The structure of peptidyl-tRNA hydrolase from F. tularensis, the causative agent of tularemia, is comparable to that of other bacterial peptidyl-tRNA hydrolases, with most residues in the active site conserved amongst the family. The resultant reagents, structural data, and analyses provide essential information for structure-based design of novel inhibitors for this class of proteins.
Keywords: peptidyl-tRNA hydrolases, Francisella tularensis
Abstract
The rational design of novel antibiotics for bacteria involves the identification of inhibitors for enzymes involved in essential biochemical pathways in cells. In this study, the cloning, expression, purification, crystallization and structure of the enzyme peptidyl-tRNA hydrolase from Francisella tularensis, the causative agent of tularemia, was performed. The structure of F. tularensis peptidyl-tRNA hydrolase is comparable to those of other bacterial peptidyl-tRNA hydrolases, with most residues in the active site conserved amongst the family. The resultant reagents, structural data and analyses provide essential information for the structure-based design of novel inhibitors for this class of proteins.
1. Introduction
Francisella tularensis is a pathogenic species of Gram-negative bacteria and is the causative agent of tularemia or rabbit fever. F. tularensis is very infectious in that a small number of organisms (10–50) can cause disease (Oyston et al., 2004 ▶). Owing to its ease of transmission by aerosol and its high virulence, F. tularensis has been classified as a Class A agent by the US government. Infection with F. tularensis can occur via several routes, including skin contact, inhalation, ingestion and inoculation of the eye. The bacteria that cause tularemia appear widely in nature and can be isolated and grown in culture, although producing an effective aerosol weapon would be difficult. Inhalation of an infectious aerosol would result in severe respiratory illness, including life-threatening pneumonia and systemic infection if left untreated. Although tularemia can be life-threatening, most infections can be treated successfully with antibiotics such as streptomycin, gentamicin, doxycycline and ciprofloxacin. However, given the organism’s potential to be used as a bioterror agent, with antibiotic-resistance genes being engineered into strains, there is a need for new classes of antibiotics.
In an effort to discover novel antibiotics for Gram-negative bacteria such as F. tularensis, essential steps in the organism’s protein-synthesis pathway have been identified as targets for potential interdiction. One such target arises during the process of protein translation, when a peptidyl-tRNA may dissociate prematurely from the ribosome before completion of mRNA decoding (Meinnel et al., 1993 ▶; Schmitt et al., 1997 ▶). Excess peptidyl-tRNA in cells leads to reduced efficiency of translation and eventually impairs translation initiation by cellular depletion of the required tRNA (Menninger, 1976 ▶, 1979 ▶). Peptidyl-tRNA hydrolase (Pth; EC 3.1.1.29) recycles peptidyl-tRNA by catalyzing the hydrolytic removal of the peptidyl moiety, allowing the reuse of the free tRNA in protein biosynthesis.
The enzyme Pth can be classified into two types: peptidyl-tRNA hydrolase 1 (Pth1) and peptidyl-tRNA hydrolase 2 (Pth2). Pth1 is found in bacteria and eukaryotes (Kössel, 1970 ▶; Brun et al., 1971 ▶; Menez et al., 2002 ▶), while Pth2 enzymes are present in eukaryotes and archaea (Rosas-Sandoval et al., 2002 ▶; Fromant et al., 2005 ▶). These two enzymes do not show any significant sequence identity or structural similarity. The first crystal structure of Pth1 from Escherichia coli (EcPth) revealed a single α/β globular domain built around a twisted mixed β-sheet (Schmitt et al., 1997 ▶). The active site resides in a channel covered by a loop and short helix (α4) and contains the catalytic residues His20 (catalytic base) and Asp93 (Goodall et al., 2004 ▶). The structure of Mycobacterium tuberculosis Pth (MtPth) revealed an open conformation of the peptide binding site of the enzyme and led to a model of structural changes associated with enzyme action, which was subsequently probed through a dynamics analysis utilizing MtPth solution structures (Selvaraj et al., 2007 ▶; Pulavarti et al., 2008 ▶). The first crystal structure of human Pth2 (hPth2; de Pereda et al., 2004 ▶) showed a mixed β-sheet sandwiched by two groups of two α-helices and lacked any three-dimensional structural similarity to the structure of Pth1.
