SecG proteins imported. Similarly, proteins are often transferred to the SecY complex via interaction with the large, soluble chaperone SecA (Chiba et al. 2002; Mori and Ito 2006; Karamanou et al. 2008; Zimmer et al. 2008). Conservation of the dibasic interaction motif in SecY is consistent with the continued presence of a SecA homolog in the mitochondria of R. americana but, if this is the case, this SecA must be imported from the cytosol. Sequencing of the nuclear genome of R. americana is required to resolve this and further aspects of mitochondrial biogenesis in these fascinating organisms.
With both a SecY complex and TIM protein translocases, R. americana provides a snapshot of evolution, reflecting a transition stage in which ancestral and derived protein transport machinery coexist in the mitochondrial inner membrane. A related jakobid protist, Jakoba libera, has proceeded just a few steps further along the gene transfer pathway having lost four more genes from its mitochondrial genome, those encoding the ribosomal proteins S10 and L18, and the RNA polymerase subunits RpoA and RpoD (Lang et al. 1999). The jakobids, therefore, can be considered as a group of “living fossils” in respect to the evolution of mitochondria (Lang et al. 1997; Gray et al. 2004).
In bacteria, multispanning membrane proteins often require both the SecY complex and a protein called YidC (Luirink et al. 2005; Kiefer and Kuhn 2007; Kol et al. 2008; Xie and Dalbey 2008). YidC plays an essential role in protein assembly into the inner membrane and physically cooperates with the SecY complex. In mitochondria that do not have a SecY, a protein called Oxa1 has evolved from the ancestral YidC, to the point that several significant features in sequence and topology now distinguish the mitochondrial-type Oxa1 proteins from the bacterial-type YidC proteins (Kiefer and Kuhn 2007). In yeast, the mitochondrial Oxa1 now cooperates with the TIM23 complex to assist the assembly of at least some, difficult, multispanning inner membrane proteins (Bohnert et al. 2010; Webb and Lithgow 2010). It will be fascinating to see whether the equivalent protein encoded in the nucleus of R. americana is more akin to a mitochondrial-type Oxa1, or a bacterial-type YidC, given the continued presence of SecY in this exceptional organelle.
Supplementary Material
Supplementary figure S1 and table S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Hans-Georg Koch who generously provided antiserum to the SecY from E. coli. We also thank Felicity Alcock, Victoria Hewitt, Georg Ramm, Ross Waller, and Chaille Webb for comments on the manuscript. J.T. is supported by a Monash International Postgraduate Scholarship, P.D. is supported by a Marie Curie Outgoing International Fellowship, T.L. is an Australian Research Council Federation Fellow. S.K.B. is supported by the Intramural Research Program of the National Institute of Health, National Institute of Diabetes and Digestive and Kidney Diseases. The project was supported by a grant from the Australian Research Council (to T.L.).
References
- Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Alcock F, Clements A, Webb C, Lithgow T. Tinkering inside the organelle. Science. 2010;327:649–650. doi: 10.1126/science.1182129. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Origins of mitochondria and hydrogenosomes. Curr Opin Microbiol. 1999;2:535–544. doi: 10.1016/s1369-5274(99)00013-2. [DOI] [PubMed] [Google Scholar]
- Archibald JM, O'Kelly CJ, Doolittle WF. The chaperonin genes of jakobid and jakobid-like flagellates: implications for eukaryotic evolution. Mol Biol Evol. 2002;19:422–431. doi: 10.1093/oxfordjournals.molbev.a004097. [DOI] [PubMed] [Google Scholar]
- Banting GS, Glerum DM. Mutational analysis of the Saccharomyces cerevisiae cytochrome c oxidase assembly protein Cox11p. Eukaryot Cell. 2006;5:568–578. doi: 10.1128/EC.5.3.568-578.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz T, Suzuki CK, Lithgow T. A toxic fusion protein accumulating between the mitochondrial membranes inhibits protein assembly in vivo. J Biol Chem. 1998;273:35268–35272. doi: 10.1074/jbc.273.52.35268. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bohnert M, Rehling P, Guiard B, Herrmann JM, Pfanner N, van der Laan M. Cooperation of stop-transfer and conservative sorting mechanisms in biogenesis of mitochondrial ABC transporter. Curr Biol. 2010;20:1227–1232. doi: 10.1016/j.cub.2010.05.058. [DOI] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Boy D, Koch HG. Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Mol Biol Cell. 2009;20:1804–1815. doi: 10.1091/mbc.E08-08-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennicke A, Grohmann L, Heisel R, Knoop V, Schuster W. The mitochondrial genome on its way to the nucleus: different stages of gene transfer in higher plants. FEBS Lett. 1993;325:140–145. doi: 10.1016/0014-5793(93)81430-8. [DOI] [PubMed] [Google Scholar]
