Abstract
Posttranslational histone modifications are a critical regulatory mechanism of gene transcription. Previous studies from our laboratory have shown that contingent on binding to its cognate promoter motifs in the Cyp1a1 gene, activation of the aryl hydrocarbon receptor (AHR) by benzo[a]pyrene (BaP) treatment induces histone modifications in the Cyp1a1 promoter that are required for activation of gene transcription. Here, we have studied different AHR ligands, including polychlorinated biphenyls (PCBs) of different toxic equivalency factors (TEF), to determine whether changes in histone modifications are linked to different levels of Cyp1a1 expression or dependent on AHR-ligand affinity. We find that all ligands lead to the same pattern of histone modifications in a relationship that parallels the strength of their AHR-ligand affinity. Thus, whereas PCB126 (TEF 0.1), 3-methylcholanthrene, β-naphthoflavone, and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) initiate a pattern of histone marks similar to those induced by BaP, PCB77 (TEF 0.0001) causes a lower level of change in the same marks and requires a longer activation time than PCB126, BaP, or TCDD. In contrast, the non–dioxin-like PCB153 recruits AHR to the Cyp1a1 enhancer causing a displacement of enhancer-associated histone H3 but does not cause the other observed histone mark changes nor does it induce transcription. These results indicate that AHR recruitment to the promoter is not sufficient to induce the histone modifications needed to activate gene expression and show that there is a good correlation between the regulatory chromatin changes associated with ligand-induced AHR target gene transcription and the resultant toxicity of the ligand.
Keywords: Ah receptor, dioxin-like compounds, PCBs, TEF, histone marks
Polychlorinated biphenyls (PCBs) are persistent, bio-accumulative environmental contaminants that were produced in the United States from the late 1920’s until the 1970’s. Due to their stability and thermal properties, PCBs were used as heat transfer and hydraulic fluids in electrical capacitors and transformers. It is estimated that more than 1.5 million metric tons of PCBs were made worldwide in the 1960’s, 650,000 metric tons in the United States alone (De Voogt and Brinkman, 1989). Estimates are that 20–30% of the PCBs manufactured entered into the environment whereas the rest remains in storage or are still in use in transformers and capacitors (Reijenders and Brasseur, 1992). Over a period of 30 years, General Electric disposed of an estimated 1.3 million pounds of PCBs in the Hudson River, with the current 6 year Phase-1 cleanup effort expected to remove only 10% of the contaminating PCBs. Contamination with PCBs has been identified in most every component of the global ecosystem, including fish, birds, and humans, in whom PCBs have been detected in adipose tissue, serum, and milk (Safe, 1995). PCBs can remain in the environment for decades and are taken up by various animals in the food chain, which constitutes the most common route of human exposure and accumulation (McLachlan, 1996; Schecter et al., 2001).
Because of their biochemical characteristics, some PCBs are classified as dioxin-like compounds (DLCs), a term that denotes that these chemicals have a similar mechanism of action and biological outcome as the most toxic prototypical congener, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; dioxin). DLCs are all hydrophobic, coplanar compounds with multiple sites of halogenation and van der Waals measurements of 12 × 14 × 5 Å (Denison et al., 2002; Gasiewicz et al., 1996). At low chronic doses, DLCs are teratogenic, immunotoxic, cause male reproductive damage, are linked to heart disease, and act as endocrine disruptors (Bertazzi et al., 2001; Bjerke and Peterson, 1994; Courtney and Moore, 1971; Kerkvliet et al., 1990; Mocarelli et al., 2008; Oughton et al., 1995; Steenland et al., 1997).
Although the in vivo effects in the literature are reasonably consistent, in vitro effects of DLC exposure are often contradictory. DLCs have been shown both to enhance and to inhibit rat hepatocyte proliferation and to be both proapoptotic and antiapoptotic (Moolgavkar et al., 1996; Wiebel et al., 1991; Worner and Schrenk, 1996). Despite their inhibitory effect on cell proliferation and DNA synthesis in rat hepatocytes, rat liver, and mouse epithelial cells, DLCs are known to be carcinogenic in animal models and in humans (Knutson and Poland, 1982; Onozuka et al., 2009; Poland et al., 1982). All the dioxin-like effects of the coplanar PCBs are the result of activation of the aryl hydrocarbon receptor (AHR), a member of the basic helix-loop-helix/PER-ARNT-SIM (bHLH/PAS) family of transcription factors. AHR resides in the cytoplasm of cells in complex with HSP90/XAP/p23 (Hankinson, 1995). Following ligand binding and nuclear translocation, the AHR dissociates from the complex and forms a heterodimer with the aryl hydrocarbon receptor nuclear translocator (ARNT) (Hoffman et al., 1991). This heterodimeric complex binds to cis-acting xenobiotic response elements (XRE; DRE, AHRE) containing the sequence 5′-TNGCGTG-3′ (Ikuta et al., 1998; Swanson et al., 1995). The activated complex recruits the basal transcription machinery and remodels chromatin to begin transcription of a battery of genes, which include several the Phase I drug metabolism enzymes as well as several Phase II drug metabolizing enzymes (Hankinson, 1995; Nebert et al., 2000; Schnekenburger et al., 2007a, 2007b; Whitlock, 1999).