In this paper, we present the crystal structure of F. tularensis Pth (FtPth) at 2.25 Å resolution. Molecular modeling has indicated that there are two proximal sites for the potential binding of small molecules on either side of the His20 residue of EcPth in the crevice containing the active site (Pulavarti et al., 2008 ▶). A structural comparison was used to determine residues unique to the binding site of FtPth which could be exploited for the development of antibacterial agents specific for F. tularensis.
2. Materials and methods
2.1. Cloning, protein expression and purification
The F. tularensis subspecies tularensis peptidyl-tRNA hydrolase (FTF0680c) gene encoding residues 1–188 was Escherichia coli optimized and synthesized by incorporating an NdeI restriction site at the N-terminus and an XhoI site at the C-terminus. The gene was subcloned into the E. coli expression vector pET-15b (Novagen) to incorporate an N-terminal hexahistidine tag. The recombinant plasmid was transformed into E. coli BL21 (DE3) CodonPlus RIPL (Invitrogen) cells. 10 ml overnight starter culture (100 ml TB with 100 mg l−1 ampicillin) was used to inoculate 1 l of the same medium in 2.5 l baffled flasks. Cultures were grown at 310 K and 220 rev min−1 to an OD600 of ∼3–3.5 and then transferred to 291 K and induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside overnight. Cells were harvested, flash-frozen in liquid nitrogen and stored at 193 K. Thawed cells were resuspended in 10 mM HEPES pH 7.5, 0.5 M NaCl buffer and lysed by sonication in the presence of 0.5% CHAPS. Cell debris was removed by centrifugation at 3500g for 40 min and the supernatant was passed through a DE52 column in the same buffer and loaded onto an Ni2+-chelate affinity column. After extensive washes with buffer containing 25 mM imidazole, the protein was eluted with 250 mM imidazole, concentrated and subjected to gel filtration on a Superdex 200 26/60 column. Initially, low-salt no-additive buffer (10 mM HEPES pH 7.5, 150 mM NaCl) was used for final storage to allow the exploration of wider crystallization space. However, FtPth could not be concentrated to above 5 mg ml−1 as it precipitated under these conditions. By increasing the ionic strength of the buffer and using Tris–HCl instead of HEPES–Na, the solubility and the protein stability were improved. Glycerol had a detrimental effect. A buffer consisting of 10 mM Tris–HCl, 250 mM NaCl, 0.2 mM TCEP was selected for the final purification step and for the storage of all subsequent protein batches. Fractions containing FtPth were subsequently concentrated, aliquoted, flash-frozen and stored at 193 K.
All samples were subjected to quality control, which included denaturing polyacrylamide gel electrophoresis (SDS–PAGE) and electrospray ionization–time of flight mass spectrometry (ESI–ToF MS). SDS–PAGE did not show additional bands upon loading 15 µg protein. The mass determined by ESI–ToF MS was within the error margin (1 Da) of the expected mass calculated from the sequence and analysis of the remaining ESI–ToF spectra did not reveal the presence of any modified protein species detrimental to crystallization such as partially proteolysed, modified or cross-linked species.
2.2. Crystallization and data collection
Initial crystallization screening was carried out at room temperature using frozen aliquots of protein at concentrations between 5 and 15 mg ml−1 in 96-well sitting-drop crystallization trays utilizing various commercially available and internally developed proprietary crystallization screens. Two crystal forms, thin hexagonal plates (50 µm) and thicker nonhexagonal plates, appeared within one week under the same conditions. Suitable crystals for diffraction studies were obtained from a crystallization solution consisting of 25% PEG 3350, 0.2 M MgCl2 and 0.1 M Bis-Tris pH 6.5 with a protein concentration of 13.8 mg ml−1. Prior to data collection, the crystals were cryoprotected by transferring them to a mixture of ethylene glycol and well solution in a 1:4 volume ratio, soaked for 120 s and flash-frozen at 100 K in a cold nitrogen-gas stream. Diffraction data were collected on a Cu Kα rotating-anode X-ray radiation source using an imaging-plate (IP) detector. The crystal diffracted to a resolution limit of 2.25 Å and a complete data set was obtained. The data were processed with HKL-2000 (Otwinowski & Minor, 1997 ▶) in space group P21212, with unit-cell parameters a = 60.7, b = 93.0, c = 33.0 Å. Data-collection statistics are summarized in Table 1 ▶.
