In this study, the CIDE domains of Drep2 and Drep3 from Drosophila melanogaster were purified in Escherichia coli, after which they formed a stable complex in vitro and were crystallized by the hanging-drop vapour-diffusion method.
Keywords: apoptosis, DNA-fragmentation factors, Drep1, Drep2, Drep3, Drep4, CIDE domains
Abstract
The DFF40–DFF45 heterodimeric complex is a primary player in apoptotic DNA fragmentation and is conserved among different species including Drosophila melanogaster. DFF40 is a novel nuclease, while DFF45 is an inhibitor that can suppress the nuclease activity of DFF40 via tight interaction. Unlike mammalian systems, apoptotic DNA fragmentation in the fly is controlled by four DFF-related proteins known as Drep1, Drep2, Drep3 and Drep4. Drep1 and Drep4 are DFF45 and DFF40 homologues, respectively. Although the exact functions of Drep2 and Drep3 are unclear, they are also involved in apoptotic DNA fragmentation via regulation of the function of Drep1 and Drep4. DFF-related proteins contain a conserved CIDE domain of ∼90 amino-acid residues that is involved in protein–protein interaction. In this study, the CIDE domains of Drep2 and Drep3 were purified in Escherichia coli, after which they formed a stable complex in vitro and were crystallized by the hanging-drop vapour-diffusion method. X-ray diffraction data were collected to a resolution of 5.8 Å.
1. Introduction
Characteristic changes in chromosomes in the nucleus, such as chromatin condensation and cleavage, are hallmarks of apoptosis that are often used as a tool for the detection of apoptotic cells (Nagata, 2000 ▶; Liu et al., 1997 ▶; Enari et al., 1998 ▶; Park et al., 2007 ▶; Jang et al., 2010 ▶). During this process, the DNA undergoes fragmentation and is eventually cleaved into regular fragments with unit lengths of ∼180 bp (Enari et al., 1998 ▶; Liu et al., 1997 ▶). This DNA fragmentation is mainly executed by the heterodimeric DNA-fragmentation factor complex known as DFF40–DFF45. The DFF40–DFF45 complex was first identified as a caspase-3-dependent activity in the human HeLa S-100 fraction (Liu et al., 1997 ▶). The orthologous mouse proteins have also been identified and designated as caspase-activated deoxyribonuclease (CAD) and its inhibitor (ICAD). DFF40 (CAD) is a novel nuclease with a nuclear localization signal, while DFF45 (ICAD) is an inhibitor that can suppress the nuclease activity of DFF40 via tight interaction with DFF40 (CAD). Interestingly, DFF45 seems to function as a chaperone for DFF40 during its synthesis (Park, 2009 ▶). When effector caspases such as caspase-3 are activated by apoptotic stimuli, they cleave DFF45, allowing DFF40 to enter the nucleus and degrade chromosomal DNA (Sakahira et al., 1999 ▶; Enari et al., 1998 ▶). The N-termini of DFF40 and DFF45 contain a conserved CIDE domain of ∼90 amino-acid residues that is involved in the interaction between the two proteins (Lugovskoy et al., 1999 ▶; Wu et al., 2008 ▶). Apoptotic DNA fragmentation is also executed by several mitochondrial proteins such as AIF (apoptosis-inducing factor) and EndoG in a caspase-independent manner. EndoG is a DNAse I-like endonuclease that translocates into the nucleus to cleave DNA once released (Li et al., 2001 ▶; Parrish et al., 2001 ▶). Conversely, AIF does not possess intrinsic nuclease activity but induces chromatin condensation and the cleavage of DNA into large fragments (Susin et al., 1999 ▶).
Apoptosis and apoptotic DNA fragmentation is conserved among different species including Drosophila melanogaster and several homologues of the DFF40–DFF45 complex have been identified (Inohara & Nuñez, 1999 ▶). Unlike the mammalian system, apoptotic DNA fragmentation in Drosophila is controlled by four DFF-related proteins known as Drep1 (dDFF45), Drep2, Drep3 and Drep4 (dDFF40) (Inohara & Nuñez, 1999 ▶). Drep1 and Drep4 are DFF45 and DFF40 homologues, respectively. Drep2 and Drep3 are known to be regulators of apoptotic DNA fragmentation in flies (Inohara & Nuñez, 1999 ▶). All four proteins have a conserved CIDE domain that enables protein–protein interaction. Two structures of a CIDE domain have been identified by NMR to date, including the complex structure between the DFF40 and DFF45 CIDE domains, which shows an α/β roll with two α-helices and five β-strands (Lugovskoy et al., 1999 ▶; Otomo et al., 2000 ▶). Despite the availability of the NMR structures, further structural studies of the CIDE domain are important in order to understand apoptotic DNA fragmentation. In the present study, we overexpressed, purified and crystallized the Drep2 CIDE–Drep3 CIDE complex as a first step toward elucidating its molecular structure and regulation mechanism. Details of the atomic structure of this domain should enable us to understand the regulation mechanism of apoptotic DNA fragmentation. Identifying binding partners and investigating the protein-interaction interfaces formed by them is always important in order to understand signalling events.