The promoter of the mouse Cyp1a1 gene has a 5′ transcriptional regulatory region, which we term hereafter the “proximal promoter,” located directly upstream of the transcription start site. The proximal promoter has no AHRE motifs but has binding motifs for several other transcription factors, including TATA-binding proteins (Jones and Whitlock, 1990). Several hundred base pairs further upstream of the proximal promoter, there is a distal “enhancer” region where the AHREs are clustered (Neuhold et al., 1989; Yanagida et al., 1990). Work in our laboratory established that, after AHR-mediated activation by a ligand such as benzo[a]pyrene (BaP), recruitment of RNA polymerase II to the promoter region of the Cyp1a1 gene requires the AHR-dependent displacement of an HDAC1/DNMT1 complex associated with the proximal promoter. Displacement of the complex allows for the initiation of posttranslational histone modifications in the nucleosomes associated with the proximal promoter region and the distal enhancer region of the gene (Schnekenburger et al., 2007a, 2007b). AHR recruitment in response to BaP therefore leads to a patterned change in histone modifications that requires displacement of the HDAC1/DMNT1 complex to take place and is associated with active transcription of the Cyp1a1 gene (Schnekenburger et al., 2007a, 2007b). The pattern of histone modifications that corresponds to active transcription after BaP treatment includes changes both at the enhancer domain where the AHR binds, and at the promoter, where the basal transcription machinery binds (Schnekenburger et al., 2007a, 2007b). These modifications include a large increase of Ser-10 phosphorylation in histone H3 and of Lys-16 acetylation in histone H4 in the Cyp1a1 enhancer domain and an increase of Lys-4 trimethylation in histone H3, Lys-14 acetylation in histone H3 and a decrease of Lys-4 dimethylation in histone H3 in the proximal promoter region (Schnekenburger et al., 2007a, 2007b). This pattern of histone modifications is in line with what is known as the “histone-code” for activation of transcription (Hon et al., 2009; Jenuwien and Allis, 2001; Zippo et al., 2009).
The work summarized above did not address the question of whether changes in histone modification occurring at the Cyp1a1 promoter are dependent on the affinity of AHR for BaP or whether other AHR ligands, including the very high affinity halogenated ligand TCDD, would cause the same pattern of histone modifications. Because PCBs are major environmental contaminants that differ in their relative toxicities, we wished to examine how the relative toxicities and AHR affinities of different coplanar PCBs affect the previously demonstrated pattern of epigenetic changes associated with gene activation. We set out to determine if the histone mark changes upstream of AHR target genes, which we had previously demonstrated to be associated with activation of transcription, are similarly affected by the AHR ligands affinity for the receptor. To do so, we compared three PCBs with different toxic equivalency factors (TEFs) and one non–dioxin-like PCB, to dioxin. TEFs are order of magnitude estimates of a DLC’s relative toxicity to TCDD; they were derived by the World Health Organization after scientific review of all the available in vivo and in vitro literature for each DLC AHR-mediated effects, to be used in risk characterization and assessment (Van den Berg et al., 1998, 2006). Here, we show that PCB77, with a TEF of 10−4, causes a different patterns of histone modifications than PCB 169, with a TEF of 10−2, and PCB126, with a TEF of 10−1. Likely to be responsible for this difference, PCB77 causes a low level of AHR recruitment to the gene enhancer that lasts for a longer period of time than activation by TCDD, PCB126, or PCB169.
MATERIALS AND METHODS
Cell culture and treatments.
Mouse Hepatoma-1c1c7 (Hepa-1) cells from the American Type Culture Collection were cultured in α-minimum essential medium (Invitrogen; Carlsbad, CA) supplemented with 5% fetal bovine serum (Sigma) and 1% by volume antibiotic–antimycotic (Invitrogen) in a 5% CO2-humidified atmosphere at 37°C. Cells were treated when they reached 70–85% confluence with 5μM BaP, 5nM TCDD, 5μM 3-methylcholanthrene (3MC), 5μM β-naphthoflavone (BNF). In experiments where cells were treated with 3,3′,4,4′,5-pentachlorobiphenyl (PCB126), 3,3′4,4′-tetrachlorobiphenyl (PCB77), 3,3′4,4′,5,5′-hexachlorobiphenyl (PCB169), or 2, 2′,4,4′,5,5′-hexachlorobiphenyl (PCB153) these compounds were dissolved in dimethyl sulfoxide (DMSO) and were added to the medium in a volume not to exceed 0.1% of the total culture medium volume. The same volume of DMSO was used to treat vehicle controls.
RNA extraction and cDNA synthesis.
Hepa-1 cells were treated as described for 8 h. After treatment, cells were lysed using Trizol reagent (Sigma-Aldrich; St Louis, MO) and RNA extracted using chloroform. RNA was precipitated by using 80% ethanol and resuspended in water. cDNA was synthesized by reverse transcription of 5 μg of total RNA with SuperScript II RNase H- reverse transcriptase (Invitrogen). An aliquot of cDNA was used as a template for quantitative real-time PCR (QRT-PCR). QRT-PCR was performed using primers for a 199 bp fragment of the mouse Cyp1a1 mRNA (Forward-5′-GTGTCTGGTTACTTTGACAAGTGG-3′; reverse-5′-AACATGGACATGCAAGGACA-3′) or a 287 bp fragment of the β-actin mRNA (forward, 5′-CATCCGTAAAGACCTCTATGCC-3′; reverse, 5′-ACGCAGCTCAGTAACAGTCC-3′). All assays were done in duplicate and repeated at least twice.
Chromatin immunoprecipitation.
