Myosin IIA and IIB synergistically generate front–back polarity through their effects on actomyosin bundling formation and stability, and adhesion maturation, which are mediated by localized Rac GEF depletion.
Abstract
Migratory front–back polarity emerges from the cooperative effect of myosin IIA (MIIA) and IIB (MIIB) on adhesive signaling. We demonstrate here that, during polarization, MIIA and MIIB coordinately promote localized actomyosin bundling, which generates large, stable adhesions that do not signal to Rac and thereby form the cell rear. MIIA formed dynamic actomyosin proto-bundles that mark the cell rear during spreading; it also bound to actin filament bundles associated with initial adhesion maturation in protrusions. Subsequent incorporation of MIIB stabilized the adhesions and actomyosin filaments with which it associated and formed a stable, extended rear. These adhesions did not turn over and no longer signal to Rac. Microtubules fine-tuned the polarity by positioning the front opposite the MIIA/MIIB-specified rear. Decreased Rac signaling in the vicinity of the MIIA/MIIB-stabilized proto-bundles and adhesions was accompanied by the loss of Rac guanine nucleotide exchange factor (GEFs), like βPIX and DOCK180, and by inhibited phosphorylation of key residues on adhesion proteins that recruit and activate Rac GEFs. These observations lead to a model for front–back polarity through local GEF depletion.
Introduction
Front–back polarization is intrinsic to directed cell migration. In most cells, the front is characterized by a region of intense actin polymerization, which occurs either in a narrow band along the leading edge of protrusions or in more localized, spike-like filopodia (Small and Resch, 2005). The rear can also adopt different morphologies. It can be extended and long, as in many fibroblasts; stubby and short, as in leukocytes; or flat and parallel to the leading edge, as in keratocytes and some cancer cells. In epithelial sheet migration, the rear of the leading cells of the cohort is defined by cell–cell contacts with cells more interior (Parent and Devreotes, 1999; Etienne-Manneville and Hall, 2003; Ridley et al., 2003; Montell, 2008; Vicente-Manzanares et al., 2009).
Recent progress has focused on the signaling pathways that regulate polarity in migrating cells. In Dictyostelium discoideum and leukocyte chemotaxis, the localized production of PIP3, via segregated activities of PI3K and PTEN, recruits machinery involved in actin polymerization to the regions of the cell sensing the highest concentration of the chemoattractant, and thereby creates a protrusion (Van Haastert and Devreotes, 2004). In epithelial cells, Par3 and Par6, aPKCζ, and GSK3 regulate polarity by positioning microtubules, the nucleus, and the Golgi apparatus (Etienne-Manneville and Hall, 2001; Gomes et al., 2005). Finally, in migrating fibroblasts and other cells, the dynamic adhesions in protrusions contain signaling complexes that locally control actin polymerization (Nayal et al., 2006; Zaidel-Bar et al., 2007), whereas signaling through ROCK is implicated in forming the rear (Kolega, 2003; Totsukawa et al., 2004; Iwanicki et al., 2008). A common feature of all of these signaling networks is their convergence on the Rho GTPases, which in turn regulate actin polymerization; nuclear, Golgi, and microtubule positioning; and myosin II (MII) activation (Etienne-Manneville and Hall, 2003; Gomes et al., 2005; Vicente-Manzanares et al., 2009).
Building on the work of others (Verkhovsky et al., 1999; Kolega, 2003; Yam et al., 2007), we have recently implicated MII in front–back polarization in fibroblasts by forming the cell rear (Vicente-Manzanares et al., 2008). MII is comprised of two myosin heavy chains (MHCII), two essential light chains (ELCs), and two regulatory light chains (RLCs) that control, through phosphorylation on Thr18 and/or Ser19, the ATPase activity in the globular head of the heavy chain (Vicente-Manzanares et al., 2009). Three isoforms of the MHC—MHCII-A, II-B, and II-C—define the three functional isoforms of MII, e.g., MIIA, MIIB, and MIIC, respectively. MIIA and MIIB are commonly found in migrating cells. MIIA decorates more anterior filaments in protrusions, whereas MIIB colocalizes with MIIA in the center and rear of migrating cells (Maupin et al., 1994; Kolega, 1998; Vicente-Manzanares et al., 2007). MII is emerging as a major protein responsible for symmetry breaking and organization of the actin bundles that define the rear (Chrzanowska-Wodnicka and Burridge, 1996; Verkhovsky et al., 1999; Xu et al., 2003; Vicente-Manzanares et al., 2008; Mseka et al., 2009; Cramer, 2010). However, the mechanism by which MII breaks and maintains symmetry and how actomyosin organization controls the signals that drive protrusion are critical, outstanding questions, and are pertinent to understanding the integrative role of MII during cell migration and morphogenesis.