- Burri L, Keeling PJ. Protein targeting in parasites with cryptic mitochondria. Int J Parasitol. 2007;37:265–272. doi: 10.1016/j.ijpara.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Burri L, Vascotto K, Gentle IE, Chan NC, Beilharz T, Stapleton DI, Ramage L, Lithgow T. Integral membrane proteins in the mitochondrial outer membrane of Saccharomyces cerevisiae. FEBS J. 2006;273:1507–1515. doi: 10.1111/j.1742-4658.2006.05171.x. [DOI] [PubMed] [Google Scholar]
- Bursać D, Lithgow T. Jid1 is a J-protein functioning in the mitochondrial matrix, unable to directly participate in endoplasmic reticulum associated protein degradation. FEBS Lett. 2009;583:2954–2958. doi: 10.1016/j.febslet.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Cardol P, Lapaille M, Minet P, Franck F, Matagne RF, Remacle C. ND3 and ND4L subunits of mitochondrial complex I, both nucleus encoded in Chlamydomonas reinhardtii, are required for activity and assembly of the enzyme. Eukaryot Cell. 2006;5:1460–1467. doi: 10.1128/EC.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr HS, Maxfield AB, Horng YC, Winge DR. Functional analysis of the domains in Cox11. J Biol Chem. 2005;280:22664–22669. doi: 10.1074/jbc.M414077200. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proc Biol Sci. 2006;273:1943–1952. doi: 10.1098/rspb.2006.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138(4):628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Pfanner N, Meisinger C. How mitochondria import hydrophilic and hydrophobic proteins. Trends Cell Biol. 2002;12:299–303. doi: 10.1016/s0962-8924(02)02310-3. [DOI] [PubMed] [Google Scholar]
- Chan NC, Likić VA, Waller RF, Mulhern TD, Lithgow T. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J Mol Biol. 2006;358:1010–1022. doi: 10.1016/j.jmb.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Chan NC, Lithgow T. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol Biol Cell. 2008;19:126–136. doi: 10.1091/mbc.E07-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Mori H, Ito K. Roles of the C-terminal end of SecY in protein translocation and viability of Escherichia coli. J Bacteriol. 2002;184(8):2243–2250. doi: 10.1128/JB.184.8.2243-2250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Perea J, Shu Y, Samatey FA, Popot JL, Jacq C. Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. The case of a cytoplasmically synthesized apocytochrome b. Eur J Biochem. 1995;228:762–771. [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Clements A, Bursac D, Gatsos X, et al. (11 co-authors) The reducible complexity of a mitochondrial molecular machine. Proc Natl Acad Sci U S A. 2009;106:15791–15795. doi: 10.1073/pnas.0908264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine PA, Pierrel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim Biophys Acta. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Cserzö M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- Daley DO, Clifton R, Whelan J. Intracellular gene transfer: reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc Natl Acad Sci U S A. 2002;99:10510–10515. doi: 10.1073/pnas.122354399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley DO, Whelan J. Why genes persist in organelle genomes. Genome Biol. 2005;6:110. doi: 10.1186/gb-2005-6-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- de Grey AD. Forces maintaining organellar genomes: is any as strong as genetic code disparity or hydrophobicity? Bioessays. 2005;27:436–446. doi: 10.1002/bies.20209. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. San Carlos (CA): DeLano Scientific; 2002. [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Edgcomb VP, Roger AJ, Simpson AG, Kysela DT, Sogin ML. Evolutionary relationships among “jakobid” flagellates as indicated by alpha- and beta-tubulin phylogenies. Mol Biol Evol. 2001;18:514–522. doi: 10.1093/oxfordjournals.molbev.a003830. [DOI] [PubMed] [Google Scholar]