Hepa-1 cells were grown and treated as described for 90 min, unless otherwise noted. Following treatment, cells were incubated for 10 min at room temperature in 1% formaldehyde. The cross-linking reaction was quenched with 0.125M glycine for 10 min at room temperature, and the cells were rinsed three times with cold 1× PBS. Cells were scraped from the plate, pelleted by centrifugation, resuspended in cold cell lysis buffer (5mM PIPES [pH 8.0], 85mM KCl, 0.5% NP-40, and 1× protease inhibitor cocktail) (Roche; Indianapolis, IN) and incubated on ice for 10 min. Nuclei were pelleted, resuspended in nuclei lysis buffer (50mM Tris [pH 8.1], 10mM EDTA, 1% SDS, and 1× protease inhibitor cocktail) and incubated on ice for 10 min. Chromatin was sheared to a range of 0.3–0.6 kb by sonication in a crushed ice–water bath with six 30-s bursts of 200 W with 30-s intervals between bursts using a Bioruptor (Diagenode, Danville, NJ). Cell debris was removed by centrifugation. Chromatin was precleared with a 50% slurry of protein-A agarose beads. Precleared lysates were diluted three times in dilution buffer (16.7mM Tris [pH 8.1], 167mM NaCl, 1.2mM EDTA, 1.1% Triton-x 100, 0.01% SDS). A 10% aliquot of the precleared lysates was used for input, 45% of the diluted lysate was incubated overnight at 4°C on a rotating platform with the specific antibody of interest, and the remaining 45% of the diluted lysate was similarly incubated with the appropriate nonspecific IgG. Antibodies used for chromatin immunoprecipitation (ChIP) were directed against AHR (Bio-Mol; Sa-210), AcK16-H4 (Upstate; 07-329), pS10-H3 (Millipore; Danvers, MA; 05-817), AcK14-H3 (Millipore; 07-353), 2MeK4-H3 (Abcam; Cambridge, MA; ab7766), 3MeK4-H3 (Abcam; ab8580), or Histone 3 (Abcam; ab1791). Immune complexes were recovered by 2-h incubation on a rotating platform at 4°C with a 50% slurry of protein A-agarose or protein G-agarose beads, depending on antibody specificity. Beads were pelleted by centrifugation and washed twice with dialysis buffer (50mM Tris-HCl [pH 8.0], 2mM EDTA, 0.2% Sarkosyl) and four times with IP wash buffer (100mM Tris-HCl [pH 9.0], 500mM LiCl, 1% NP-40, 1% deoxycholic acid). Immune complexes were eluted from the beads by incubation with elution buffer (50mM NaHCO3, 1% sodium dodecyl sulfate) while mildly vortexing. Elution was repeated and eluates combined. Cross-linking was reversed by increasing the NaCl concentration to 0.3M and incubating overnight at 65°C with RNase A. Proteins were digested with proteinase K at 45°C for 90 min. DNA was purified using QIAquick affinity chromatography columns (Qiagen) and eluted in distilled water. Primers specific for either the AHRE-cluster containing distal enhancer (forward: 5′-AGGCTCTTCTCACGCAACTC-3′; reverse: 5′-TAAGCCTGCTCCATCCTCTG-3′) or proximal promoter (forward: 5′-TATCCGGTATGGCTTCTTGC-3′; reverse: 5′-CACCTTCAGGGTTAGGGTGA-3′) region of the mouse Cyp1a1 gene were used for QRT-PCR. Duplicate ChIPs were performed at least twice, and antibodies from the same lot were used for the replicates.
QRT-PCR analysis.
QRT-PCR was performed at least in duplicate in a reaction mix containing 1× SYBR green PCR master mix (Applied Biosystems; Carlsbad, CA) and 0.1μM of each primer. Samples were heated to 95°C followed by 35 cycles of a denaturing step at 95°C for 15 s and an annealing/elongation step of 60°C for 60 s using an ABI 7500 real-time PCR system (Applied Biosystems). Single product formation was confirmed by melting curve analysis after amplification. Analysis of results was performed using sequence detection software (SDS software version 1.3.1; Applied Biosystems).
Data analyses.
The ΔCT for each sample was determined using the mean cycle threshold (CT) of replicates from the input DNA, to normalize ChIP assay results, or the β-actin signal, to normalize gene expression assays. ΔΔCT values were determined by subtracting the ΔCT of control from the corresponding experimental ΔCT. The resulting values were converted to fold changes over control by raising 2 to the power of −ΔΔCT.
RESULTS
Different AHR Ligands Induce Similar Histone Modifications
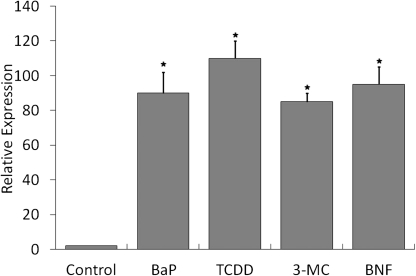
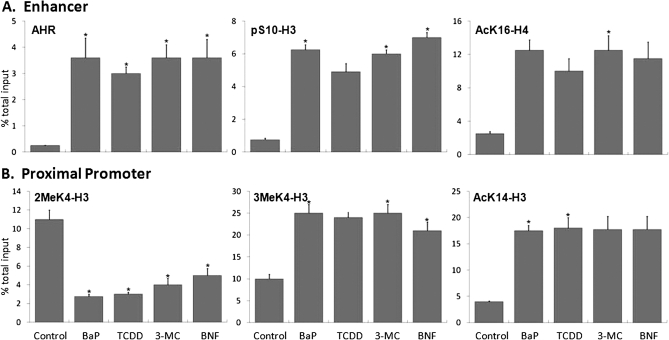
To determine if AHR activation by other AHR ligands induce the same histone marks as those found for BaP, Hepa-1 cells were treated with concentrations of 3MC, BNF, TCDD, and BaP known to induce Cyp1a1 expression maximally. All ligands substantially induced Cyp1a1 mRNA expression, measured at maximal induction time of 8 h after treatment (Fig. 1). Histone marks were analyzed after a 90-min treatment, the maximal induction time previously determined for AHR (Schnekenburger et al., 2007a, 2007b). At this time point, concomitant with more than 10-times the increase in AHR recruitment to the enhancer region of Cyp1a1, there was a 3-fold increase of AcK16-H4 and greater than 6-fold increase of pS10-H3 at the enhancer in response to all ligands relative to DMSO (Fig. 2A). At the same time, all ligands increased the levels of AcK14-H3 associated with the Cyp1a1 proximal promoter by at least 4-fold over DMSO and each more than doubled the amount of 3MeK4-H3 and decreased by at least half the amount of 2MeK4-H3 associated with the Cyp1a1 promoter, compared with DMSO-treated controls (Fig. 2B). We conclude that all four ligands activated and completed AHR translocation and binding to its cognate sites in the Cyp1a1 enhancer equally well, initiating a common wave of changes in histone marks that resulted in the increase in Cyp1a1 transcription.
FIG. 1.
Activation of target gene transcription by different AHR ligands. Hepa-1 cells were treated for 8 h with 5μM BaP, 5nM TCDD, 5μM 3MC, 5μM BNF, or vehicle. RNA was isolated, and duplicate QRT-PCR was performed to determine the amount of Cyp1a1 mRNA relative to β-actin. Error bars represent SE. The * indicates statistical significance when compared with DMSO (p value < 0.05).
