5-(Hydroxyethyl)-4-methylthiazole kinase from S. aureus, which is essential to vitamin B1 metabolism, has been crystallized in space group P1. The crystals diffracted to 2.1 Å resolution.
Keywords: ThiM, Staphylococcus aureus, 5-(hydroxyethyl)-4-methylthiazole kinase, vitamin B1 metabolism
Abstract
ThiM [5-(hydroxyethyl)-4-methylthiazole kinase; EC 2.7.1.50] from Staphylococcus aureus is an essential enzyme of thiamine or vitamin B1 metabolism and has been crystallized by the vapour-diffusion method. The crystals belonged to the primitive space group P1, with unit-cell parameters a = 62.06, b = 62.40, c = 107.82 Å, α = 92.25, β = 91.37, γ = 101.48° and six protomers in the unit cell, corresponding to a packing parameter V M of 2.3 Å3 Da−1. Diffraction data were collected to 2.1 Å resolution using synchrotron radiation. The phase problem was solved by molecular replacement.
1. Introduction
Staphylococcus aureus is a commensally existing bacterium that colonizes 20% of healthy adults permanently and up to 50% transiently. Its pathogenicity plays an important role in nosocomial infections affecting immunosuppressed patients. Symptoms caused by S. aureus range from superficial skin lesions to life-threatening pneumonia or endocarditis (Lowy, 1998 ▶). In 2005, S. aureus re-emerged as a major human pathogen owing to methicillin-resistant S. aureus (MRSA) strains and caused more than 18 000 deaths in the USA. Staphylococcal pneumonia contributed to more than 75% of these deaths (Klevens et al., 2007 ▶; Lowy, 1998 ▶).
The active form of vitamin B1 is thiamine pyrophosphate (TPP), which is a cofactor for several key enzymes of carbohydrate and amino-acid metabolism such as the pyruvate dehydrogenase complex, the 2-oxoglutarate dehydrogenase complex and transketolase (Pohl et al., 2004 ▶; Begley et al., 1999 ▶). A lack of vitamin B1 can result in Wernicke’s disease and the disease known as beriberi (Ogershok et al., 2002 ▶; Platt & Lu, 1936 ▶). Current antibiotics for the treatment of S. aureus infections mainly target cell-wall synthesis or interfere with protein synthesis at a transcriptional or translational level (Ruhe et al., 2005 ▶; Apodaca & Rakita, 2003 ▶; Nguyen & Graber, 2010 ▶). The occurrence of multidrug resistance in bacterial pathogens such as S. aureus necessitates novel chemotherapeutic interventions. Ideal drugs target metabolic pathways that are absent in the host organism such as vitamin B1 biosynthesis. Recently, the vitamin B1 metabolism of S. aureus has been investigated biochemically (Müller et al., 2009 ▶), but structural information about the S. aureus enzymes involved in vitamin B1 metabolism is so far absent. Such information is most useful for structure-based development of prodrugs. When introduced into the bacterial thiamine metabolism they lead to a toxic TPP derivative which will poison vitamin B1-dependent enzymes and their host, the pathogen.
The synthesis of vitamin B1 includes two branches leading to a thiazole (THZ) moiety and a pyrimidine (HMP) moiety. THZ has to be phosphorylated by the THZ kinase ThiM (Jurgenson et al., 2009 ▶). This pathway is conserved in all kingdoms, whereas only lower eukaryotes and plants are able to synthesize thiamine de novo. Pyrococcus horikoshii and Bacillus subtilis express a ThiM analogue called ThiK to phoshorylate THZ. Both proteins show a trimeric assembly in the crystal structure (Zhang et al., 1997 ▶; PDB entry 3dzv; Joint Center for Structural Genomics, unpublished work). In yeast, ThiM and the thiamine phosphate synthase ThiE reside on the bifunctional protein Thi4-p (Nosaka et al., 1993 ▶).
Here, we report the crystallization and data collection of the enzyme SaThiM, which is one of five enzymes required for vitamin B1 biosynthesis in S. aureus.
