Abstract
Virulence and antibiotic resistance of the dangerous human pathogen Staphylococcus aureus are to large extent determined by the acquisition of mobile genetic elements (MGEs). Up to now, these elements were known to comprise either resistance or virulence determinants, but not a mixture of the two. Queck et al. now found a cytolysin gene of the phenol-soluble modulin (PSM) family within SCCmec elements, which contain methicillin resistance genes and are largely responsible for the spread of methicillin-resistant S. aureus (MRSA). The novel gene, called psm-mec, had a significant impact on virulence in MRSA strains that do not produce high levels of genome-encoded PSMs. This first example of a combination of toxin and resistance genes on one staphylococcal MGE shows that such bundling is possible and may lead to an even faster acquisition of toxin and resistance genes by S. aureus and other staphylococcal pathogens.
Key words: Staphylococcus aureus, Staphylococcus epidermidis, MRSA, MRSE, toxin, antibiotic resistance, mobile genetic element, methicillin
Staphylococcus aureus is a dangerous human pathogen, causing moderate to severe skin infections such as furunculosis, impetigo, and abscesses, organ infections such as endocarditis or osteomyelitis, and a variety of toxinoses such as toxic shock syndrome.1 Pathogenicity of the organism in these diseases is mainly determined by a series of secreted virulence factors, which include degradative exoenzymes and toxins. While some S. aureus toxins are encoded on the core genome, for instance the channel-forming cytolysin alpha-toxin, many secreted virulence factors are found on accessory genetic elements such as transposons, prophages, and pathogenicity islands (PIs). The most important toxins encoded on such elements are the superantigens, which comprise a series of enterotoxins, the exfoliative toxins A and B, and toxic shock syndrome toxin (TSST).2
Carriage on accessory genetic elements implies that virulence genes have been acquired by horizontal gene transfer from other strains, although such transmission cannot always be directly shown in the laboratory. Nonetheless, these elements are usually called mobile genetic elements (MGEs). Important MGEs in S. aureus include the pathogenicity islands SaPI1 through 4, SaPIbov (found in a strain of bovine origin), prophages, and plasmids encoding exfoliative toxins and enterotoxins. The best studied PI is SaPI1, which contains the gene for TSST.3
The second important family of MGEs in S. aureus consists of elements that encode resistance factors. Among those, the staphylococcal chromosome cassette methicillin-resistance islands (SCCmecs) have gained most attention, as they represent the key determinant conferring resistance to methicillin and other beta-lactam antibiotics and are thus responsible for the occurrence of the infamous MRSA (methicillin-resistant S. aureus).4 Remarkably, pathogenicity islands such as the SaPIs have so far been not known to contain resistance factors, while the SCCmec elements were assumed to contain only resistance but no toxin genes.2
Antibiotic resistance of S. aureus is a major concern to public health. While antibiotic resistance may in some cases be acquired by mutation—such as resistance to quinolone antibiotics by mutation in the DNA gyrase or topoisomerase genes—the most common mechanism that S. aureus uses to become resistant to a given antibiotic is the acquisition of the corresponding resistance determinant on an MGE by horizontal gene transfer. Especially under selective pressure, i.e. when an antibiotic is used at considerable levels and frequency in the population, the development and spread of resistant strains can be extremely fast. The most famous examples, perhaps among all cases of bacterial antibiotic resistance, are the development of penicillin- and later methicillin-resistant strains of S. aureus. Penicillin was introduced in 1941 and proved a very efficacious means to combat S. aureus infections. However, penicillin-resistant, penicillinase-containing strains were detected only 4 years later5 and the switch to other antibiotics was followed by the rise of multi-drug-resistant S. aureus. Penicillinase-resistant methicillin was introduced in 1961, but methicillinresistant strains, the first MRSA, were already found within a year.6 Considerable use of methicillin and its derivatives in the following decades led to global spread of MRSA and the fact that methicillin is nowadays not a usable drug for S. aureus infections in many countries.
Recent advances in deciphering the entire genomic information of many S. aureus strains has also allowed detailed insight in the composition and distribution of SCCmec elements. SCCmec elements are grouped into main allotypes (I through VIII) according to the type of the essential recombinase (ccr) and resistance (mec) genes. Subtypes are further classified according to the composition of the non-essential J regions, which may carry additional genes such as transposons and further resistance genes.4
The archaic MRSA clones that arose in the UK in the 1960s carry SCCmec type I.7,8 SCCmec types II and III carry additional antibiotic resistance genes, conferring resistance for example to tetracycline and erythromycin.7 Today, there are five major clonal types of MRSA with two different genetic backgrounds, one of which is characterized by carrying SCCmec types I or III and the other, SCCmec type II.9 SCCmec type IV is characteristic for the recently emerged community-associated (CA-) MRSA strains,10 which are known to have high virulence and cause infections in otherwise healthy patients outside hospital settings.11 The SCCmec type IV element is shorter than the other SCCmec types and there is evidence to suggest that as a result, it does not lead to a measurable fitness cost for the bacteria, facilitating the spread of CA-MRSA strains.12 Strains with other SCCmec types are very rare.