FIG. 2.
Different AHR ligands elicit similar patterns of histone modifications concomitant with activation of Cyp1a1 transcription. Hepa-1 cells were treated for 90 min with 5μM BaP, 5nM TCDD, 5μM 3MC, 5μM BNF, or vehicle. After treatment, antibodies against AHR or specific histone modifications were used for ChIP analyses. The amount of AHR or specifically modified histone associated with specific regions upstream of the Cyp1a1 gene promoter was determined by QRT-PCR targeted for either the enhancer (A) or the promoter (B) region. The ordinate represents the percent of the total input immunoprecipitated by each specific antibody. Error bars represent SEs. The * indicates statistical significance when compared with DMSO (p value < 0.05).
Ligand Toxicity Correlates with the Pattern of Histone Modifications
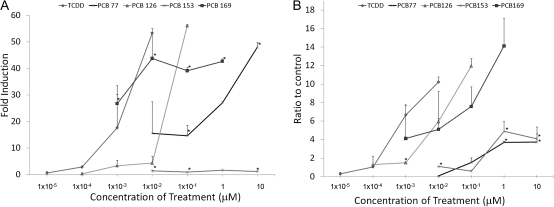
Next, we wished to determine whether the quality or quantity of the histone marks elicited by an AHR ligand would correlated to the toxicity of that particular ligand. For this purpose, we chose to use AHR ligands with different TEFs and determine whether they would cause patterns of histone modifications similar to those induced by TCDD or BaP. The ligands studied were the coplanar DLCs PCB126, PCB169, and PCB77 and the noncoplanar, non-DLC PCB153, with TEFs of 10−1, 10−2, 10−4 and 0, respectively (Van den Berg et al., 2006). Hepa-1 cells were treated with identical toxic equivalent (TEQ) levels of all three DLCs, TCDD, PCB126, and PCB77, and a concentration-response curve was determined using four concentrations of each compound with equivalent TEQ. The fold induction of Cyp1a1 expression induced by PCB126 did significantly differ from that of an equivalent TEQ of TCDD across all the tested concentrations (Fig. 3A), confirming the equivalency of the chosen concentrations. PCB77 induced greater than 10-fold higher Cyp1a1 expression than a similarly toxic level of TCDD at the lower concentration but at the highest concentration TCDD induced expression was 7-fold higher than PCB77 (Fig. 3A). PCB169 induces high levels of Cyp1a1 expression even at low concentrations (Fig. 3A). As expected from a noncoplanar PCB, the non-DLC PCB153 caused no change in Cyp1a1 mRNA expression across all tested concentrations (Fig. 3A). On the other hand, ChIP assays on Hepa-1 cells treated with PCB169, PCB126, and PCB77 showed significant differences in their effectiveness to promote AHR recruitment to the Cyp1a1 enhancer. PCB126 recruited nearly 20% more AHR than TCDD, whereas PCB77 recruited 60% less (Fig. 3B). PCB169-induced AHR recruitment was higher than TCDD when compared with a similar TEQ at any concentration (Fig. 3B).
FIG. 3.
Different DLCs induce Cyp1a1 expression in proportion to their TEF. (A) Hepa-1 cells were treated for 8 h with the indicated concentrations. After treatment, RNA was extracted and used for QRT-PCR determination of the amount of Cyp1a1 mRNA relative to β-actin. (B) Hepa-1 cells were treated with the indicated concentrations of TCDD, DMSO, or PCBs for 90 min. Cells were then fixed, and ChIP was performed for AHR. Error bars represent SE. An * indicates statistical significance when compared with the similar TEQ of TCDD (p value < 0.05).
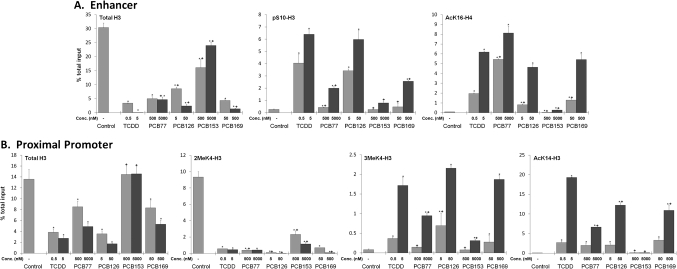
In order to determine which histone modifications occurred in response to the dioxin-like PCBs, we treated Hepa-1 cells with two concentrations of each compound differing by one order of magnitude. We treated cells with TCDD at 0.5nM and 5nM or the corresponding toxic equivalent concentrations of PCB77 (500 and 5000nM), PCB126 (5 and 50nM), or PCB169 (50 and 500nM). We also treated cells with the non–dioxin-like PCB153 at concentrations equivalent to that of PCB77 (500 and 5000nM), the dioxin-like PCB with the lowest TEF that we used. All PCB treatments lowered the total amount of histone H3 associated with the distal enhancer to some extent, though a large decrease was only seen when cells were treated with the dioxin-like PCBs (Fig. 4A). When treated with TCDD or the PCB with the next highest TEF, PCB126, the amount of enhancer-associated phosphorylation of Ser10 in histone 3 increased more than 12-fold over control at the lowest tested concentrations and more than 20-fold when treated with the higher toxic equivalent concentrations (Fig. 4A). PCB77 and PCB169 caused double the amount of pSer10-H3 to be associated with enhancer after treatment with the lower tested concentration, and when cells were treated with higher concentrations of PCB77 and PCB126, the amount of pSer10-H3 was nearly an order of magnitude higher than control (Fig. 4A). As far as the acetylation of Lys16 in histone H4, another mark previously demonstrated to be associated with AHR-dependent Cyp1a1 induction, PCB126, PCB169, and TCDD caused similar increases at both tested toxic equivalent concentrations, whereas the lower concentration of PCB77 treatment led to more than double the amount of acetylation at this position in the Cyp1a1 enhancer than a similar treatment with TCDD (Fig. 4A). The higher dose of PCB77 lead to an increase of acetylated histone 4 that was also higher than that of cells treated with a similarly toxic concentration of TCDD (Fig. 4A). Interestingly, the non-DLC, noncoplanar PCB153 led to an increase in AHR binding and presumably activation of just greater than 50% that of TCDD, even greater than the level of binding induced by PCB77 (Fig. 4A). This PCB153-induced increase in AHR recruitment was dependent on concentration and caused the displacement of at least a small amount of the histone H3 associated with the enhancer region (Fig. 4A). Despite its apparent effect on AHR binding to its cognate sites and displacement of enhancer-associated monosomes, PCB153 treatment had a minimal effect on phosphorylation of S10-H3 or acetylation of K16-H4 in the enhancer region of Cyp1a1, consistent with its inability to induce Cyp1a1 gene expression (Fig. 4A).
