Abstract
The interplay between C-C chemokine receptor type 5 (CCR5) host genetic background, disease progression, and intrahost HIV-1 evolutionary dynamics remains unclear because differences in viral evolution between hosts limit the ability to draw conclusions across hosts stratified into clinically relevant populations. Similar inference problems are proliferating across many measurably evolving pathogens for which intrahost sequence samples are readily available. To this end, we propose novel hierarchical phylogenetic models (HPMs) that incorporate fixed effects to test for differences in dynamics across host populations in a formal statistical framework employing stochastic search variable selection and model averaging. To clarify the role of CCR5 host genetic background and disease progression on viral evolutionary patterns, we obtain gp120 envelope sequences from clonal HIV-1 variants isolated at multiple time points in the course of infection from populations of HIV-1–infected individuals who only harbored CCR5-using HIV-1 variants at all time points. Presence or absence of a CCR5 wt/Δ32 genotype and progressive or long-term nonprogressive course of infection stratify the clinical populations in a two-way design. As compared with the standard approach of analyzing sequences from each patient independently, the HPM provides more efficient estimation of evolutionary parameters such as nucleotide substitution rates and dN/dS rate ratios, as shown by significant shrinkage of the estimator variance. The fixed effects also correct for nonindependence of data between populations and results in even further shrinkage of individual patient estimates. Model selection suggests an association between nucleotide substitution rate and disease progression, but a role for CCR5 genotype remains elusive. Given the absence of clear dN/dS differences between patient groups, delayed onset of AIDS symptoms appears to be solely associated with lower viral replication rates rather than with differences in selection on amino acid fixation.
Keywords: CCR5, envelope, HIV-1, hierarchical phylogenetic models, disease progression, Bayesian inference
Introduction
The high mutation rate and rapid viral turnover that characterize HIV-1 infection (Ho et al. 1995; Wei et al. 1995) generate a highly diverse genetic viral population within an HIV-1–infected individual (Shankarappa et al. 1999). Continuous emergence of new HIV-1 variants facilitates rapid viral adaptation to humoral and cellular immune responses of the host (Borrow et al. 1997; Goulder et al. 1997; Wei et al. 2003; Jones et al. 2004), escape from antiretroviral drugs (Coffin 1995), and the selection for optimal biological properties such as replication capacity and use of the entry complex (Koning et al. 2003; Kwa et al. 2003; Sterjovski et al. 2007; Repits et al. 2008).
Following primary infection, an asymptomatic phase with a gradual loss of CD4+ T cells and T-cell function characterizes the clinical course of HIV-1 infection (Lane et al. 1985; Polk et al. 1987; Miedema et al. 1988), resulting eventually in the development of AIDS. The duration of this asymptomatic phase in the absence of antiretroviral therapy varies among patients, from several months to more than two decades, and determines their rate of disease progression (Veugelers et al. 1994; Munoz et al. 1997). Many selective forces may play a role in intrahost viral evolution and disease progression such as neutralizing antibodies and cytotoxic T-cell lymphocyte (CTL) responses, immune activation, target cell availability, coreceptor expression levels, and emergence of C-X-C chemokine receptor type 4 (CXCR-4) using viruses among others. The severity of HIV infection may be further complicated by co-infections and heritable viral genetic factors (Hollingsworth et al. 2010). Largely stimulated by a comprehensive longitudinal analysis demonstrating common patterns of sequence divergence, diversity, and emergence of CXCR4-using variants in chronic HIV-1 infections (Shankarappa et al. 1999), phylogenetic analyses have been widely used as a means of elucidating how host factors impact HIV within-host dynamics. More specific evolutionary parameters such as evolutionary rate (Lemey et al. 2007; Lee et al. 2008), adaptation rates (Williamson 2003), positively selected sites (Ross and Rodrigo 2002), compartmentalization (Kemal et al. 2003), and recombination (Carvajal-Rodriguez et al. 2008) have been scrutinized, but consistent associations with disease progression have rarely been revealed.
Here, we focus on a polymorphism in the C-C chemokine receptor type 5 (CCR5) gene, which is a host factor known to influence disease progression. The CCR5 gene encodes one of the main coreceptors required for HIV-1 entry, and a heterozygous genotype for a 32 bp deletion (CCR5 wt/Δ32) associates with a lower viral load set point, defined as the viral load between 18 and 24 months after seroconversion (SC) which is stable in most HIV-1–infected individuals and predictive for clinical course of infection (Mellors et al. 1996; de Wolf et al. 1997), and a slower HIV-1 disease progression (de Roda Husman et al. 1997; Ioannidis et al. 2001). Given the reported lower percentages of CCR5-expressing target cells and higher levels of RANTES production in HIV-1–infected individuals with a CCR5 wt/Δ32 genotype (de Roda Husman, Blaak, et al. 1999; Blaak et al. 2000), it is likely that target cell and CCR5 availability influence HIV-1 intrapatient evolution and contribute to the progression to AIDS.
To investigate these influences, we compared the evolution of CCR5-using HIV-1 variants (R5) in individuals with either a CCR5 wt/wt or CCR5 wt/Δ32 genotype who only harbored CCR5-using HIV-1 variants in their progressive or long-term nonprogressive course of infection. Such comparisons require asking questions across multiple populations of individuals about the evolutionary histories that occur within each individual. Traditional modeling of evolutionary histories across individuals generally assumes that within-individual processes vary independently and are fit separately from individual to individual (Shankarappa et al. 1999; Ross and Rodrigo 2002; Potter et al. 2006; Lemey et al. 2007; Carvajal-Rodriguez et al. 2008). Often, this approach results in poor estimates of the underlying evolutionary parameters, as the information content within a single intrahost data set is sparse. Not surprisingly, Carvajal-Rodriguez et al. (2008) arrived at the conclusion that the statistical characterization of HIV within-host evolutionary processes in relationship to disease progression is a difficult task and suffers from a lack of power. To overcome the data sparsity, one may enforce strict equality between within-individual evolutionary parameters (Rodrigo et al. 2003). In both cases, however, the ability to formally assess similarities or differences between populations of individuals is lost. Hierarchical modeling (Laird and Ware 1982; Gelman et al. 1995) and in particular hierarchical phylogenetic models (HPMs) (Suchard et al. 2003) furnish an advantageous statistical framework in which to consider drawing conclusions across populations of individuals about the evolutionary processes within individuals. In general, the Bayesian hierarchical framework allows different evolutionary histories of the intrahost variants and different pressures driving their evolution from individual to individual while providing overall or across-individual summaries of important evolutionary measures, such as the DNA sequence mutation rate or synonymous/nonsynonymous substitution rate ratio (dN/dS) identifying positive selection. Critically, the HPM allows the within–individual-level parameters to vary about, for example, an unknown common mean for each population. This occurs through the employment of a hierarchical prior distribution on the parameters that are in turn characterized by unknown estimable hyperparameters. Then conveniently, hypothesis testing reduces to asking if these common mean parameters differ between populations. Fortuitously, the hierarchical prior embedded in the HPM also affords a borrowing of strength of information from one individual by another, providing more precise within–individual-level estimates (Suchard et al. 2003; Kitchen et al. 2004, 2006, 2009).