2. Materials and methods
2.1. Protein expression and purification
The CIDE domains of Drep2 (Gene ID 35955) and Drep3 (Gene ID 36292) were expressed in Escherichia coli. Drep2 CIDE (amino-acid residues 1–84) was amplified by PCR using gene-specific primers containing NdeI and XhoI sites. PCR fragments were subsequently digested and ligated into the pET26b vector containing a C-terminal hexahistidine tag. PCR fragments for Drep3 CIDE corresponding to amino acids 112–195 were prepared in a similar fashion and cloned into a homemade pOKD5 vector containing a C-terminal hexahistidine tag (Dzivenu et al., 2004 ▶). The sequences of the cloned genes were verified by DNA sequencing. Both vector constructions add an eight-residue tag that includes six C-terminal histidine residues (LEHHHHHH).
The two proteins were expressed and purified in a similar fashion. The resulting plasmids were transformed into BL21 (DE3) E. coli competent cells individually. The E. coli competent cells were plated onto Luria–Bertani (LB) medium and incubated for 18 h at 310 K. A single colony was inoculated in 5 ml LB medium and incubated overnight at 310 K in a shaking incubator. Cultured cells were then moved to 1 l LB medium and incubated for 4 h at 310 K in a shaking incubator. Expression was then induced by treating the bacteria with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 20 h at 293 K. The bacteria were then collected, resuspended and lysed by sonication in 50 ml lysis buffer (20 mM Tris–HCl pH 7.9, 500 mM NaCl, 10 mM imidazole). The bacterial lysate was then centrifuged at 28 000g for 30 min at 277 K. The supernatant fraction was applied onto a gravity-flow column (Bio-Rad) packed with 2 ml Ni–NTA affinity resin (Qiagen). The unbound bacterial proteins were subsequently removed from the column using 100 ml washing buffer (20 mM Tris–HCl pH 7.9, 500 mM NaCl, 60 mM imidazole). The C-terminally His-tagged Drep2 CIDE and Drep3 CIDE were eluted from the column using elution buffer (20 mM Tris–HCl pH 7.9, 500 mM NaCl, 250 mM imidazole) and 1 ml elution fractions were collected over a total of 10 ml. Fractions containing more than 90% homogenous protein, as shown by SDS–PAGE, were selected and combined. The protein purity was further improved using a Superdex 200 10/30 gel-filtration column (GE Healthcare) pre-equilibrated with 20 mM Tris–HCl pH 8.0 and 150 mM NaCl.
2.2. In vitro reconstitution of the CIDE-domain complex
The quantified Drep2 CIDE and Drep3 CIDE, which had been purified by affinity and gel-filtration chromatography, were mixed and pre-incubated at room temperature for 1 h with a molar excess of Drep3 CIDE. The incubation and storage buffers were the same as that used for gel-filtration chromatography (20 mM Tris–HCl pH 8.0 and 150 mM NaCl). The mixture was concentrated to 15–20 mg ml−1 using a Centricon (Millipore, 10 kDa cutoff). The concentrated solution containing the two proteins was then applied onto a Superdex 200 10/30 gel-filtration column (GE Healthcare) that had been pre-equilibrated with a solution consisting of 20 mM Tris pH 8.0 and 150 mM NaCl. The complex eluted at around 16 ml and was collected and concentrated to 8–10 mg ml−1. The complex peak was then confirmed to contain both Drep2 CIDE and Drep3 CIDE by SDS–PAGE.
Formation of the complex between Drep2 CIDE and Drep3 CIDE was assayed by native (nondenaturing) PAGE conducted on a PhastSystem (GE Healthcare) with pre-made 8–25% acrylamide gradient gels (GE Healthcare). Coomassie Brilliant Blue was used for staining and detection of shifted bands. To produce a stable complex, we mixed the proteins and then incubated the mixture for 1 h before further analysis.