- Flavin M, Nerad TA. Reclinomonas americana N.G., N. Sp., a new freshwater heterotrophic flagellate. J Eukaryot Microbiol. 1993;40:172–179. doi: 10.1111/j.1550-7408.1993.tb04900.x. [DOI] [PubMed] [Google Scholar]
- Fox TD. Mitochondrial genes in the nucleus. Nature. 1983;301:371–372. doi: 10.1038/301371a0. [DOI] [PubMed] [Google Scholar]
- Gabriel K, Egan B, Lithgow T. Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J. 2003;22:2380–2386. doi: 10.1093/emboj/cdg229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle IE, Perry AJ, Alcock FH, et al. (15 co-authors) Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial intermembrane space. Mol Biol Evol. 2007;24:1149–1160. doi: 10.1093/molbev/msm031. [DOI] [PubMed] [Google Scholar]
- Glick BS, Brandt A, Cunningham K, Müller S, Hallberg RL, Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- Gray MW. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria. Genome Biol. 2001;2:1018. doi: 10.1186/gb-2001-2-6-reviews1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu Rev Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- Gross J, Bhattacharya D. Mitochondrial and plastid evolution in eukaryotes: an outsiders’ perspective. Nat Rev Genet. 2009;10:495–505. doi: 10.1038/nrg2610. [DOI] [PubMed] [Google Scholar]
- Hauth AM, Maier UG, Lang BF, Burger G. The Rhodomonas salina mitochondrial genome: bacteria-like operons, compact gene arrangement and complex repeat region. Nucleic Acids Res. 2005;33:4433–4442. doi: 10.1093/nar/gki757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulett JM, Lueder F, Chan NC, Perry AJ, Wolynec P, Likić VA, Gooley PR, Lithgow T. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J Mol Biol. 2008;376:694–704. doi: 10.1016/j.jmb.2007.12.021. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Molecular genetics of yeast: a practical approach. Oxford: IRL Press Ltd; 1994. Oxford University Press. [Google Scholar]
- Kadowaki K, Kubo N, Ozawa K, Hirai A. Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J. 1996;15:6652–6661. [PMC free article] [PubMed] [Google Scholar]
- Karamanou S, Bariami V, Papanikou E, Kalodimos CG, Economou A. Assembly of the translocase motor onto the preprotein-conducting channel. Mol Microbiol. 2008;70(2):311–322. doi: 10.1111/j.1365-2958.2008.06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J Biol Chem. 1974;249:3297–3303. [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J Cell Biol. 1975;65:1–14. doi: 10.1083/jcb.65.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Ostermann K, Rödel G. Evidence for the association of yeast mitochondrial ribosomes with Cox11p, a protein required for the Cu(B) site formation of cytochrome c oxidase. Curr Genet. 2005;47:223–233. doi: 10.1007/s00294-005-0569-1. [DOI] [PubMed] [Google Scholar]
- Kiefer D, Kuhn A. YidC as an essential and multifunctional component in membrane protein assembly. Int Rev Cytol. 2007;259:113–138. doi: 10.1016/S0074-7696(06)59003-5. [DOI] [PubMed] [Google Scholar]
- Koehler CM. New developments in mitochondrial assembly. Annu Rev Cell Dev Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057. [DOI] [PubMed] [Google Scholar]
- Kol S, Nouwen N, Driessen AJ. Mechanisms of YidC-mediated insertion and assembly of multimeric membrane protein complexes. J Biol Chem. 2008;283(46):31269–31273. doi: 10.1074/jbc.R800029200. [DOI] [PubMed] [Google Scholar]
- Kurland CG, Andersson SG. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 2000;64:786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, Stroud DA, Wiedemann N, Pfanner N. Evolution of mitochondrial protein biogenesis. Biochim Biophys Acta. 2009;1790:409–415. doi: 10.1016/j.bbagen.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Lang BF, Burger G, O'Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. (13 co-authors) ClustalW and ClustalX version 2. Bioinformatics. 2007;23(21):2947–2958. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Likic VA, Dolezal P, Celik N, Dagley M, Lithgow T. Using hidden markov models to discover new protein transport machines. Methods Mol Biol. 2010;619:271–284. doi: 10.1007/978-1-60327-412-8_16. [DOI] [PubMed] [Google Scholar]