In this study, we extend the HPM across multiple populations of individuals through the introduction of population-specific fixed effects. These effects allow the expected evolutionary parameter estimated within a population to potentially vary across populations. We then exploit ideas from Bayesian model averaging (Hoeting et al. 1999) and selection (Suchard et al. 2001) to formally ask if these effects statistically differ between populations. We use this approach to estimate viral evolutionary rates and selective pressures within hosts and to evaluate whether these quantities differ with respect to CCR5 wt/Δ32 host genetic background and disease progression.
Materials and Methods
Study Subjects
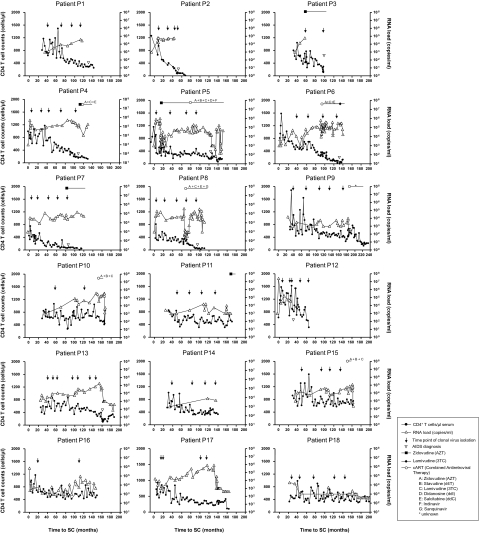
We selected eighteen men who have sex with men (MSM) participants in the Amsterdam Cohort Studies on HIV and AIDS, 11 with a WT genotype (patients P1–P11), and 7 with a CCR5 wt/Δ32 genotype (patients P12–P18), who at all times tested during follow-up harbored only R5 HIV-1 variants. All patients were either seropositive at entry in the cohort studies (seroprevalent cases with an average imputed seroconversion [SC] date of 18 months before entry in the cohort; Geskus 2000) or seroconverted during active follow-up in the cohort studies. Nine individuals were classified as long-term nonprogressors (LTNP) (defined as HIV-1–infected patients who at the end of follow-up [April 1997] had an asymptomatic seropositive follow-up of at least 11 years with relatively stable CD4+ T-cell counts that were still above 400 cells per μl in the ninth year of follow-up in the absence of antiretroviral therapy). The remaining nine individuals progressed to AIDS during the study period (median time to AIDS = 8.2 [2.7–10.8] years after SC or imputed SC date) and were classified as progressors (P). Individuals included in this study did not receive effective antiretroviral therapy during the study period. Clinical parameters and time points of virus isolation are shown per patient in figure 1.
FIG. 1.
CD4+ T-cell numbers, viral loads, and antiretroviral treatments of 18 participants from the Amsterdam Cohort Studies who were selected for this study. Time points of clinical AIDS diagnosis are indicated with open downward triangles. Arrows indicate time points of clonal virus isolation. The length and type of antiretroviral therapy are indicated in the top part of the panels.
The Amsterdam Cohort Studies are conducted in accordance with the ethical principles set out in the declaration of Helsinki, and written informed consent was obtained prior to data collection. The study was approved by the Academic Medical Center institutional medical ethics committee.
Isolation of Clonal HIV-1 Variants
Clonal HIV-1 variants were isolated by cocultivation of serial dilutions of patient peripheral blood mononuclear cells (PBMC) from 2 to 8 time points in the course of their infection and expanded to viral stocks for further study as described previously (Schuitemaker et al. 1992; van 't Wout et al. 2008). For each patient, time points of virus isolation and number of clonal HIV-1 variants per time point are summarized in supplementary table S1 (Supplementary Material online). The R5 phenotype of all clonal HIV-1 variants that were isolated was confirmed by inability to replicate in the MT2 cell line, in phytohemagglutinin-stimulated PBMC from a donor with a CCR5Δ32 homozygous genotype, and in astroglioma cells transfected with CD4 and CCR3 or CXCR4 (de Roda Husman, van Rij, et al. 1999) and predicted coreceptor use based on the V3 amino acid sequence using the syncytium-inducing/non-syncytium-inducing (sinsi) position-specific scoring matrix (http://indra.mullins.microbiol.washington.edu/webpssm/) (Jensen et al. 2003).
DNA Isolation, Polymerase Chain Reaction, and Sequencing
Total DNA was isolated from PBMCs infected with clonal HIV-1 variants using a modification of the L6 isolation method (Kootstra and Schuitemaker 1999). Precipitated DNA was dissolved in 100 μl of distilled water, and 5 μl were used for polymerase chain reaction (PCR) amplification of the gp120 (C1–C4) region corresponding to HXB2 nucleotide positions 6444–7595. Amplification was performed by PCR with primers TB3 forward (5′-GGCCTTATTAGGACACATAGTTAGCC-3′) and OFM19 reverse (5′-GCACTCAAGGCAAGCTTTATTGAGGCTTA-3′) using the expand high-fidelity Taq polymerase kit (Roche) and the following amplification cycles: 2 min 30 s at 94 °C, 9 cycles of 15 s at 94 °C, 45 s at 50 °C, 6 min at 68 °C, 30 cycles of 15 s at 94 °C, 45 s at 53 °C, 6 min at 68 °C, followed by a 10-min extension at 68 °C and subsequent cooling to 4 °C. Nested PCR was performed with two different inner PCR primer combinations: Seq1 forward (5′-TACATAATGTTTGGGCCACACATGCC-3′), Seq4 reverse (5′-CTTGTATTGTTGTTGGGTCTTGTAC-3′), Seq5 forward (5′-GTCAACTCAACTGCTGTTAAATGGC-3′), and Seq2 reverse (5′-TCCTTCATATCTCCTCCTCCAGGTC-3′). Nested PCRs were performed using Promega Taq polymerase in the presence of 2 mM MgCl2 using the following amplification cycles: 5 min at 94 °C, 40 cycles of 15 s at 95 °C, 30 s at 59 °C, 2 min at 72 °C, followed by a 10-min extension at 72 °C and subsequent cooling to 4 °C.