FIG. 4.
Different DLCs induce different levels of histone marks depending on their affinity for AHR. Hepa-1 cells were treated with 0.5 or 5nM TCDD, 5 or 50nM PCB126, 50 or 500nM PCB169, 500 or 5000nM PCB77, 500 or 5000nM PCB-153, or vehicle for 90 min. ChIP analyses were performed using antibodies against AHR, histone H3, or specific modified histones. QRT-PCR using specific primers for either the enhancer region (A) or the promoter region (B) of the Cyp1a1 gene was performed to determine the amount of chromatin target associated with each region. The ordinate represents the percent of the total input immunoprecipitated by each specific antibody. Error bars represent SE. An * indicates statistical significance when compared with control (p value < 0.05). A + symbol above a bar indicates statistical significance when compared with the equivalent TEQ of TCDD (p value < 0.05).
In the proximal promoter domain of the Cyp1a1 gene, PCB126, PCB169, and PCB77 decreased the levels of 2MeK4-H3 associated with the promoter by an amount equivalent to the decrease caused in response to TCDD treatment (Fig. 4B). It should be noted that this decrease in 2MeK4-H3 is concomitant with a loss of promoter-associated histone 3 (Fig. 4B). The non–dioxin-like PCB153 also decreased the amount of dimethylated Lys-4 in H3 associated with the promoter proximal sequences but did so by an amount slightly less than that seen after the treatment with the dioxin-like PCBs, and this decrease was seen in the absence of a loss of promoter associated total histone H3 (Fig. 4B). A concomitant increase in trimethylation of lysine 4 of histone H3 was only detected after PCB77, PCB126, PCB 169, and TCDD treatments (Fig. 4B). PCB77 caused an increase in trimethylation of Lys-4 in H3 that was nearly half that caused by TCDD, whereas PCB153 did not cause an increase in 3MeK4-H3 levels in comparison to DMSO at the lower tested concentration and increased 3MeK4-H3 only (Fig. 4B). Acetylation of Lys-14 in histone 3, another mark of gene induction by BaP-activated AHR, was evident in the Cyp1a1 proximal promoter domain of TCDD-treated cells and to a lesser extent in PCB126- and PCB169-treated cells (Fig. 4B). PCB77 induced increase in the acetylation of Lys-14 in histone associated with the promoter was less than half that of either PCB126 or PCB169 and nearly one-third that of TCDD-treated cells (Fig. 4B). PCB153, in agreement with its inability to activate transcription showed no increase in AcK14 of histone 3 associated with the Cyp1a1 promoter (Fig. 4B).
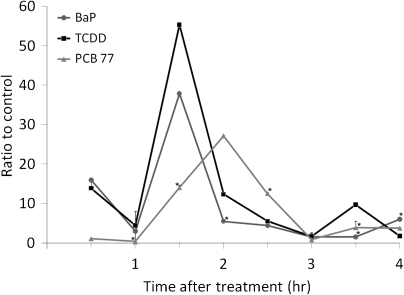
PCB77-Induced AHR Recruitment to the Cyp1a1 Enhancer Is Delayed Relative to TCDD or BaP
Our results indicated that PCB77 induces Cyp1a1 mRNA to an extent comparable to that of TCDD or BaP but recruits significantly lower AHR levels to its enhancer domain. One possible explanation for this apparent discrepancy is that there might be timing differences in AHR activation between TCDD and a PCB of such low TEF as PCB77. To determine if PCB77 initiated or sustained AHR recruitment to the Cyp1a1 enhancer in a manner substantially different from TCDD, BaP or the higher TEF DLCs, we treated Hepa-1 cells with 5000nM PCB77, 5nM TCDD, 5μM BaP, or vehicle and measured AHR recruitment to the Cyp1a1 enhancer at 30-min intervals for 4 h. TCDD and BaP showed an association kinetics consistent with prior determinations (Schnekenburger et al., 2007a, 2007b), with a sharp peak of maximum binding at 90 min. In contrast, the kinetics in PCB77-treated cells was slower and did not reach peak association until 2 h after initiation of treatment (Fig. 5). In addition, the extent of AHR promoter recruitment induced by PCB77 at peak value was only 50% of the recruitment induced by TCDD or BaP (Fig. 5). Whereas TCDD and BaP induced a similar sharp peak of AHR recruitment that returned to baseline levels within 30 min of peak value, PCB77 induced a broader, more prolonged activation status that did not return to baseline for at least 1 additional hour, twice the time that it took the cells treated with BaP or TCDD to return to baseline (Fig. 5). We conclude that the lower level of activation over a longer period of time resulting from PCB77 exposure leads to the same final expression level seen in cells treated with TCDD or BaP.
FIG. 5.
PCB 77 induces AHR enhancer recruitment at a lower level but over a longer period of time. Hepa-1 cells were treated with 5μM BaP, 5nM TCDD, 5000nM PCB77, or vehicle. At 30-min intervals, cells were fixed, and ChIP was performed using AHR-specific antibodies. QRT-PCR with specific primers was performed to determine the amount AHR recruited to the enhancer. PCR was done in duplicate, and the experiment was repeated twice. Each sample’s percent total input was set relative to vehicle control and plotted as a function of time. Error bars represent SEs. An * denotes statistical significance when compared with the TCDD at the same time point (p value < 0.05).