Phenol-soluble modulins (PSMs) are a class of peptides that have been named according to their behavior during hot phenol extraction.13 Although considerably different in amino acid sequence, they share an amphipathic, α-helical structure, which likely plays a major role in determining their manifold demonstrated or putative biological activities. Lysis of human immune cells, particularly neutrophils, is the most intensely investigated feature of PSMs.14 This activity is mainly found in the shorter PSMs of the α-type, while the longer β-type PSMs are not cytolytic. Furthermore, cytolytic activity can dramatically differ between members of the α-type PSMs, with some PSMs found in the psm-α operon of S. aureus being the most active in that regard. In accordance with the key importance of neutrophils for host defense against S. aureus infection, deletion of the psm-α operon of S. aureus strains led to dramatically impaired virulence in bacteremia and abscess infection models.14 PSM genes are found in all S. aureus and do not substantially differ between strains. Increased production of PSMs and other key virulence determinants, such as α-toxin, thus has prompted researchers to explain the increased virulence potential of CA-MRSA compared to traditional HA-MRSA by differential gene expression and a potentially unique arrangement of gene regulatory networks.15
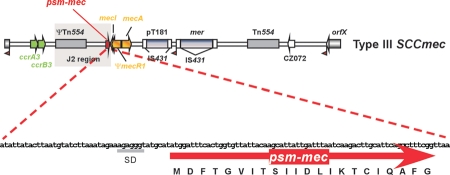
Queck et al. have recently described a novel psm gene, termed psm-mec, which differs from all other known psm genes in being encoded on an MGE rather than the S. aureus core genome.16 Specifically, the psm-mec gene is found in SCCmec types II and III very near to the mecI gene in the J2 region (Fig. 1). While cytolytic activity of PSM-mec was not as pronounced as in the most potent PSMs, production levels of strains harboring the psm-mec gene were frequently extremely high, giving reason to believe that PSM-mec contributes considerably to the neutrophil-lytic capacity as well as virulence of these strains. Indeed, a significant impact on neutrophil lysis and virulence in an abscess model was detected in an MRSA strain whose production of core genome-encoded PSMs was low, indicating that acquisition of the psm-mec gene on an SCCmec element may substitute for the lack of such intrinsic PSM production and significantly increase pathogenicity.
Figure 1.
Location of the psm-mec gene in SCCmec elements. The graph shows the location of psm-mec in the SCCmec type III element, near the mecI gene, within the J2 segment. It is also present in the corresponding part of SCCmec type II and some rare elements that contain a similar region, but absent from SCCmec types I and IV. Essential parts of SCCmec are in color: green, recombinase genes, orange, resistance genes.
Coagulase-negative staphylococci, particularly S. epidermidis, have often been discussed as potential sources of SCCmec and there is evidence indicating that methicillin resistance was acquired from S. epidermidis to create MRSA.17 In accordance with the high frequency of MRSE and the distribution of type II and III SCCmec types in S. epidermidis, the frequency of PSM-mec production was found to be very high among S. epidermidis strains, with production levels reaching similar levels as in strong MRSA producers.16 Cytolytic activities of S. epidermidis culture filtrates in general are low compared to S. aureus (unpublished). Furthermore, most PSMs of S. epidermidis are either only moderately or not cytolytic and more potent PSMs are not produced at a considerable level, a feature that is consistent with the non-aggressive lifestyle of S. epidermidis.18 On that ground, it can be assumed that PSM-mec production makes a difference in S. epidermidis regarding its pathogenic potential that is even higher than in S. aureus, a hypothesis that is currently under investigation.
Remarkably, PSM-mec is the first staphylococcal toxin to be found that is encoded on MGEs that primarily encode resistance factors. SCCmec types II and III thus bundle antibiotic resistance and virulence determinants, representing the hitherto only of their kind known to do so. While this may represent the exception rather than the rule, which appears quite likely considering information from the many available staphylococcal genomes, it shows that there is no physiological or other reason that would categorically prevent the bundling of these two different classes of accessory genes on a transferable element. Thus, it is quite likely that—given the speed of MGE rearrangements—MGEs that harbor both antibiotic resistance and virulence determinants may arise in the foreseeable future.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), NIH.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/10453
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Novick RP, Schlievert P, Ruzin A. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 2001;3:585–594. doi: 10.1016/s1286-4579(01)01414-9. [DOI] [PubMed] [Google Scholar]
- 3.Novick RP, Subedi A. The SaPIs: mobile pathogenicity islands of Staphylococcus. Chem Immunol Allergy. 2007;93:42–57. doi: 10.1159/000100857. [DOI] [PubMed] [Google Scholar]
- 4.Ito T, Okuma K, Ma XX, Yuzawa H, Hiramatsu K. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist Updat. 2003;6:41–52. doi: 10.1016/s1368-7646(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 5.Barber M, Rozwadowska-Dowzenko M. Infection by penicillin-resistant staphylococci. Lancet. 1948;2:641–644. doi: 10.1016/s0140-6736(48)92166-7. [DOI] [PubMed] [Google Scholar]
- 6.Jevons MP, Parker MT. The Evolution of New Hospital Strains of Staphylococcus aureus. J Clin Pathol. 1964;17:243–250. doi: 10.1136/jcp.17.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart GT, Holt RJ. Evolution of natural resistance to the newer penicillins. Br Med J. 1963;1:308–311. doi: 10.1136/bmj.1.5326.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira DC, Tomasz A, de Lencastre H. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb Drug Resist. 2001;7:349–361. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 10.Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002;46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diep BA, Otto M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 2008;16:361–369. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages SA, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 13.Mehlin C, Headley CM, Klebanoff SJ. An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J Exp Med. 1999;189:907–918. doi: 10.1084/jem.189.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Diep BA, Villaruz AE, Braughton KR, Jiang XG, DeLeo FR, et al. Evolution of virulence in epidemic community-associated MRSA. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Queck SY, Khan BA, Wang R, Bach TH, Kretschmer D, Chen L, et al. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 2009;5:e1000533. doi: 10.1371/journal.ppat.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanssen AM, Kjeldsen G, Sollid JU. Local variants of Staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative Staphylococci: evidence of horizontal gene transfer? Antimicrob Agents Chemother. 2004;48:285–296. doi: 10.1128/AAC.48.1.285-296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto M. Staphylococcus epidermidis - the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]