PCR products were purified using ExoSAP-IT (USB, Cleveland, OH) according to manufacturer's protocol. Sequencing conditions consisted of 5 min at 94 °C, 30cycles of 15s at 94 °C, 10 s at 50 °C, 2 min at 60 °C, and a 10 min extension at 60 °C. Sequencing was performed using BigDye Terminator v1.1 Cycle Sequencing kit (ABI Prism, Applied Biosystems, Warrington, UK) according to the manufacturer's protocol using the nested PCR primers. Sequences were analyzed on the Applied Biosystems 3130 xl Genetic Analyzer. The nucleotide sequences are available from GenBank under the accession numbers EU743973.1–EU44009.1, EU744014.1–EU744046.1, EU744055.1–EU744093.1, EU744097.1–EU744129.1, EU744146.1–EU744175.1, GU455514–GU455525, and HQ644787–HQ645012.
Bayesian Inference of Within-Host HIV Evolutionary Rates and Selection Pressures
Nucleotide gp120 (C1–C4) sequences for all clonal HIV-1 variants isolated from the individual patients were aligned using ClustalW (Thompson et al. 1994) and manually edited. Cross-contamination was excluded using phylogenetic analysis.
Independent Estimates of Within-Host Evolutionary Rates
Nucleotide substitution rates were estimated for each patient using strict and relaxed (uncorrelated lognormal) molecular clock models implemented in BEAST v.1.4.8 (Drummond et al. 2006; Drummond and Rambaut 2007). We used a general time-reversible (GTR) model of nucleotide substitution with discrete gamma-distributed rate variation among sites. Posterior distributions were obtained using Bayesian Markov chain Monte Carlo (MCMC) analysis. MCMC chains were run sufficiently long to ensure stationarity and adequate effective sample sizes > 100 as diagnosed using Tracer (http://tree.bio.ed.ac.uk/software/tracer/). The uncertainty of continuous parameter estimates is expressed as 95% highest posterior density (HPD) intervals.
Hierarchical Estimates of Evolutionary Parameters
To draw inference about different evolutionary patterns across populations of patients, we implement a novel HPM in BEAST (Suchard et al. 2003). HPMs analyze viral sequence data from multiple patients simultaneously and have found extensive use in uncovering common patterns of intrahost HIV evolution (Kitchen et al. 2004, 2006, 2009). At the heart of the HPM lies a Bayesian mixed effects model that pools information across patients. Pooling information through random effects affords more precise individual–patient parameter estimates when the data are sparse for a patient. Furthermore, unique to the work here, the introduction of fixed effects (see below) offers a formal hypothesis-testing framework from which to identify differences in evolutionary process between patient population groups.
Let θi for i = 1, …, N patients represent the evolutionary process parameter of interest; this could be, for example, the overall rate of nucleotide substitution or the nonsynonymous/synonymous substitution rate ratio (dN/dS) in a codon substitution process across the unknown genealogy relating the sequences from within patient i. In the analysis of four different patient groups: Progressors, Long-term non-progressors (LTNP), CCR5 wt/wt (WT), and CCR5 wt/Δ32 (Δ32), we assume that either log θi or θi is drawn from an underlying normal distribution where the mean and variance of this underlying prior distribution are also unknown and simultaneously estimated along with all sequence data. The choice of a log transform is convenient for modeling strictly positive parameters. Importantly, fixing this mean and variance to known values does not return a hierarchical model but rather results in complete independence across individuals. On the other hand, estimating the mean or variance imparts both an approach to make comparisons across populations and the borrowing of strength for poorly informed, within-individual model parameters.
For nucleotide analyses, we apply this hierarchical setup to the strict clock evolutionary rate (on the log scale), the mean evolutionary rate parameter of the lognormal relaxed clock (log), the constant population size (log) of the demographic prior, the GTR substitution parameters (log), and the shape parameter (log) of the discrete gamma distribution modeling rate variation among sites. For codon model analyses, a hierarchical transition/transversion rate parameter and a hierarchical dN/dS rate ratio (Goldman and Yang 1994) replace the GTR model parameters.
Hierarchical Estimation with Population-Specific Fixed Effects
For hypothesis-testing purposes, we extend the HPM to include across-population fixed effects. Each patient belongs to one of four fixed population groups that we can designate using two indicator factors: LTNPi = 0(1) for short (long)-term progressors and Δ32i = 0(1) for deletion 32 absent (present) patients. Our HPM assumes
where β0 is an unknown grand mean, δLTNP and δΔ32 are binary indicator variables, βLTNP and βΔ32 are conditional effective sizes, and εi are independent and normally distributed random variables with mean 0 and an estimable variance. The inclusion of the indicator variables follows from a Bayesian stochastic search variable selection approach (Kuo and Mallick 1998; Chipman et al. 2001) that simultaneously estimates the posterior probabilities of all possible linear models that may or may not include LTNP or Δ32 status effects. When an indicator equals 1, this effect is included in the model, demonstrating that the evolutionary process parameter differs with high probability between patient population groups. Lemey et al. (2009) discuss Bayesian stochastic search variable selection in further detail.