2.3. Crystallization
The initial conditions for crystallization were screened at 293 K by the hanging-drop vapour-diffusion method using screening kits from Hampton Research (Crystal Screen, Crystal Screen 2, Index HT, SaltRX, Natrix, MembFac and Crystal Screen Cryo) and Emerald BioStructures (Wizard I, II, III and IV). Initial crystals were grown on a siliconized cover slip by equilibrating a mixture consisting of 1 µl protein solution (8–10 mg ml−1 protein in 20 mM Tris pH 8.0, 150 mM NaCl) and 1 µl reservoir solution consisting of 0.1 M Tris pH 8.0, 5% PEG 8000 and 35% glycerol (condition No. 36 of Crystal Screen Cryo) against 0.5 ml reservoir solution. This crystal only diffracted poorly to 8–10 Å resolution. The crystals were further refined using Additive Screen (Hampton Research) and 200 mM lithium chloride gave a nicely shaped crystal. Crystals appeared in 3 d and grew to maximum dimensions of 0.1 × 0.1 × 0.3 mm. These crystals diffracted to 5.8 Å resolution.
2.4. Crystallographic data collection
For data collection, the crystals were frozen in liquid nitrogen. 35% glycerol in the crystallization solution was sufficient for use as a cryoprotectant. Diffraction data sets were collected on beamline BL-4A of the Pohang Accelerator Laboratory (PAL), Republic of Korea. The data set was indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Diffraction data statistics are given in Table 1 ▶.
Table 1. Diffraction data statistics for Drep2 CIDE–Drep3 CIDE complex crystals.
Values in parentheses are for the highest resolution shell.
| No. of crystals | 1 |
| Beamline | BL-4A, PAL |
| Wavelength (Å) | 1.0000 |
| Detector | ADSC Quantum 315r |
| Crystal-to-detector distance (mm) | 300 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 180 |
| Exposure time per image (s) | 10 |
| Resolution range (Å) | 50–5.8 |
| Space group | P41 |
| Unit-cell parameters (Å) | a = 109.0, b = 109.0, c = 46.2 |
| Mosaicity (°) | 0.8 |
| Total No. of measured intensities | 90452 |
| No. of unique reflections | 3649 |
| Multiplicity | 2.8 (2.8) |
| Mean I/σ(I) | 13.9 (2.6) |
| Completeness (%) | 99.2 (92.4) |
| Rmerge† (%) | 6.5 (34.4) |
| Overall B factor from Wilson plot (Å2) | 72 |
R
merge = 
 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations of reflection hkl.
3. Results and discussion
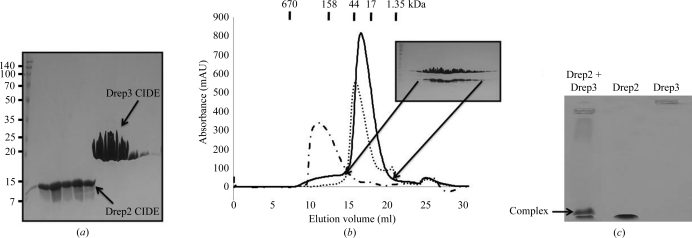
Affinity chromatography followed by gel-filtration chromatography produced 95% pure Drep2 CIDE and 90% pure Drep3 CIDE, which were analyzed by SDS–PAGE (Figs. 1 ▶ a and 1 ▶ b). Although Drep2 CIDE domains are highly oligomeric, eluting at around 200 kDa, and Drep3 CIDE domains are monomeric in solution, when they were mixed together a complex containing both Drep2 CIDE and Drep3 CIDE eluted at around 20 kDa from a Superdex 200 gel-filtration column (Fig. 1 ▶ b). No contaminating bands were visible on SDS–PAGE analysis of the complex. Assuming that the complex bands contain ∼20 µg protein and the detection limit of SDS–PAGE is 0.1 µg, the purity of the complex was >99%. Complex formation was confirmed by native PAGE. Separately purified Drep2 CIDE and Drep3 CIDE were mixed for 60 min at room temperature, after which the mixture was subjected to native PAGE. A newly appearing band clearly indicated that Drep2 CIDE and Drep3 CIDE form a stable complex (Fig. 1 ▶ c). As the calculated molecular weights of monomeric Drep2 CIDE and Drep3 CIDE are 10 864.4 and 11 038.4 Da, respectively, the calculated molecular weight of a 1:1 Drep2 CIDE–Drep3 CIDE complex is 21 902.8 Da, which agrees well with the molecular weight measured by gel-filtration chromatography (Fig. 1 ▶ b).
Figure 1.