Table 1. Summary of crystallographic data for F. tularensis Pth.
Values in parentheses are for the highest shell.
| Data collection | |
| Wavelength (Å) | 1.54 |
| Resolution (Å) | 50.0–2.25 (2.33–2.25) |
| Space group | P21212 |
| Unit-cell parameters | a = 60.7, b = 93.0, c = 33.0 |
| Unique reflections | 9412 |
| Total no. of reflections | 62322 |
| Multiplicity | 7 |
| Completeness (%) | 100 (100) |
| 〈I/σ(I)〉 | 14.4 (4.0) |
| Rmerge (%) | 14.0 (49.0) |
| Structure refinement | |
| No. of amino-acid chains | 1 |
| No. of protein atoms | |
| Main chain | 758 |
| Side chain | 706 |
| Total | 1458 |
| No. of solvent molecules | 51 |
| No. of other atoms | 21 |
| Resolution range (Å) | 33.0–2.25 |
| R factor (%) | 19.0 |
| Rfree (%) | 24.8 |
| Average protein-chain B factor (Å2) | |
| Main chain | 24.3 |
| Side chain | 25.7 |
| Total | 25.0 |
| Average solvent B factor (Å2) | 24.2 |
| Average B factor for other atoms (Å2) | 34.3 |
| R.m.s.d. covalent bond lengths (Å) | 0.013 |
| R.m.s.d. bond angles (°) | 1.291 |
| Missing residues | N-terminal hexahistidine tag, Met1 |
2.3. Structure determination
The FtPth structure was solved by molecular replacement using the E. coli Pth coordinates (PDB entry 2pth; Schmitt et al., 1997 ▶) as a search model with the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). Alternate cycles of restrained refinement and manual rebuilding in the program Coot (Emsley & Cowtan, 2004 ▶) completed the structure for residues 2–188. The entire N-terminal hexahistidine tag was not observed. During the last cycles of refinement, water molecules were built automatically into spherical peaks of the F o − F c difference electron density and verified by manual inspection. Clear difference densities also appeared for one chloride ion and five ethylene glycol molecules. Table 1 ▶ contains a summary of the structure-refinement statistics.
3. Results and discussion
3.1. Overall structure of FtPth
We have established the cloning, expression, purification and crystallization of peptidyl-tRNA hydrolase from F. tularensis (FtPth) and determined the crystal structure. Since the C-terminus was found to be disordered in previous Pth structures, the FtPth protein construct was truncated by three residues in an attempt to improve the probability of crystallization. The Ramachandran plot (not shown) indicates 97.3% of residues in the most favored region, 2.2% of residues in allowed regions and 0.5% of residues as outliers. Electron density for the N-terminal residues of the His tag and the terminal Met are missing, probably owing to disorder, and these were not built into the final model, consisting of residues 2–188. In addition, 51 water molecules, a chloride ion and five ethylene glycol molecules were modeled.
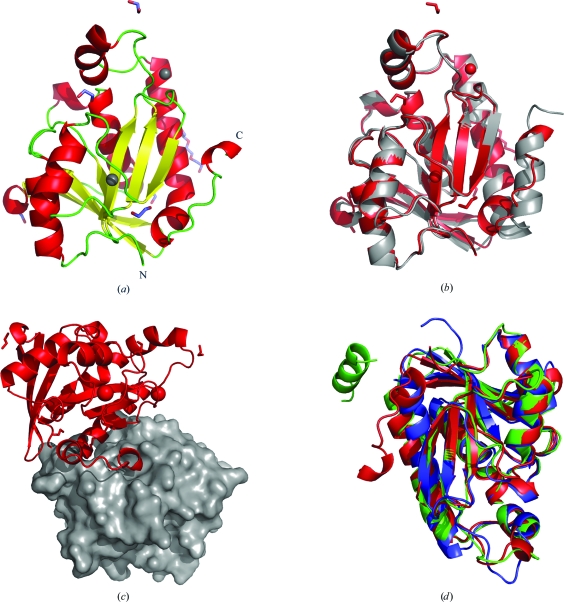
FtPth adopts an α/β fold with a twisted seven-stranded β-sheet flanked by helices, similar to other Pth proteins (Figs. 1a–1c). The mixed β-sheet has four parallel strands flanked by antiparallel strands, with another strand parallel to the central four on the end. The mixed β-sheet is packed between two long α-helices (α1 and α5) and two shorter α-helices (α2 and α3). Another short mobile α-helix (α4) completes the structure. When the crystal packing of FtPth was examined, the C-terminus of each molecule was found to reside within the active site of a neighboring symmetry mate (Fig. 1 ▶ c). The last residue, Leu188, makes contact with the catalytic residue His22 in a symmetry mate.