We complete this HPM model with variable selection through assigning independent Bernoulli prior probability distributions on δLTNP and δΔ32. These distributions place equal probability on each factor's inclusion and exclusion. We further assume diffuse priors on the unknown grand mean and error variance and specify that a priori βLTNP and βΔ32 are normally distributed with mean 0 and a variance of 1/2. We choose 1/2, as, before seeing the data, we believe that if a factor does result in different evolutionary parameters across population groups, process parameters should differ by at most an order of magnitude on their original scale. The introduction of HPMs into BEAST necessitates the development of MCMC transition kernels to efficiently explore that space of the grand mean and effect size, model indicator, and random-effects variance parameters. Given our judicious prior choices, the full conditional distributions of these parameters are in standard form: multivariate normal, binomial, and inverse gamma, respectively. This enables us to build highly effective Gibbs samplers (Casella and George 1992; Suchard et al. 2003) over the joint space of these parameters. Suchard et al. (2003) provide detailed derivations of the full condition distributions and their Gibbs samplers (Suchard et al. 2003). We implement these Gibbs samplers as regular BEAST “operators” that are now accessible to interested readers through BEAST's XML model specification language. Supplementary Material online to this paper reports the transition kernels’ XML syntax and gives examples on their use to implement HPMs.
To assign statistical significance to differences between population groups, we employ Bayes factors (BFs) (Jeffreys 1998; Suchard et al. 2001) that report how much the data change our prior opinion (here, 1:1 odds) about the inclusion of each factor. These BFs are straightforward to estimate through the variable selection procedure, as the BF equals the posterior odds that a factor indicator equals 1 divided by the corresponding prior odds. The posterior odds follow immediately from the marginal posterior probability that a factor indicator equals 1 that we estimate through the posterior expectation of the factor indicator. In cases where an estimate of this expectation approaches very closely to 0 or 1, an estimator based on a Rao-Blackwellization procedure is available (Casella and Robert 1996).
Results
Independent versus Hierarchical Estimation of Evolutionary Parameters
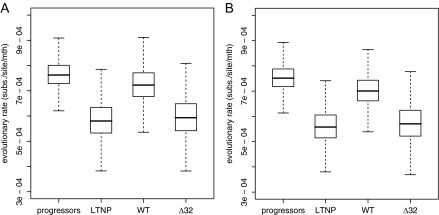
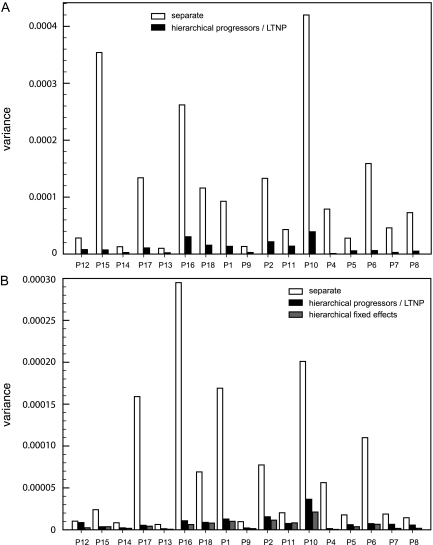
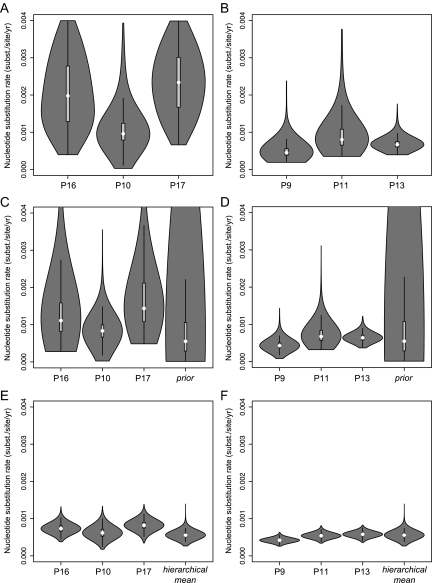
We first explored the nucleotide substitution rate as a hierarchical parameter estimated across patients in four separate patient groups: progressors, LTNP, WT, and Δ32. Using a strict clock model, a higher mean evolutionary rate was estimated in the progressors group (mean = 7.65 × 10−4 substitutions/site/mth [95% HPD = 6.45 × 10−4, 8.84 × 10−4]) compared with the LTNP group (5.87 × 10−4 [4.30 × 10−4, 7.55 × 10−4] substitutions/site/mth) (fig. 2A). Although these estimates demonstrate overlapping marginal posterior credible intervals (CIs), immediately concluding that their difference is not significant, ignores the correlation between the rates, we return to a formal test later. A less pronounced difference in evolutionary rate was estimated between the WT (7.27 × 10−4 [5.74 × 10−4, 8.75 × 10−4] substitutions/site/mth) and Δ32 (6.00 × 10−4 [4.21 × 10−4, 7.89 × 10−4] substitutions/site/mth) groups. Similar rate differences, with somewhat less overlapping CIs between progressors (7.57 × 10−4 [6.49 × 10−4, 8.67 × 10−4] substitutions/site/mth) and LTNPs (5.63 × 10−4 [4.19 × 10−4, 7.06 × 10−4] substitutions/site/mth), were observed using a relaxed-clock model (fig. 2B), in which the log of the mean evolutionary rate across all branches in a patient genealogy is drawn from an underlying normal distribution. For both strict and relaxed evolutionary rate estimates (fig. 3A and B), as well as other substitution model and population genetic parameters (data not shown), we observed significant shrinkage in uncertainty under the standard hierarchical fit, which clearly demonstrates the HPM improvement. Moreover, separate fit of parameter-rich models such as the uncorrelated relaxed clock required informative priors to achieve efficient sampling. To demonstrate the impact of such priors on our posterior rate estimates obtained by separate model fitting and compare these with the hierarchical estimates that did not require such priors, we plot the marginal posterior rate estimates for the three least informative (lowest number of time points and/or sequences per time point) and three most informative patients within the LTNP group (P10, P16, and P17 vs. P9, P11, and P13, respectively) as violin plots in figure 4. Violin plots are box plots overlaid with (rotated) kernel density estimates in order to show to the probability density at different parameter values. The patients for which only 2 or 3 time points were available resulted in rate estimates that only weakly diverged from their respective prior (uniform [0,0.004] or lognormal [−7.5,1]; fig. 4A and C, respectively), whereas many time points provide sufficient information to dominate over these priors (fig. 4B and D). Under the hierarchical model, even weakly informative patient-specific data sets with extremely diffuse priors on the rate yield relative precise posteriors (fig. 4E), and the individual patient estimates are only marginally higher than for the three most informative patients (fig. 4F). This demonstrates that comparing the mean rates for individual estimates would be inappropriate to assess differences among patient groups. Weakly informative patients result in relatively high mean rates, but their high variances ensure that the contribution to the population rate (LTNP group) in the hierarchical model remains low.