In this study, we address these questions. We find that MIIA and MIIB function cooperatively to regulate adhesion signaling and thereby form the rear. MIIA forms localized proto-bundles that break symmetry, upstream of microtubules, and define the nascent rear. However, these bundles, their associated adhesions, and the location of the rear, are unstable without MIIB. Adhesions connected to MIIB-containing actomyosin bundles at the rear are stable, do not localize two Rac guanine nucleotide exchange factors (GEFs)—βPIX and DOCK180—and exhibit attenuated tyrosine phosphorylation, including sites that are implicated in recruiting the GEFs.
Results
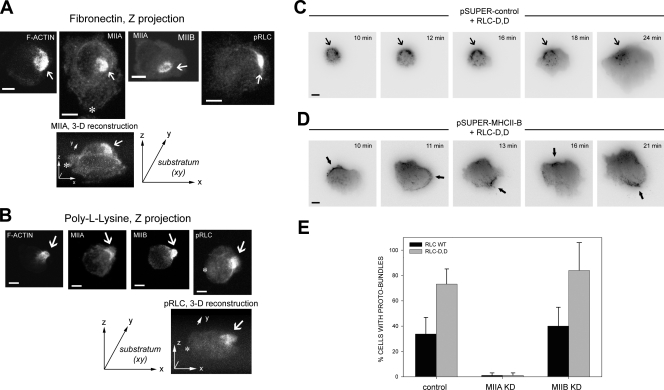
MIIA and MIIB coordinately define and assemble the rear of migrating cells
Our previous evidence points to MII and particularly MIIB as a critical symmetry breaking element in establishing the cell rear (Vicente-Manzanares et al., 2008). To determine how MII forms the rear, we imaged the organization of actin and MII in newly adhering CHO.K1 cells as they spread and polarized. Just after plating on fibronectin (<10 min), most cells are round and have not yet spread. However, ∼50% of them display a discrete accumulation of actomyosin “proto-bundles.” The rest neither display these bundles nor go on to polarize (unpublished data). The bundles contain F-actin, and MHCII-A, MHCII-B, and phosphorylated RLC, as visualized by staining with rhodamine-conjugated phalloidin (for actin) or immunostaining (Fig. 1 A). These proto-bundles are also present in primary human endothelial cells (HUVECs) spreading on fibronectin (unpublished data). In a small fraction (∼10–20%) of the proto-bundle–containing cells, the proto-bundles remain highly localized and evolve to form extended “rears” (Fig. 1 C and Fig. S1 B). Conversely, in the rest of the cells displaying proto-bundles, they do not remain localized in one area, undergoing assembly in different regions of the cell; these cells do not go on to display prominent extended rears (Fig. S1). Similar actomyosin proto-bundles are also seen in cells adhering to poly-l-lysine (PLL) for 10–180 min (shown at 45 min in Fig. 1 B). PLL supports cell attachment, but not spreading; consequently, these cells remain round and do not display visible adhesions, leading edges, or rears (Fig. 1 B), which suggests that the formation of proto-bundles may be integrin independent. Furthermore, there is no discrete accumulation of adhesion proteins, e.g., paxillin, integrin (α5-GFP), or secreted fibronectin in the vicinity of the myosin-containing proto-bundle (Fig. S2).
Figure 1.