- Lister R, Whelan J. Mitochondrial protein import: convergent solutions for receptor structure. Curr Biol. 2006;16(6):R197–R199. doi: 10.1016/j.cub.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Lithgow T. Targeting of proteins to mitochondria. FEBS Lett. 2000;476:22–26. doi: 10.1016/s0014-5793(00)01663-x. [DOI] [PubMed] [Google Scholar]
- Lucattini R, Likic VA, Lithgow T. Bacterial proteins predisposed for targeting to mitochondria. Mol Biol Evol. 2004;21:652–658. doi: 10.1093/molbev/msh058. [DOI] [PubMed] [Google Scholar]
- Luirink J, von Heijne G, Houben E, de Gier JW. Biogenesis of inner membrane proteins in Escherichia coli. Annu Rev Microbiol. 2005;59:329–355. doi: 10.1146/annurev.micro.59.030804.121246. [DOI] [PubMed] [Google Scholar]
- Mandon EC, Trueman SF, Gilmore R. Translocation of proteins through the Sec61 and SecYEG channels. Curr Opin Cell Biol. 2009;21:501–507. doi: 10.1016/j.ceb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Ito K. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci U S A. 2006;103(44):16159–16164. doi: 10.1073/pnas.0606390103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale P, Brüser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- O'Brien EA, Koski LB, Zhang Y, Yang L, Wang E, Gray MW, Burger G, Lang BF. TBestDB: a taxonomically broad database of expressed sequence tags (ESTs) Nucleic Acids Res. 2007;35:D445–D451. doi: 10.1093/nar/gkl770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kelly CJ. The jakobid flagellates: structural features of Jakoba, Reclinomonas and Histiona and implications for the early diversification of eukaryotes. J Eukaryot Microbiol. 1993;40:627–636. [Google Scholar]
- Ogawa S, Yoshino R, Angata K, et al. (11 co-authors) The mitochondrial DNA of Dictyostelium discoideum: complete sequence, gene content and genome organization. Mol Gen Genet. 2000;263:514–519. doi: 10.1007/pl00008685. [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq MP, Loiseaux-de Goër S, Stam WT, Olsen JL. Complete mitochondrial genomes of the three brown algae (Heterokonta: Phaeophyceae) Dictyota dichotoma, Fucus vesiculosus and Desmarestia viridis. Curr Genet. 2006;49:47–58. doi: 10.1007/s00294-005-0031-4. [DOI] [PubMed] [Google Scholar]
- Palmer T, Sargent F, Berks BC. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 2005;13:175–180. doi: 10.1016/j.tim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Popot JL, de Wry C. On the microassembly of integral membrane proteins. Annu Rev Biophys Biophys Chem. 1990;19:369–403. doi: 10.1146/annurev.bb.19.060190.002101. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol. 1999;286:105–120. doi: 10.1006/jmbi.1998.2455. [DOI] [PubMed] [Google Scholar]
- Rehling P, Pfanner N, Meisinger C. Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—a guided tour. J Mol Biol. 2003;326:639–657. doi: 10.1016/s0022-2836(02)01440-7. [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Bursać D, Lithgow T. The direct route: a simplified pathway for protein import into the mitochondrion of trypanosomes. Trends Cell Biol. 2008;18:12–18. doi: 10.1016/j.tcb.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Simpson AG, Inagaki Y, Roger AJ. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol Biol Evol. 2006;23:615–625. doi: 10.1093/molbev/msj068. [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The mitochondrial genome of Chara vulgaris: insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants. Plant Cell. 2003;15(8):1888–1903. doi: 10.1105/tpc.013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Why mitochondria need a genome. FEBS Lett. 1986;198:1–4. doi: 10.1016/0014-5793(86)81172-3. [DOI] [PubMed] [Google Scholar]
- Waller RF, Keeling PJ, van Dooren GG, McFadden GI. Comment on “A green algal apicoplast ancestor”. Science. 2003;301:49. doi: 10.1126/science.1084684. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CT, Lithgow T. Mitochondrial biogenesis: sorting mechanisms cooperate in ABC transporter assembly. Curr Biol. 2010;20:R564–R567. doi: 10.1016/j.cub.2010.05.024. [DOI] [PubMed] [Google Scholar]
- Xie K, Dalbey RE. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol. 2008;6(3):234–244. doi: 10.1038/nrmicro3595. [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- Zara V, Ferramosca A, Robitaille-Foucher P, Palmieri F, Young JC. Mitochondrial carrier protein biogenesis: role of the chaperones Hsc70 and Hsp90. Biochem J. 2009;419:369–375. doi: 10.1042/BJ20082270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.