FIG. 2.
Evolutionary rate estimates using an HPM applied separately to four patient groups (progressors, LTNP, WT, and Δ32). Evolutionary rate estimated under strict clock model (A). Mean evolutionary rate estimated under relaxed-clock model (B). LTNP (Long-term non-progressors); WT (CCR5 wt/wt); Δ32 (CCR5 wt/Δ32).
FIG. 3.
Improved statistical efficiency (shrinkage effect) of the HPM. Strict clock (A). Relaxed clock (B). Posterior variance of estimated evolutionary rate from the independent analyses of each patient (white); evolutionary rate variance from the hierarchical analysis of LTNPs and progressors (black); evolutionary rate variance from the hierarchical analysis of LTNPs and progressors incorporating fixed effects (gray).
FIG. 4.
Marginal posterior rate distributions for LTNP patients with different numbers of sampling time points. Least informative patients (lowest number of time points or sequences per time point): P10, P16, and P17. Most informative patients: P9, P11, and P13. (A and B) Assuming a uniform (0,0.004) rate prior. (C and D) Lognormal (−7.5,1) rate prior. (E and F) HPM with unknown mean and variance and diffuse priors.
Although the application of relaxed-clock models to individual data sets with few time points or sequences may be questionable, analysis under an HPM, in which information is pooled between patients, enables us to side step this limitation. Marginal likelihood estimates for the both strict and relaxed-clock analyses of the different patient groups (supplementary table S2, Supplementary Material online) indicate a better fit of the relaxed-clock model, with ln BFs of 7.8, 6.1, 4.4, and 4.2 in favor of the relaxed clock for progressors, LTNP, WT, and Δ32, respectively. The fact that a strict clock could often not be rejected for individual patient analysis also indicates the HPM draws on increased statistical power of HPMs to reject simpler models. Because of the increased model fit, we employ relaxed clocks in further codon model analyses and hypothesis tests incorporating fixed effects.
Analyses using a codon model revealed comparable codon substitution rate differences between progressors/LTNP and between WT and Δ32 compared with the nucleotide analyses (fig. 2B vs. supplementary fig. 1A, Supplementary Material online). Hierarchical dN/dS estimates, however, were comparable for the four patient groups (supplementary fig. 1B, Supplementary Material online).
Hypothesis Testing Using HPMs Incorporating Across-Population Fixed Effects
The four different groups considered previously are not comprised of independent patient sets; some patients fall in more than one group. Hence, direct comparison of the marginal parameter estimates fit to each group independently does not generate independent estimates. For more appropriate hypothesis testing of difference, the HPM for the evolutionary rate was extended to accommodate fixed effects (see Materials and Methods), enabling estimation of hierarchical parameters across all patients. Successfully, hierarchical estimation with fixed effects across all patients resulted in even further shrinkage of individual patient estimates compared with hierarchal models applied to separate groups (fig. 3B). BF comparison of the fixed-effects HPM model with a model that assumes either completely linked or unlinked parameters (ln BF of 51.7 and 57.2, respectively) provides strong evidence that the shrinkage is accompanied by improved goodness of fit. The main results of the fixed-effect HPM analyses are listed in table 1. For the nucleotide analysis, the LTNP versus progressor and WT versus Δ32 effects were employed to model the evolutionary rates. Through examining the posterior distribution of the rate indicators (δeffect), we estimate the posterior probability for including the LTNP versus progressor effect at 0.72 resulting in a moderate BF support of 2.6 in agreement with the group-by-group hierarchical rate estimates obtained above. Importantly, the rate decrease attributable to this fixed effect returns a CI that does not include 0. This approach appropriately controls for the nonindependence missed in the group-by-group analyses and rejects the null hypothesis of no difference between LTNP and progressor patients.
Table 1.
Estimates of the LTNP and Δ32 Effects on Nucleotide Substitution Rates, Codon Substitution Rates, and dN/dS.
| Evolutionary Parameter | Effect Support/Size | LTNP Effect | Δ32 Effect |
| Nucleotide substitution rate | Posterior probability δeffect = 1 | 0.72 | 0.27 |
| BFeffect | 2.6 | 0.4 | |
| βeffect|δeffect = 1a | −0.275 (−0.524, −0.016) | −0.007 (−0.940,0.920) | |
| Codon substitution rate | Posterior probability δeffect = 1 | 0.726 | 0.324 |
| BFeffect | 2.6 | 0.5 | |
| βeffect|δeffect = 1a | −0.265 (−0.523,0.019) | −0.012 (−0.700,0.692) | |
| dN/dS | Posterior probability δeffect = 1 | 0.502 | 0.393 |
| BFeffect | 1.0 | 0.6 | |
| βeffect|δeffect = 1a | 0.083 (−0.101,0.25) | −0.005 (−0.228,0.242) |
These are effective sizes conditional on the effect being included (the binary effect indicator δeffect being 1). For the rates, these effective sizes are in log space. LTNP (Long-term non-progressor); Δ32 (CCR5 wt/Δ32).
There was no support in favor of a Δ32 effect. Even after conditioning on the effect indicator equaling 1 to estimate the potential effect size, the posterior Δ32 effect-size parameter distribution remained centered close to 0 with symmetric CIs. In the codon analysis, the same effects were tested on both the substitution rate and dN/dS. A very similar LTNP effect was observed for the codon substitution rate, although the CIs now included 0. Interestingly, the conditional effect size of LTNP versus progressor on codon substitution rate remains very similar to the effect size on nucleotide substitution rate. Furthermore, there was more support against than in favor of a Δ32 effect. Finally, no support for an LTNP effect or Δ32 effect was observed on the hierarchical dN/dS estimates.
Discussion
In this study, we adopted an HPM approach to estimate within-host HIV evolutionary parameters and test evolutionary hypotheses regarding host susceptibility and disease progression. We sought to investigate whether the CCR5 wt/Δ32 genotype, which is associated with a lower viral load set point and a slower HIV-1 disease progression (de Roda Husman et al. 1997; Ioannidis et al. 2001), also impacts the evolutionary rate of the virus by limiting target cell or CCR5 availability. Furthermore, we wanted to evaluate the contribution of CCR5 availability and CCR5 use on the selection pressure directed against the viral envelope protein by estimating dN/dS.