(a) Purification of Drep2 CIDE and Drep3 CIDE by His-tag affinity chromatography. (b) Purification of the Drep2 CIDE–Drep3 CIDE complex by gel-filtration chromatography: Superdex S-200 gel-filtration column analysis of Drep2 CIDE (dotted/dashed line), Drep3 CIDE (dotted line) and the Drep2 CIDE–Drep3 CIDE complex (unbroken line). An SDS–PAGE of fractions from the peak corresponding to the Drep2 CIDE–Drep3 CIDE complex is shown. (c) Formation of the Drep2 CIDE–Drep3 CIDE complex. Separately purified Drep2 CIDE and Drep3 CIDE were mixed for 60 min at room temperature, after which the mixture was subjected to native PAGE. The arrow indicates the newly appearing complex band.
Diffracting crystals of the complex were successfully obtained as a result of making various different construct lengths. Many different Drep2 CIDE and Drep3 CIDE constructs were attempted, most of which led to crystals that did not diffract. The best pair of Drep2 CIDE and Drep3 CIDE constructs reported here produced a long rectangular-shaped crystal that diffracted to a resolution of 5.8 Å (Fig. 2 ▶). The native crystal belonged to space group P41, with unit-cell parameters a = b = 109.0, c = 46.2 Å. Assuming the presence of two heterodimers in the crystallographic asymmetric unit, the Matthews coefficient (V M) was calculated to be 3.43 Å3 Da−1, which corresponds to a solvent content of 64.13% (Matthews, 1968 ▶). Such a high solvent content might explain the poor diffraction of the complex crystal. The diffraction data statistics are given in Table 1 ▶. The data set was indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Phasing was performed using the molecular-replacement method with the program CNS (Brünger et al., 1998 ▶) and a CIDE domain of human CIDE-B (PDB code 1d4b; Lugovskoy et al., 1999 ▶), which has 29% and 31% amino-acid sequence homology to Drep2 CIDE and Drep3 CIDE, respectively, as a search model.
Figure 2.
Crystal of the Drep2 CIDE–Drep3 CIDE complex. The crystal grew in 3 d in the presence of 5% PEG 8000, 0.2 M lithium chloride, 35% glycerol and 0.1 M Tris pH 8.0. The approximate dimensions of the crystal were 0.1 × 0.1 × 0.3 mm.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0020962). We are grateful to Dr Yeon Gil Kim of BL-4A at the Pohang Accelerator Laboratory for help during data collection.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Dzivenu, O. K., Park, H. H. & Wu, H. (2004). Protein Expr. Purif. 38, 1–8. [DOI] [PubMed]
- Enari, M., Sakahira, H., Yokoyama, H., Okawa, K., Iwamatsu, A. & Nagata, S. (1998). Nature (London), 391, 43–50. [DOI] [PubMed]
- Inohara, N. & Nuñez, G. (1999). Cell Death Differ. 6, 823–824. [DOI] [PubMed]
- Jang, T.-H., Bae, J. Y., Park, O. K., Kim, J. H., Cho, K.-H., Jeon, J.-H. & Park, H. H. (2010). Biochim. Biophys. Acta, 1804, 1557–1563. [DOI] [PubMed]
- Li, L. Y., Luo, X. & Wang, X. (2001). Nature (London), 412, 95–99. [DOI] [PubMed]
- Liu, X., Zou, H., Slaughter, C. & Wang, X. (1997). Cell, 89, 175–184. [DOI] [PubMed]
- Lugovskoy, A. A., Zhou, P., Chou, J. J., McCarty, J. S., Li, P. & Wagner, G. (1999). Cell, 99, 747–755. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Nagata, S. (2000). Exp. Cell Res. 256, 12–18. [DOI] [PubMed]
- Otomo, T., Sakahira, H., Uegaki, K., Nagata, S. & Yamazaki, T. (2000). Nature Struct. Biol. 7, 658–662. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Park, H. H. (2009). BMB Rep. 42, 713–718. [DOI] [PubMed]
- Park, H. H., Lo, Y.-C., Lin, S.-C., Wang, L., Yang, J. K. & Wu, H. (2007). Annu. Rev. Immunol. 25, 561–586. [DOI] [PMC free article] [PubMed]
- Parrish, J., Li, L., Klotz, K., Ledwich, D., Wang, X. & Xue, D. (2001). Nature (London), 412, 90–94. [DOI] [PubMed]
- Sakahira, H., Enari, M. & Nagata, S. (1999). J. Biol. Chem. 274, 15740–15744. [DOI] [PubMed]
- Susin, S. A. et al. (1999). Nature (London), 397, 441–446.
- Wu, C., Zhang, Y., Sun, Z. & Li, P. (2008). BMC Evol. Biol. 8, 159. [DOI] [PMC free article] [PubMed]