DISCUSSION
Previous work from our laboratory has established that a pattern of histone modification changes occurs upstream of the Cyp1a1 when mouse cells are treated with the AHR agonist BaP (Schnekenburger et al., 2007a, 2007b). This pattern is dependent on AHR recruitment to the distal enhancer region of the gene and displacement of an HDAC1/DMNT1 complex at the promoter (Schnekenburger et al., 2007a, 2007b). The results of the present study show that the pattern of histone modifications triggered by ligand-dependent AHR activation in the promoter of the Cyp1a1 gene is common to all AHR ligands studied so far, including the various DLCs tested in this work, TCDD, BaP, 3-MC, and βNF. It is reasonable to conclude that this pattern is a conserved signature of gene expression induction by the AHR, as previously hypothesized. Despite the fact that the different types of AHR agonists tested induce the same pattern of nucleosome modifications, treatment with PCB77, with a low TEF of 10−4, does not elicit these modifications to the same extent as the stronger AHR agonists, such as PCB126 with a TEF of 10−1 or TCDD (TEF 1), even after treatment with concentrations of equal TEQ, which induce gene transcription to similar levels. The main difference between high- and low-TEF ligands is the kinetics at which the modifications are established, with the low-TEF ligand being considerably slower than the high-TEF ligands. For example, the higher concentration PCB77 treatment induces a similar level of the transcription-associated histone H4 Lys-16 acetylation in the distal enhancer region as the other dioxin-like treatments, although PCB77 recruits much lower levels of AHR to that region (Figs. 3B and 4B). Also at the distal enhancer, PCB77 treatment leads to an increase of phosphorylation of S10 of histone 3 that is half that of either TCDD or PCB126 but similar in intensity as that of PCB169, which recruited the highest levels of AHR to the enhancer (Figs. 3B and 4A). Similar stark differences in proximal promoter region histone marks are also evident in response to PCB77 treatment. PCB77 showed a decrease in the 2MeK4-H3 mark accompanied by a decrease in total H3 associated with the proximal promoter in much the same way the other dioxin-like treatments did but caused an increase in 3MeK4-H3 and Ack14-H3 that was nearly half that induced by the high-TEF ligands (Fig. 4B). Our data suggest that part of this difference may be due to the kinetics of AHR-ligand binding and the related AHR activation, as PCB77 does not reach maximum AHR activation until after 120 min of treatment, compared with 90 min for both TCDD and BaP (Fig. 5). The histone mark modifications seen in response to PCB77 could be a snapshot of the early steps of chromatin modification that precede transcription, opening up new questions about the temporal–spatial kinetics of histone modification. It is likely that early AHR recruitment leads to histone modifications at the enhancer domain allowing for AHR to come into contact with the proximal promoter region, either by a sliding mechanism or by enabling chromatin looping (Steenland et al., 1997; Tian et al., 2003). This could facilitate later modification of histone marks at the proximal promoter ultimately leading to recruitment of the basal transcription machinery. Whether by a sliding or looping mechanism, this concept would agree with previous data showing that when the HDAC1/DMNT1 complex poised at the proximal promoter nucleosome was cross-linked to the promoter chromatin, AHR binding to the distal enhancer was retained, but its binding to the proximal promoter was blocked as were the changes in histone marks and the subsequent recruitment of RNA pol II associated with transcription (Schnekenburger et al., 2007a, 2007b). Similarly, suppression of the initial histone modifications at the distal enhancer would block the AHR from coming in contact with the proximal promoter and would inhibit the changes of histone marks that allow RNA pol II to be recruited. In this case, PCB77 and PCB126 (or TCDD) treatments would induce similar levels of Cyp1a1 expression albeit PCB77 would reach that level at a slower rate, which would explain the lower levels of histone mark modifications (Fig. 4) and RNA pol II recruitment (Supplementary fig. 1). If this hypothesis were correct, histone mark changes at the enhancer domain, such as acetylation of lysine 16 on histone 4, and loss of the 2MeK4-H3 mark might be early events occurring when levels of activated AHR are still low and function to facilitate subsequent changes. This would suggest that a cascade of changes must take place before Cyp1a1 transcription can proceed, requiring the recruitment of an appropriate amount of AHR to the distal enhancer and altering histone marks at the distal enhancer before changes can occur at the proximal promoter. For the FOSL gene enhancer, it has been shown that pS10-H3 is required to initiate acetylation of K16-H4, and that both are required for FOSL gene expression (Zippo et al., 2009). Therefore, a mechanism requiring first one set of histone mark changes to occur before a second set can be established would not be surprising. Further studies are needed to determine definitively the order in which these changes occur and which changes are necessary for the later changes to happen. It is possible that some of the changes that we observe are not necessary for later changes to take place.
The non-DLC PCB153 induced a low but significant increase of AHR enhancer recruitment without increasing Cyp1a1 expression. Previous work from other groups has shown that PCB153 can function as an antagonist of AHR-induced ethoxyresorufin-O-deethylase (Sanderson et al., 1996) and Cyp1a1 gene transcription (Suh et al., 2003). The mechanism for this antagonism was not well characterized, although it was shown that PCB153 antagonism of AHR activation was dependent on the agonist affinity and concentration, perhaps by direct competition with the agonist for binding (Suh et al., 2003). If PCB153 were to bind to the AHR ligand binding domain, it could induce nuclear translocation of the AHR complex and facilitate binding to its cognate sites in DNA, while simultaneously inhibiting association with other transcriptional cofactors required to complete transcription by assuming an improper tertiary structure or causing steric hindrance. Although PCB153 did recruit AHR to the enhancer of the Cyp1a1 gene, and this recruitment did cause a displacement of histone H3 from the enhancer region, it did not activate Cyp1a1 transcription at any tested concentration, and correspondingly, none of the activating histone marks that accompany transcription changed. However, PCB153 caused a decrease in the mark 2MeK4-H3 without further methylating this same site to the active transcription-associated mark 3MeK4-H3, which indicates that the methylation state of this site could have been changed to either the mono-methyl substitution or to a complete loss of methylation. It may be that recruitment of AHR to the enhancer could displace a histone lysine2 methyltransferase that maintains the 2MeK4-H3 mark during inactive transcription, or alternatively, that PCB153 treatment activates a histone demethylase, such as a JARID1 family member, that removes the dimethylation mark. However, this loss is not reflective of a complete loss of the monosomes at the promoter region, as the amount of total histone H3 in PCB153 treated is similar to that of control (Fig. 4B). This suggest that the mechanism by which PCB153 removes the dimethylation of lysine 4 of histone 3 may be different than that which results in the loss of this mark by the DLCs, as each DLC significantly decreased the total H3 associated with the promoter region (Fig. 4B).