HPMs have been used for HIV evolutionary enquiry before, but this is the first study that develops HPMs to estimate evolutionary rate, dN/dS, and demographic parameters. In an HPM framework, we assume that the patient-specific HIV-1 evolutionary parameters can be drawn from a population distribution. Estimations of the evolutionary process based on a limited sample from each patient are riddled with noise, and the improvement of an HPM follows from the reduced uncertainty on individual patient estimates. BF comparison further confirms a considerable improvement in goodness of fit of the HPM with respect to a completely linked and unlinked model. This can be explained by the fact that the completely linked model inappropriately ignores any difference among patients on the one hand and a completely linked model suffers from an unnecessarily high effective number of parameters (Spiegelhalter et al. 2002) arising from the independent prior specifications on the other hand. The HPM sits in between these two extremes and reduces the effective number of parameters without sacrificing fit to the data. Furthermore, we demonstrate that the HPM is more powerful in rejecting simpler evolutionary models, like the constant rate assumption, which is frequently violated for HIV.
The hierarchical estimates for the progressors, LTNP, WT, and Δ32 groups indicated a pronounced strict and relaxed-clock rate difference between the progressors and the LTNP, whereas differences between WT and Δ32 rates were less pronounced. The same patterns were observed for relaxed codon substitution rates, but no real differences were noted in terms of dN/dS. These comparisons are based on nonindependent data because patients will be part of two different groups. For more appropriate hypothesis testing, we incorporated fixed effects and employed Bayesian stochastic search variable selection to estimate the posterior probability that different patient group characteristics influence within-host evolutionary parameters. The advantage of a Bayesian model averaging approach that simultaneously explores the space of models and regression coefficients is the opportunity to distinguish between the relative size of an effect and its importance, which can be formalized in terms of standard BF support. The latter effectively becomes independent of the scale of the predictors, which otherwise may confound drawing conclusions on the effect sizes only. Because both predictors we considered only achieve 0 or 1, controlling for scale is not an issue in the current study, but it does contribute to a more general framework for evolutionary hypothesis testing. Although the statistical support is not decisive, the fixed-effects HPM approach produces substantially more efficient parameter estimates and conditional effect sizes confirm rate differences among LTNP and progressors. Despite the elevated power, more elaborate sampling in terms of numbers of patients, within-host time points, or maybe even larger genome regions would be desirable.
The HPM estimates suggest an association between evolutionary rate and disease progression, but the CCR5 genotype does not account for the rate differences. Given the absence of clear dN/dS differences—if anything, they are slightly higher in LTNP—we cannot attribute the rate nuances to differences in selection on amino acid fixation. Therefore, we conclude that these differences are due to variations in the product of mutation rate and generation time. In particular, lower replication rates may be associated with delayed onset of AIDS symptoms. In agreement with this, a codon model extension of the Bayesian relaxed-clock analysis of more extensively sampled patients has shown that absolute synonymous substitutions are correlated with disease progression (Lemey et al. 2007). These authors argued that synonymous substitutions were a marker of replication rate and most probably reflect the action of immune activation, which in itself is a marker of disease progression. In the current study, we employed standard codon model implementation in the Bayesian framework rather than evaluating genealogies under nucleotide models as a proxy. This approach comes at a computational expense, and further extensions—such as codon models to estimate absolute rates of synonymous and nonsynonymous substitutions (Seo et al. 2004)—may prove even more computationally intensive. Fortunately, recent advances in graphics processing unit computation provide significant increases in computation speed for high state-space models (Suchard and Rambaut 2009). These advances promise to stimulate further development of various codon models in the Bayesian framework, the parameters of which could be efficiently estimated in hierarchical models.
CCR5 genotype has a measurable impact on disease progression (de Roda Husman et al. 1997; Ioannidis et al. 2001), but there appears to be no absolute relationship (not all CCR5 wt/Δ32 HIV-1 infected individuals are LTNP). This implies a more complex scenario, in which the combination of CCR5 availability with other host genetic factors, in particular cellular and humoral immune pressures, and immune activation, will determine the viral replication rate and progression of the disease in a patient. Although lower CCR5 availability does not appear to exert selection pressure on the viral envelope during the chronic phase of infection, it cannot be excluded that in HIV-1–infected individuals with CCR5 WT/Δ32 genotype, in whom CCR5+ target cells and CCR5 expression are already limiting in the acute phase, selection for viruses with optimal CCR5 use occurs in a very early stage. Moreover, we performed analyses on sequences in which ambiguously aligned hypervariable regions were deleted, which may play an important role in both humoral immune responses (Cao et al. 1997; Chackerian et al. 1997; Stamatatos and Cheng-Mayer 1998; Pinter et al. 2004; Sagar et al. 2006; Gray et al. 2007) and selection for optimal CCR5 use (Hubert and Arabie 1985; Stamatatos et al. 1998; Wang et al. 1999; Sagar et al. 2006; Repits et al. 2008).
Studying evolutionary dynamics within hosts has become an integral part of HIV research but one that still faces the challenge of fully unraveling the relationship between evolutionary parameters and clinical outcome. There may be several reasons for the difficulty in establishing the role of evolutionary processes in disease progression. Within-host dynamics appear to be highly complex, with many host-specific and environmental (co-infections) factors interacting with various evolutionary processes such as hypermutation, diversifying and directional selection, recombination, and compartmentalization. Untangling this complex interplay requires accurate measurement of all host factors involved and evolutionary models that explicitly accommodate the relevant evolutionary forces. Without the latter, many simplifying assumptions are at risk of being violated when considering HIV evolution. Parameter-rich models may be limited by current sampling as they require highly informative data. To our knowledge, the most elaborate sampling dates back to over a decade ago (Shankarappa et al. 1999), which, differently from this study, included patients with HIV populations harboring CXCR4-using variants. Next generation sequencing may offer new opportunities for within-host HIV genetic analyses but produces data with particular challenges for comparative analyses (Vrancken et al. 2010). Here, we have adopted a modeling approach that efficiently pools the information from multiple individuals, and we demonstrate how this can be employed for rigorous testing across patient populations. We hope that this stimulates further model-based inference of evolutionary processes, which ultimately may lead to more profound insights into persistent viral infections.