2. Materials and methods
2.1. Cloning, expression and purification
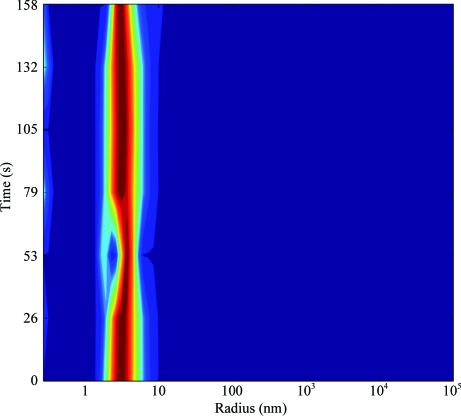
The open reading frame encoding SaThiM was amplified by PCR from S. aureus genomic DNA using sequence-specific antisense (5′-GCGCGCGGTCTCAGCGCTTTAATGATGATGATGATGATGGCCCTGAAAATAAAGATTCTCTTCCACCTCTTGAATGCGAATCCG-3′) and sense (5′-GCGCGCGGTCTCGAATGAATTATCTAAATAACATACGTATTG-3′) oligonucleotides. The PCR for the construct was performed using Taq polymerase (Invitrogen, USA) and the following PCR program: denaturation for 5 min at 368 K followed by 30 cycles of 45 s at 368 K, 60 s at 321 K and 60 s at 345 K. The generated PCR product was cloned via BsaI restriction sites into the Escherichia coli expression vector pASK-IBA3 previously digested with the same enzyme, resulting in the expression construct SaThiM-IBA3. The C-terminus of the native protein is supplemented with a TEV protease cleavage site and a 6×His tag (ENLYFQGHHHHHH) that allows purification of the recombinant fusion protein using Ni–NTA agarose (Qiagen, Germany). The final construct consists of 276 amino acids and has a molecular weight of 29 744 Da. The nucleotide sequence was verified by automated sequencing (Seqlab, Germany). Nucleotide and amino-acid analyses were performed with the Gene Runner software (Hastings Software Inc.). E. coli BLR (DE3) (Stratagene, Germany) was transformed with the SaThiM-IBA3 construct. Single colonies were picked and grown overnight in Luria–Bertani medium. The bacterial culture was diluted 1:100 and grown at 310 K until the A 600 reached 0.5. Expression was initiated with 200 ng ml−1 anhydrotetracycline and the cells were grown for 4 h at 310 K before being harvested. The cell pellet was resuspended in buffer A (100 mM Tris buffer pH 8, 150 mM NaCl) with 10 mM imidazole, sonicated and centrifuged at 75 000g for 1 h at 277 K. The soluble fraction of the lysate was applied onto an Ni–NTA column previously incubated in buffer A with 20 mM imidazole. The bound sample was washed extensively in the same buffer, eluted with buffer A with 250 mM imidazole and subsequently dialyzed overnight against buffer A. The purity of the sample was also assessed on SDS–PAGE (Laemmli, 1970 ▶; Fig. 1 ▶). Samples were centrifuged for 1 h at 277 K and 50 000g. The supernatant was then concentrated to 20 mg ml−1. The affinity tag was not removed. The solution was monitored by dynamic light scattering (DLS) with a Spectroscatter 201 (Molecular Dimensions, UK) over a suitable period of time and a stable particle radius of approximately 3.2 nm (Fig. 2 ▶) was observed.
Figure 1.
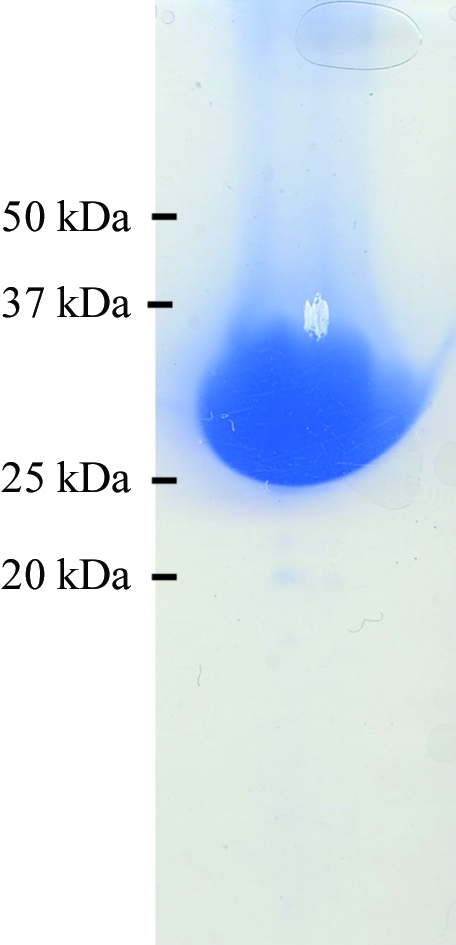
SDS–PAGE analysis of SaThiM after elution from the Ni–NTA column. The fraction was analysed on a 12.5% SDS gel and stained with Coomassie Blue. A single band with an approximate molecular weight of 30 kDa was observed.