Our results support the view that AHR activation by ligand is a prerequisite for induction of a wave of chromatin modifications that are required for active transcription. Induction of this wave is not the exclusive effect of exposure to halogenated or polycyclic aromatic hydrocarbons but is the result of AHR activation by all ligands tested so far. The epigenetic regulatory changes associated with induction of AHR target genes by ligands of differing TEFs, particularly those located at the proximal promoter region, show a good correlation to their resulting toxicity and might be useful parameters in the assignment of toxic equivalency.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health National Institute of Environmental Health Sciences (NIEHS) grants R01 ES06273, R01 ES10807; the NIEHS Center for Environmental Genetics grant P30 ES06096; NIEHS Training Grant 5T32 ES016646 on Gene–Environment Interactions (to J.L.O.).
Acknowledgments
The authors declare no conflicts of interest.
References
- Bertazzi PA, Consonni D, Bachetti S, Rubagotti M, Bacarelli A, Zocchetti C, Pesatori AC. Health effects of dioxin exposure: a 20-year mortality study. Am. J. Epidemiol. 2001;153:1031–1044. doi: 10.1093/aje/153.11.1031. [DOI] [PubMed] [Google Scholar]
- Bjerke DL, Peterson RE. Reproductive toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in male rats: different effects of in vitro vs lactational exposure. Toxicol. Appl. Pharmacol. 1994;127:241–249. doi: 10.1006/taap.1994.1158. [DOI] [PubMed] [Google Scholar]
- Courtney KD, Moore JA. Teratolgy studies with 2,4,5-trichlorophenoxyacetic acid and 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Appl. Pharmacol. 1971;20:396–403. doi: 10.1016/0041-008x(71)90282-1. [DOI] [PubMed] [Google Scholar]
- De Voogt P, Brinkman U. Halogenated Biphenyls, Terphenyls, Naphthalenes, Dibenzodioxins, and Related Products. Amsterdam: Elsevier Science; 1989. [Google Scholar]
- Denison MS, Pandini A, Nagy S, Baldwin EP, Bonati L. Ligand binding to the Ah receptor. Chem. Biol. Interact. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- Gasiewicz TA, Kende AS, Rucci G, Whitney B, Willey JJ. Analysis of structural requirements for Ah receptor antagonist activity: ellipticines, flavones, and related compounds. Biochem. Pharmacol. 1996;52:1787–1803. doi: 10.1016/s0006-2952(96)00600-4. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hoffman E, Reyes H, Chu FF, Sander F, Conley LH, Brooks BA, Hankinson O. Cloning for a factor required for activity of the Ah receptor. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- Hon GL, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet. 2009;18(2):R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T, Eguchi H, Tachibana T, Yoneda Y, Kawajiri K. Nuclear localization and export signals of the human aryl hydrocarbon receptor. J. Biol. Chem. 1998;273:2895–2905. doi: 10.1074/jbc.273.5.2895. [DOI] [PubMed] [Google Scholar]
- Jenuwien T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones KW, Whitlock JP. Functional analysis of the transcriptional promoter for the Cyp1a1 gene. Mol. Cell. Biol. 1990;10:5098–5105. doi: 10.1128/mcb.10.10.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkvliet NI, Steppan LB, Smith BB, Youngberg JA. Role of the Ah locus in suppression of cytotoxic T lymphocyte activity by halogenated aromatic hydrocarbons (Pcbs and Tcdd): structure-activity relationships and effects in congenic C57bl/6 mice. Fundam. Appl. Toxicol. 1990;14:532–541. doi: 10.1016/0272-0590(90)90257-k. [DOI] [PubMed] [Google Scholar]
- Knutson JC, Poland A. Response of murine epidermis to 2,3,7,8-tetrachlorodibenzo-p-dioxin: interaction of the Ah and Hr loci. Cell. 1982;30:225–234. doi: 10.1016/0092-8674(82)90028-9. [DOI] [PubMed] [Google Scholar]
- McLachlan MS. Bioaccumulation of hydrophobic chemicals in agricultural food chains. Environ. Sci. Technol. 1996;30:252–259. [Google Scholar]
- Mocarelli P, Pier MG, Patterson DG, et al. Dioxin exposure, from infancy through puberty, produces endocrin disruption and affects human semen quality. Environ. Health Perspect. 2008;116(1):70–77. doi: 10.1289/ehp.10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolgavkar S, Luebeck EG, Buchmann A, et al. Quantitative analysis of enzyme-altered liver foci in rats initiated with diethylnitrosamine and promoted with 2,3,7,8-tetrachlorodibenzo-p-dioxin or 1,2,3,4,6,7,8 heptachlorodibenzo-p-dioxin. Toxicol. Appl. Pharmacol. 1996;138(1):31–42. doi: 10.1006/taap.1996.0094. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Roe AL, Dieter MZ, et al. Role of the aromatic hydrocarbon receptor and Ah gene battery in the oxidative stress response. Biochem. Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- Neuhold LA, Shirayoshi Y, Ozato K, Jones JE, Nebert DW. Regulation of mouse Cyp1a1 gene expression by dioxin: requirement of two cis-acting elements during induction. Mol. Cell. Biol. 1989;9:2378–2386. doi: 10.1128/mcb.9.6.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onozuka D, Yoshimura T, Kaneko S, Furue M. Mortality after exposure to polychlorinated biphenyls and polychlorinated dibenzofurans: a 40-year follow-up study of Yusho patients. Am. J. Epidemiol. 2009;169(1):86–95. doi: 10.1093/aje/kwn295. [DOI] [PubMed] [Google Scholar]