Supplementary Material
Supplementary materials are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Public Health Service of Amsterdam, the Academic Medical Center of the University of Amsterdam, the Sanquin Blood Supply Foundation, the University Medical Center Utrecht, and the Jan van Goyen Medical Center, are part of The Netherlands HIV Monitoring Foundation and financially supported by the Center for Infectious Disease Control of The Netherlands National Institute for Public Health and the Environment. Netherlands AIDS fund (6006); The European Community's Seventh Framework Programme NGIN (FP7/2007-2013) under grant agreement no. (201433); Research Foundation-Flanders (“Fonds voor Wetenschappelijk Onderzoek-Vlaanderen”, FWO) to P.L.; National Institutes of Health R01 675 grant (GM86887) to M.A.S. and J.A.T.; European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. (260864).
References
- Blaak H, Ran LJ, Rientsma R, Schuitemaker H. Susceptibility of in vitro stimulated PBMC to infection with NSI HIV-1 is associated with levels of CCR5 expression and beta-chemokine production. Virology. 2000;267:237–246. doi: 10.1006/viro.1999.0111. [DOI] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Wei X, et al. 11 co-authors. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Rodriguez A, Posada D, Perez-Losada M, Keller E, Abrams EJ, Viscidi RP, Crandall KA. Disease progression and evolution of the HIV-1 env gene in 24 infected infants. Infect Genet Evol. 2008;8:110–120. doi: 10.1016/j.meegid.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Casella G, George EI. Explaining the Gibbs sampler. Am Stat. 1992;46:167–174. [Google Scholar]
- Casella G, Robert C. Rao-Blackwellisation of sampling schemes. Biometrika. 1996;83:81–94. [Google Scholar]
- Chackerian B, Rudensey LM, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman H, George E, McCulloch R. The practical implementation of Bayesian model selection. IMS Lect Notes Monogr Ser. 2001;38:67–134. [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- de Roda Husman AM, Blaak H, Brouwer M, Schuitemaker H. CC chemokine receptor 5 cell-surface expression in relation to CC chemokine receptor 5 genotype and the clinical course of HIV-1 infection. J Immunol. 1999;163:4597–4603. [PubMed] [Google Scholar]
- de Roda Husman AM, Koot M, Cornelissen M, et al. 14 co-authors. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann Intern Med. 1997;127:882–890. doi: 10.7326/0003-4819-127-10-199711150-00004. [DOI] [PubMed] [Google Scholar]
- de Roda Husman AM, van Rij RP, Blaak H, Broersen S, Schuitemaker H. Adaptation to promiscuous usage of chemokine receptors is not a prerequisite for human immunodeficiency virus type 1 disease progression. J Infect Dis. 1999;180:1106–1115. doi: 10.1086/314987. [DOI] [PubMed] [Google Scholar]
- de Wolf F, Spijkerman I, Schellekens PT, Langendam M, Kuiken C, Bakker M, Roos M, Coutinho R, Miedema F, Goudsmit J. AIDS prognosis based on HIV-1 RNA, CD4+ T-cell count and function: markers with reciprocal predictive value over time after seroconversion. AIDS. 1997;11:1799–1806. doi: 10.1097/00002030-199715000-00003. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian data analysis. New York: Chapman & Hall/CRC; 1995. [Google Scholar]
- Geskus RB. On the inclusion of prevalent cases in HIV/AIDS natural history studies through a marker-based estimate of time since seroconversion. Stat Med. 2000;19:1753–1769. doi: 10.1002/1097-0258(20000715)19:13<1753::aid-sim487>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Phillips RE, Colbert RA, et al. 12 co-authors. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- Gray ES, Moore PL, Choge IA, et al. 13 co-authors. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol. 2007;81:6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- Hoeting J, Madigan D, Raftery A, Volinsky C. Bayesian model averaging. Stat Sci. 1999;14:382–401. [Google Scholar]
- Hollingsworth TD, Laeyendecker O, Shirreff G, et al. 14 co-authors. HIV-1 transmitting couples have similar viral load set-points in Rakai, Uganda. PLoS Pathog. 2010;6:e1000876. doi: 10.1371/journal.ppat.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert L, Arabie P. Comparing partitions. J Classification. 1985;2:193. [Google Scholar]
- Ioannidis JP, Rosenberg PS, Goedert JJ, et al. 32 co-authors. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3'A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann Intern Med. 2001;135:782–795. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- Jeffreys H . Theory of probability. New York: Oxford University Press; 1998. [Google Scholar]
- Jensen MA, Li FS, van 't Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NA, Wei X, Flower DR, Wong M, Michor F, Saag MS, Hahn BH, Nowak MA, Shaw GM, Borrow P. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J Exp Med. 2004;200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal KS, Foley B, Burger H, et al. 17 co-authors. HIV-1 in genital tract and plasma of women: compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc Natl Acad Sci U S A. 2003;100:12972–12977. doi: 10.1073/pnas.2134064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen CM, Lu J, Suchard MA, Hoh R, Martin JN, Kuritzkes DR, Deeks SG. Continued evolution in gp41 after interruption of enfuvirtide in subjects with advanced HIV type 1 disease. AIDS Res Hum Retroviruses. 2006;22:1260–1266. doi: 10.1089/aid.2006.22.1260. [DOI] [PubMed] [Google Scholar]
- Kitchen CM, Philpott S, Burger H, Weiser B, Anastos K, Suchard MA. Evolution of human immunodeficiency virus type 1 coreceptor usage during antiretroviral therapy: a Bayesian approach. J Virol. 2004;78:11296–11302. doi: 10.1128/JVI.78.20.11296-11302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen CMR, Marconi V, Kuritzkes DR, Bloomquist EW, Deeks SG, Suchard MA. Two-way Bayesian hierarchical phylogenetic models: an application to the co-evolution of gp120 and gp41 during partial treatment interruptions of enfuvirtide. Comput Stat Data Anal. 2009;53:766–775. doi: 10.1016/j.csda.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Kwa D, Boeser-Nunnink B, Dekker J, Vingerhoed J, Hiemstra H, Schuitemaker H. Decreasing sensitivity to RANTES (regulated on activation, normally T cell-expressed and -secreted) neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. J Infect Dis. 2003;188:864–872. doi: 10.1086/377105. [DOI] [PubMed] [Google Scholar]