Figure 2.
DLS measurement of a 20 mg ml−1 SaThiM solution, showing a monodisperse protein solution and a hydrodynamic radius (R h) of approximately 3.2 nm.
2.2. Crystallization of SaThiM
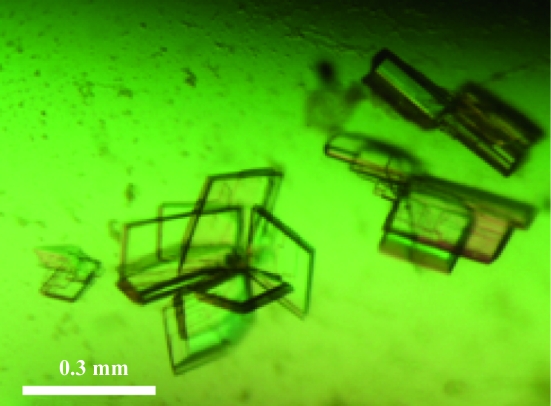
A total of 384 crystallization conditions were screened using a Honeybee 961 dispensing robot (Zinsser Analytic GmbH, Frankfurt, Germany) at 293 K in 96-well crystallization plates (NeXtal QIA1 µplates, Qiagen) using the sitting-drop vapour-diffusion method, based on the commercially available JCSG+, ComPAS, Classics and Cryos Suites (NeXtal, Qiagen). A 300 nl droplet of 20 mg ml−1 protein solution in buffer A was mixed with the same volume of reservoir solution and equilibrated against 35 µl reservoir solution. Crystals appeared after 4 d in well A5 from the JCSG+ Suite, consisting of 0.2 M magnesium formate, 20% polyethylene glycol (PEG) 3350 in the reservoir. This condition was further optimized and lamella-shaped crystals were reproducibly obtained using 0.2 M magnesium formate, 18% PEG 3350 and 5% 2-propanol in 24-well Linbro plates (ICN Biomedicals, USA) sealed with 20 × 20 mm siliconized cover slips (Marienfeld, Germany) applying the hanging-drop technique (Fig. 3 ▶). A 1 µl droplet of 20 mg ml−1 protein solution in buffer A was mixed with the same volume of reservoir solution and equilibrated against 1 ml reservoir solution at 293 K. Crystals grew to maximum dimensions of approximately 0.3 × 0.2 × 0.02 mm after 5 d (Fig. 3 ▶). The crystals were separated with microtools (Hampton Research, USA) prior to data collection and were harvested in nylon loops.
Figure 3.
SaThiM crystals grew to maximum dimensions after 5 d. A single lamella (0.3 × 0.2 × 0.02 mm) was prepared by separating the bundle of crystals with microtools prior to data collection.
2.3. Diffraction experiment
Diffraction data were collected to a resolution of 2.1 Å from a flash-frozen crystal at 100 K on the consortium’s fixed-wavelength beamline X13 (HASYLAB/DESY) in Hamburg, Germany at a wavelength of 0.8081 Å using a MAR CCD detector system. Addition of cryoprotectant to the crystal was not required since the PEG concentration in the crystallization solution was sufficient to protect the crystal from cryogenic damage. The oscillation angle was 0.5° and the exposure time was 45 s per frame. Initial crystal characterization and space-group assignment were performed using the DENZO software (Otwinowski, 1993 ▶) and scaling was performed using SCALEPACK (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
SaThiM was cloned with an affinity tag and expressed in E. coli BLR (DE3) cells. The protein consists of 276 amino acids with a molecular weight of 29 744 Da as calculated from the amino-acid sequence. The purified protein showed a single band of 30 kDa on SDS–PAGE and the crystals grew to dimensions of 0.3 × 0.2 × 0.02 mm after 5 d (Fig. 3 ▶). The crystals belonged to the triclinic space group P1, with unit-cell parameters a = 62.06, b = 62.40, c = 107.82 Å, α = 92.25, β = 91.37, γ = 101.48°. A Matthews coefficient of 2.3 Å3 Da−1 and a corresponding solvent content of 46.3% were calculated assuming the presence of six molecules in the unit cell. A native data set consisting of 84 534 unique reflections with a completeness of 98.7% was collected in the resolution range 50.0–2.15 Å (Table 1 ▶).