- Oughton JA, Pereira CB, Dekrey GK, Collier JM, Frank AM, Kerkvliet NI. Phenotypic analysis of spleen, thymus, and peripheral blood cells in aged C57b1/6 mice following long-term exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Fundam. Appl. Toxicol. 1995;25(1):60–69. doi: 10.1006/faat.1995.1040. [DOI] [PubMed] [Google Scholar]
- Poland A, Palen D, Glover E. Tumour promotion by Tcdd in skin of Hrs/J hairless mice. Nature. 1982;300:271–273. doi: 10.1038/300271a0. [DOI] [PubMed] [Google Scholar]
- Reijenders P, Brasseur S. Chemically-Induced Alterations in Sexual and Functional Development: The Wildlife/Human Connection. Princeton, NJ: Princeton Scientific; 1992. [Google Scholar]
- Safe S. Modulation of gene expression and endocrine response by 2, 4, 7, 8-tetrachlorodibenzo-p-dioxin. Pharmacol. Ther. 1995;67:247–281. doi: 10.1016/0163-7258(95)00017-b. [DOI] [PubMed] [Google Scholar]
- Sanderson JT, Aarts JM, Brouwer A, Froese KL, Denison MS, Geisy JP. Comparison of Ah receptor-mediated luciferase and ethoxyresorufin-O-deethylase induction in H4IIE cells: implications for their use as bioanalytical tools for the detection of polyhalogenated aromatic hydrocarbons. Toxicol. Appl. Pharmacol. 1996;137:316–325. doi: 10.1006/taap.1996.0086. [DOI] [PubMed] [Google Scholar]
- Schecter A, Cramer P, Boggess K, Stanley J, Papke O, Olson J, Silver A, Schmitz M. Intake of dioxins and related compounds from food in the US population. J. Toxicol. Environ. Health. 2001;63(1):1–18. doi: 10.1080/152873901750128326. [DOI] [PubMed] [Google Scholar]
- Schnekenburger M, Peng L, Puga A. Hdac1 Bound to He Cyp1a1 Promoter Blocks Histone Acetylation Associated with Ah Receptor Mediated Trans-Activation. Biochemica et Biophysica Acta. 2007a;1769:569–578. doi: 10.1016/j.bbaexp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnekenburger M, Talaska G, Puga A. Chromium cross-links histone deacetylase 1-DNA methyltransferase-1 complexes to chromatin, inhibiting histone remodeling marks critical for transcriptional activation. Mol. Cell. Biol. 2007b;27:7089–7101. doi: 10.1128/MCB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenland K, Piacetelli L, Deddens J, Fingerhut M, Cheng LI. Cancer, heart disease, and diabetes in workers exposed to 2,3,7,8-tetracholorodibenzo-p-dioxin. J. Natl. Cancer Inst. 1997;91:779–786. doi: 10.1093/jnci/91.9.779. [DOI] [PubMed] [Google Scholar]
- Suh J, Kang JS, Yang K-H, Kaminski NE. Antagonism of aryl hydrocarbon receptor-dependent induction of CYP1A1 and inhibition of Igm expression by di-ortho-substituted polychlorinated biphenyls. Toxicol. Appl. Pharmacol. 2003;187(1):11–21. doi: 10.1016/s0041-008x(02)00040-6. [DOI] [PubMed] [Google Scholar]
- Swanson H, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J. Biol. Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Chen M, Sheng T. Interactions between aryl hydrocarbon receptor and P-TEFb. Sequential recruitment of transcription factors and differential phosphorylation of C-terminal domain of RNA polymerase II at cyp1a1 promoter. J. Biol. Chem. 2003;278:44041–44048. doi: 10.1074/jbc.M306443200. [DOI] [PubMed] [Google Scholar]
- Van den Berg M, Birnbaum LS, Bosveld A, Brunstrom B, Cook P, Feely M, Geisy JP, Hanberg A, Hasegawa R, Kennedy SW, et al. Toxic equivalency factors for PCBs, PCDD, PCDFs for humans and wildlife. Environ. Health Perspect. 1998;106:755–792. doi: 10.1289/ehp.98106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg M, Birnbaum LS, Denison MS, De Vito M, Farland W, Feely M, Fiedler H, Hakansson H, Hanberg A, Haws L, et al. The 2005 World Health Organization reevaluation of human and mammalian toxic equivalency factors for dioxins and diosin like compounds. Toxicol. Sci. 2006;93:233–241. doi: 10.1093/toxsci/kfl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock JP. Induction of cytochrome P450 1A1. Annu. Rev. Pharmacol. Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- Wiebel F, Klose U, Kiefer F. Toxicity of 2,3,7,8-tetracholordibenzo-p-dioxin in vitro: H4IIIEC3-derived 5l hepatoma cells as a model system. Toxicol. Lett. 1991;55(2):161–169. doi: 10.1016/0378-4274(91)90130-x. [DOI] [PubMed] [Google Scholar]
- Worner W, Schrenk D. Influence of liver tumor promoters on apoptosis in rat hepatocytes induced by 2-acetylaminoflourene, ultraviolet light, or transforming growth factor. Cancer Res. 1996;56:1272–1278. [PubMed] [Google Scholar]
- Yanagida A, Sogawa K, Yasumoto K, Fuji-Kuriyama Y. A novel cis-acting DNA element required for a high level of inducible expression of the rat p-450c gene. Mol. Cell. Biol. 1990;10:1470–1475. doi: 10.1128/mcb.10.4.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3s10ph and H4k16ac generates a histone code that mediates transcription elongation. Cell. 2009;138(6):1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.