- Kootstra NA, Schuitemaker H. Phenotype of HIV-1 lacking a functional nuclear localization signal in matrix protein of gag and Vpr is comparable to wild-type HIV-1 in primary macrophages. Virology. 1999;253:170–180. doi: 10.1006/viro.1998.9482. [DOI] [PubMed] [Google Scholar]
- Kuo L, Mallick B. Variable selection for regression models. Sankhya B. 1998;60:65–81. [Google Scholar]
- Kwa D, Vingerhoed J, Boeser B, Schuitemaker H. Increased in vitro cytopathicity of CC chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates correlates with a progressive clinical course of infection. J Infect Dis. 2003;187:1397–1403. doi: 10.1086/374650. [DOI] [PubMed] [Google Scholar]
- Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- Lane HC, Depper JM, Greene WC, Whalen G, Waldmann TA, Fauci AS. Qualitative analysis of immune function in patients with the acquired immunodeficiency syndrome. Evidence for a selective defect in soluble antigen recognition. N Engl J Med. 1985;313:79–84. doi: 10.1056/NEJM198507113130204. [DOI] [PubMed] [Google Scholar]
- Lee HY, Perelson AS, Park SC, Leitner T. Dynamic correlation between intrahost HIV-1 quasispecies evolution and disease progression. PLoS Comput Biol. 2008;4:e1000240. doi: 10.1371/journal.pcbi.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Kosakovsky Pond SL, Drummond AJ, Pybus OG, Shapiro B, Barroso H, Taveira N, Rambaut A. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol. 2007;3:e29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellors JW, Rinaldo CR, Jr., Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- Miedema F, Petit AJ, Terpstra FG, et al. 11 co-authors. Immunological abnormalities in human immunodeficiency virus (HIV)-infected asymptomatic homosexual men. HIV affects the immune system before CD4+ T helper cell depletion occurs. J Clin Invest. 1988;82:1908–1914. doi: 10.1172/JCI113809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz A, Sabin CA, Phillips AN. The incubation period of AIDS. AIDS. 1997;11(Suppl A):S69–S76. [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78:5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polk BF, Fox R, Brookmeyer R, Kanchanaraksa S, Kaslow R, Visscher B, Rinaldo C, Phair J. Predictors of the acquired immunodeficiency syndrome developing in a cohort of seropositive homosexual men. N Engl J Med. 1987;316:61–66. doi: 10.1056/NEJM198701083160201. [DOI] [PubMed] [Google Scholar]
- Potter SJ, Lemey P, Dyer WB, Sullivan JS, Chew CB, Vandamme AM, Dwyer DE, Saksena NK. Genetic analyses reveal structured HIV-1 populations in serially sampled T lymphocytes of patients receiving HAART. Virology. 2006;348:35–46. doi: 10.1016/j.virol.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Repits J, Sterjovski J, Badia-Martinez D, et al. 13 co-authors. Primary HIV-1 R5 isolates from end-stage disease display enhanced viral fitness in parallel with increased gp120 net charge. Virology. 2008;379:125–134. doi: 10.1016/j.virol.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Rodrigo AG, Goode M, Forsberg R, Ross HA, Drummond A. Inferring evolutionary rates using serially sampled sequences from several populations. Mol Biol Evol. 2003;20:2010–2018. doi: 10.1093/molbev/msg215. [DOI] [PubMed] [Google Scholar]
- Ross HA, Rodrigo AG. Immune-mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration. J Virol. 2002;76:11715–11720. doi: 10.1128/JVI.76.22.11715-11720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuitemaker H, Koot M, Kootstra NA, Dercksen MW, de Goede RE, van Steenwijk RP, Lange JM, Schattenkerk JK, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo TK, Kishino H, Thorne JL. Estimating absolute rates of synonymous and nonsynonymous nucleotide substitution in order to characterize natural selection and date species divergences. Mol Biol Evol. 2004;21:1201–1213. doi: 10.1093/molbev/msh088. [DOI] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, et al. 12 co-authors. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, Van Der Linde A. Bayesian measures of model complexity and fit. J R Stat Soc Series B Stat Methodol. 2002;64:583–639. [Google Scholar]
- Stamatatos L, Cheng-Mayer C. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J Virol. 1998;72:7840–7845. doi: 10.1128/jvi.72.10.7840-7845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Wiskerchen M, Cheng-Mayer C. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV type 1 isolate on viral envelope structure, cell entry, and replication. AIDS Res Hum Retroviruses. 1998;14:1129–1139. doi: 10.1089/aid.1998.14.1129. [DOI] [PubMed] [Google Scholar]
- Sterjovski J, Churchill MJ, Ellett A, et al. 16 co-authors. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology. 2007;4:89. doi: 10.1186/1742-4690-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Kitchen CM, Sinsheimer JS, Weiss RE. Hierarchical phylogenetic models for analyzing multipartite sequence data. Syst Biol. 2003;52:649–664. doi: 10.1080/10635150390238879. [DOI] [PubMed] [Google Scholar]
- Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370–1376. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Wout AB, Schuitemaker H, Kootstra NA. Isolation and propagation of HIV-1 on peripheral blood mononuclear cells. Nat Protoc. 2008;3:363–370. doi: 10.1038/nprot.2008.3. [DOI] [PubMed] [Google Scholar]
- Veugelers PJ, Page KA, Tindall B, et al. 11 co-authors. Determinants of HIV disease progression among homosexual men registered in the Tricontinental Seroconverter Study. Am J Epidemiol. 1994;140:747–758. doi: 10.1093/oxfordjournals.aje.a117322. [DOI] [PubMed] [Google Scholar]
- Vrancken B, Lequime S, Theys K, Lemey P. Covering all bases in HIV research: unveiling a hidden world of viral evolution. AIDS Rev. 2010;12:89–102. [PubMed] [Google Scholar]
- Wang WK, Dudek T, Essex M, Lee TH. Hypervariable region 3 residues of HIV type 1 gp120 involved in CCR5 coreceptor utilization: therapeutic and prophylactic implications. Proc Natl Acad Sci U S A. 1999;96:4558–4562. doi: 10.1073/pnas.96.8.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, et al. (15 co-authors) Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, et al. 12 co-authors. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- Williamson S. Adaptation in the env gene of HIV-1 and evolutionary theories of disease progression. Mol Biol Evol. 2003;20:1318–1325. doi: 10.1093/molbev/msg144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.