Table 1. Summary of data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Data-collection parameters | |
| Wavelength (Å) | 0.8081 |
| Temperature (K) | 100 |
| Oscillation range (°) | 0.5 |
| Crystal-to-detector distance (mm) | 195.02 |
| Data-integration statistics | |
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 62.06, b = 62.40, c = 107.82, α = 92.25, β = 91.37, γ = 101.48 |
| Resolution limits (Å) | 50–2.15 |
| Total No. of reflections | 1271580 |
| No. of unique reflections | 84534 |
| Multiplicity | 3.6 (3.4) |
| Completeness (%) | 98.7 (97.8) |
| Rmerge† | 0.076 (0.353) |
| Mean I/σ(I) | 13.4 (2.8) |
| Molecules in the unit cell | 6 |
| VM (Å3 Da−1) | 2.3 |
| Solvent content (%) | 46.3 |
R
merge is defined as 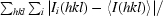
 , where Ii(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is the average intensity from multiple observations.
, where Ii(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is the average intensity from multiple observations.
The structure of ThiM from B. subtilis (BsThiK; PDB entry 1c3q; Campobasso et al., 2000 ▶) was used as a search model for initial molecular-replacement calculations. The sequence identity between the two proteins is 39%. A protomer of BsThiK was successfully placed into the unit cell six times using the program Phaser (McCoy et al., 2005 ▶), with a corresponding final Z score of 15.5 and a log-likelihood gain value of 849.7. The six protomers appear to assemble into two trimers in the unit cell. Model building and refinement are in progress.
Acknowledgments
This project was supported by the Hamburg Ministry of Science and Research and Joachim Herz Stiftung as part of the Hamburg Initiative for Excellence in Research (LEXI) and the Hamburg School for Structure and Dynamics (SDI) is gratefully acknowledged.
References
- Apodaca, A. A. & Rakita, R. M. (2003). N. Engl. J. Med. 348, 86–87. [DOI] [PubMed]
- Begley, T. P., Downs, D. M., Ealick, S. E., McLafferty, F. W., Van Loon, A. P., Taylor, S., Campobasso, N., Chiu, H.-J., Kinsland, C., Reddick, J. J. & Xi, J. (1999). Arch. Microbiol. 171, 293–300. [DOI] [PubMed]
- Campobasso, N., Mathews, I. I., Begley, T. P. & Ealick, S. E. (2000). Biochemistry, 39, 7868–7877. [DOI] [PubMed]
- Jurgenson, C. T., Begley, T. P. & Ealick, S. E. (2009). Annu. Rev. Biochem. 78, 569–603. [DOI] [PMC free article] [PubMed]
- Klevens, R. M. et al. (2007). JAMA, 298, 1763–1771. [DOI] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Lowy, F. D. (1998). N. Engl. J. Med. 339, 520–532. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Müller, I. B., Bergmann, B., Groves, M. R., Couto, I., Amaral, L., Begley, T. P., Walter, R. D. & Wrenger, C. (2009). PLoS One, 4, e7656. [DOI] [PMC free article] [PubMed]
- Nguyen, H. M. & Graber, C. J. (2010). J. Antimicrob. Chemother. 65, 24–36. [DOI] [PubMed]
- Nosaka, K., Kaneko, Y., Nishimura, H. & Iwashima, A. (1993). J. Biol. Chem. 268, 17440–17447. [PubMed]
- Ogershok, P. R., Rahman, A., Nestor, S. & Brick, J. (2002). Am. J. Med. Sci. 323, 107–111. [DOI] [PubMed]
- Otwinowski, Z. (1993). Proceedings of the CCP4 Study Weekend. Data Collection and Processing, edited by L. Sawyer, N. Isaacs & S. Bailey, pp. 56–62. Warrington: Daresbury Laboratory.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Platt, B. & Lu, G. (1936). Q. J. Med. 5, 355–374.
- Pohl, M., Sprenger, G. A. & Müller, M. (2004). Curr. Opin. Biotechnol. 15, 335–342. [DOI] [PubMed]
- Ruhe, J. J., Monson, T., Bradsher, R. W. & Menon, A. (2005). Clin. Infect. Dis. 40, 1429–1434. [DOI] [PubMed]
- Zhang, Y., Taylor, S. V., Chiu, H.-J. & Begley, T. P. (1997). J. Bacteriol. 179, 3030–3035. [DOI] [PMC free article] [PubMed]