Figure 1.
(a) Ribbon diagram of the secondary structure of F. tularensis peptidyl-tRNA hydrolase (FtPth), with N- and C-termini labeled. The structure comprises a monomer with a single α/β globular domain built around a twisted β-sheet. The coloring scheme for the secondary-structure elements is as follows: α-helices are colored red, β-strands yellow and loops green. Chloride ions are represented by gray spheres, while PEG and ethylene glycol molecules are shown as sticks colored by atom type. (b) Superposition with the published E. coli structure (PDB entry 2pth). This figure was generated using PyMOL (DeLano, 2002 ▶). (c) Crystal-packing diagram for the F. tularensis Pth apo structure. The C-terminus of each molecule (red ribbon) resides in the active site of a neighbouring symmetry mate (gray surface). (d) Superposition of the F. tularensis (colored red), E. coli (colored green; PDB entry 3ofv; R. Lam, T. E. McGrath, V. Romanov, S. A. Gothe, S. R. Peddi, E. Razumova, R. S. Lipman, A. A. Branstrom & N. Y. Chirgadze, unpublished work) and M. tuberculosis (colored blue; PDB entry 2z2i; Selvaraj et al., 2007 ▶) Pth crystal structures. The core secondary-structure elements and protein fold are conserved in all species. As shown by the superposition, the highly mobile C-terminal helix and adjacent hinge region represent opportunities for designing new constructs (for the identification of feasible crystal forms).
3.2. Structural differences between FtPth and other Pth-family members
The sequence similarity between FtPth and other Pth-family members was investigated using a BLAST search (Altschul et al., 1997 ▶). FtPth (gi:3191676) shares 44% sequence identity (84/188) and an E value of 4 × 10−34 with E. coli Pth (EcPth; gi:157835761), 37% sequence identity (71/190) and an E value of 2 × 10−28 with M. tuberculosis Pth (MtPth; gi:189095922) and 37% sequence identity (62/166) and an E value of 4 × 10−26 with chloroplast group II intron splicing factor Crs2 (CRS2; gi:40889732).
The three-dimensional structure of FtPth was compared with other structures of the Pth family and the structurally similar protein chloroplast RNA-splicing factor 2 (CRS2) from the PDB using the DALI server (Holm & Rosenström, 2010 ▶). FtPth shows high structural similarity to EcPth (PDB code 2pth, Z score 32.6, r.m.s.d. 1.3 Å), CRS2 (PDB code 1ryb, Z score 28.5, r.m.s.d. 1.9 Å; PDB code 1ryn, Z score 26.3, r.m.s.d. 2.0 Å) and MtPth (PDB code 2z2j, Z score 27.6, r.m.s.d. 1.8 Å; PDB code 2jrc, Z score 20.3, r.m.s.d. 2.7 Å) (Fig. 1 ▶ d). The core α/β fold of the structures overlay well, but the flexible C-terminus is found in a number of positions. To reduce flexibility in the molecule and possibly improve the ability of these proteins to crystallize, residues from the C-terminus and adjacent hinge region could be deleted as long as there was little detrimental effect on function. The catalytic residue His22 is conserved in all structurally similar structures as well as most of the residues lining the substrate-recognition and catalytically active site, including those in the functionally divergent CRS2 (Ostheimer et al., 2005 ▶). This similarity in the active site could be exploited to develop broader spectrum antimicrobial agents.
Peptidyl-tRNA hydrolase is considered to be a good target for antimicrobial agents owing to the differences in sequence and structure between the human and bacterial enzymes. To date, no small-molecule inhibitors for the Pth family of proteins have been discovered even though the enzyme activity and structures of several bacterial Pths have been characterized (Payne et al., 2007 ▶). The availability of a variety of protein constructs and crystals of Pth from F. tularensis and E. coli (unpublished results) enables future studies examining the binding of small molecules by fragment screening, which could be developed into potential inhibitors.
Supplementary Material
PDB reference: peptidyl-tRNA hydrolase, 3nea
Acknowledgments
This work was supported by the Transformational Medical Technologies program, contract HDTRA1-06-0041 from the Department of Defense Chemical and Biological Defense program through the Defense Threat Reduction Agency (DTRA). About the Defense Threat Reduction Agency: the Defense Threat Reduction Agency (DTRA) was founded in 1998 to integrate and focus the capabilities of the Department of Defense (DoD) that address the threat by weapons of mass destruction (WMD). DTRA’s mission is to safeguard the United States and its allies from chemical, biological, radiological, nuclear and high-yield explosive WMDs by providing capabilities to reduce, eliminate and counter the threat and mitigate its effects. DTRA combines DoD resources, expertise and capabilities to ensure the United States remains ready and able to address present and future WMD threats. For more information on DTRA, visit http://www.dtra.mil. About Transformational Medical Technologies (TMT): TMT was created by DoD to protect the warfighter from emerging and genetically engineered biological threats by discovering and developing a wide range of medical countermeasures through enhanced medical research, development and test and evaluation programs. The TMT Program Office is matrixed from the Joint Science and Technology Office-DTRA and Joint Program Executive Office-Chemical and Biological Defense with oversight from the Office of the Secretary of Defense. For more information on TMT, visit http://www.tmti-cbdefense.org.
References
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Brun, G., Paulin, D., Yot, P. & Chapeville, F. (1971). Biochimie, 53, 225–231. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- De Pereda, J. M., Waas, W. F., Jan, Y., Ruoslahti, E., Schimmel, P. & Pascual, J. (2004). J. Biol. Chem. 279, 8111–8115. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fromant, M., Schmitt, E., Mechulam, Y., Lazennec, C., Plateau, P. & Blanquet, S. (2005). Biochemistry, 44, 4294–4301. [DOI] [PubMed]
- Goodall, J. J., Chen, G. J. & Page, M. G. (2004). Biochemistry, 43, 4583–4591. [DOI] [PubMed]
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res, 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Kössel, H. (1970). Biochim. Biophys. Acta, 204, 191–202. [DOI] [PubMed]
- Meinnel, T., Mechulam, Y. & Blanquet, S. (1993). Biochimie, 75, 1061–1075. [DOI] [PubMed]
- Menez, J., Buckingham, R. H., de Zamaroczy, M. & Campelli, C. K. (2002). Mol. Microbiol. 45, 123–129. [DOI] [PubMed]
- Menninger, J. R. (1976). J. Biol. Chem. 251, 3392–3398. [PubMed]
- Menninger, J. R. (1979). J. Bacteriol. 137, 694–696. [DOI] [PMC free article] [PubMed]
- Ostheimer, G. J., Hadjivassiliou, H., Kloer, D. P., Barkan, A. & Matthews, B. W. (2005). J. Mol. Biol. 345, 51–68. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Oyston, P., Sjostedt, A. & Titball, R. (2004). Nature Rev. Microbiol. 2, 967–978. [DOI] [PubMed]
- Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. (2007). Nature Rev. Drug Discov. 1, 29–40. [DOI] [PubMed]
- Pulavarti, S. V. S. R. K., Jain, A., Pathak, P. P., Mahmood, A. & Arora, A. (2008). J. Mol. Biol. 378, 165–177. [DOI] [PubMed]
- Rosas-Sandoval, G., Ambrogelly, A., Rinehart, J., Wei, D., Cruz-Vera, L. R., Graham, D. E., Stetter, K. O., Guarneros, G. & Söll, D. (2002). Proc. Natl Acad. Sci. USA, 99, 16707–16712. [DOI] [PMC free article] [PubMed]
- Schmitt, E., Mechulam, Y., Fromant, M., Plateau, P. & Blanquet, S. (1997). EMBO J. 16, 4760–4769. [DOI] [PMC free article] [PubMed]
- Selvaraj, M., Roy, S., Singh, N. S., Sangeetha, R., Varshney, U. & Vijayan, M. (2007). J. Mol. Biol. 372, 186–193. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: peptidyl-tRNA hydrolase, 3nea