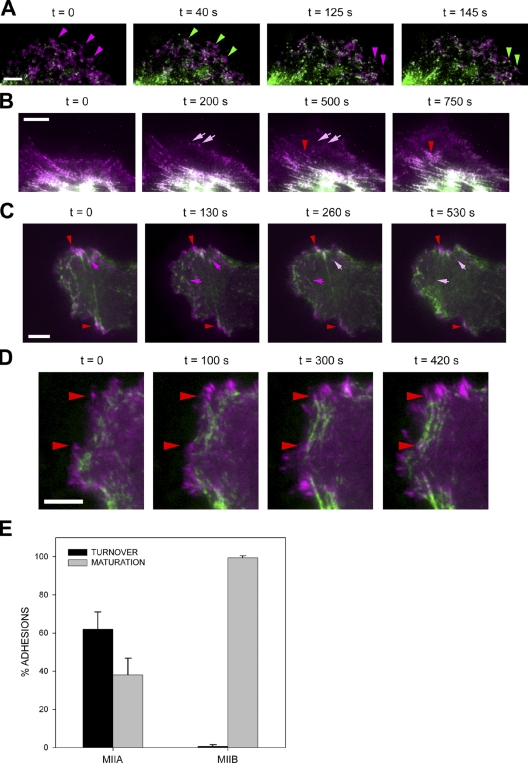
Localized actomyosin bundles that are generated by MIIA and stabilized by MIIB define the cellular region that forms the rear. (A) CHO.K1 cells were plated on fibronectin and allowed to attach for 10 min. The cells were then fixed and stained for phosphorylated RLC (pRLC), MIIA (using a MHCII-A antibody), MIIB (MHCII-B antibody), or F-actin (rhodamine-phalloidin). Images were captured using a confocal microscope (FV300; Olympus). Z projections are shown. Representative examples are shown of cells exhibiting an accumulation of these proteins (“proto-bundles,” marked with arrows). The asterisks mark the leading edge in the Z projection and corresponding 3D reconstruction. (B) CHO.K1 cells were plated on PLL and allowed to attach for 45 min. The cells were stained as in A, and they display similar proto-bundles (marked with arrows). Representative examples are shown. (C and D) Cells were transfected with RLC-D,D–GFP and either control (C) or MHCIIB-shRNA (D) to inhibit MIIB expression. Representative time points of their spreading on fibronectin are shown. In C, arrows point to the region of the cell initially primed by RLC-D,D to form the rear, where protrusion does not occur. The complete movie is shown in Video 1. In D, arrows point to clusters, or proto-bundles, denoted by the accumulation of MIIA (the only isoform present in the knockdowns, labeled with the RLC-D,D–GFP) that form and disassemble rapidly. Images were captured in TIRF mode using an inverted microscope (IX70; Olympus) coupled to a CCD camera (Retiga Exi; Qimaging). The complete movie is shown in Video 2. (E) CHO.K1 cells knocked down for MIIA or MIIB were transfected with wild-type RLC (control, black bars) or RLC-D,D (gray bars) coupled to GFP, adhered for 10 min to fibronectin, fixed, and quantified for presence of proto-bundles as for those shown in A. Data are the mean ± SD of three experiments (error bars) with >200 cells scored in each experiment. Bars, 10 µm.
We have previously reported that expression of a phospho-mimetic mutant of the RLC (RLC-D,D) in wild-type cells increases formation of extended, stable rears in migrating cells (Vicente-Manzanares et al., 2008). Cells expressing this mutant, when plated on either fibronectin or PLL, show a modest increase in the fraction of round and early spreading cells with a proto-bundle; whereas pretreatment with a MII ATPase inhibitor, blebbistatin, inhibits its formation (Table I). Interestingly, the majority of RLC-D,D–containing proto-bundles remain relatively static while the cell protrudes away from them (mainly opposite the rear) and thereby form an extended rear (Fig. 1 C, Fig. S1 B, and Video 1). Thus, the effect of RLC-D,D is to increase the frequency that the proto-bundles remain localized and stabilized to form an extended rear.
Table I.
Effect of RLC-D,D–GFP, 50 μM blebbistatin, and 5 μM nocodazole on proto-bundle formation in cells spreading on fibronectin or attached to PLL
| Condition | Fibronectin (10 min) | PLL |
| Control (wild-type RLC) | 48 ± 1 | 52 ± 10 |
| RLC-D,D | 60 ± 7a | 73 ± 12a |
| Blebbistatin | 1.5 ± 0.5 | 1 ± 2 |
| Nocodazole | 41 ± 8 | 59 ± 5 |
Data represent the percentage of cells displaying an F-actin–enriched proto-bundle, and are expressed as mean ± SD of three experiments. >200 cells were scored in each experiment.
p < 0.05, Student’s t test (RLC-D,D vs. control).
To parse the role of the different MII isoforms, we first plated cells, depleted of MIIA or MIIB by RNAi, on fibronectin or PLL. Cells depleted of MIIA do not display proto-bundles (Fig. 1 E), even when coexpressing RLC-D,D. In contrast, proto-bundles are observed in MIIB-deficient cells on either fibronectin or PLL (Fig. 1 E). Time-lapse imaging of MIIB-deficient cells expressing RLC-D,D reveals that proto-bundles form randomly and then disassemble rapidly (Fig. 1 D and Video 2), in contrast to RLC-D,D–expressing controls cells, where they are stable (Fig. 1 C). This results in cells lacking a well-defined front and back. These observations suggest that front–back polarization is driven by the formation of a MIIA-initiated and MIIB-stabilized actomyosin proto-bundle that evolves into large actomyosin bundles that form extended rears.
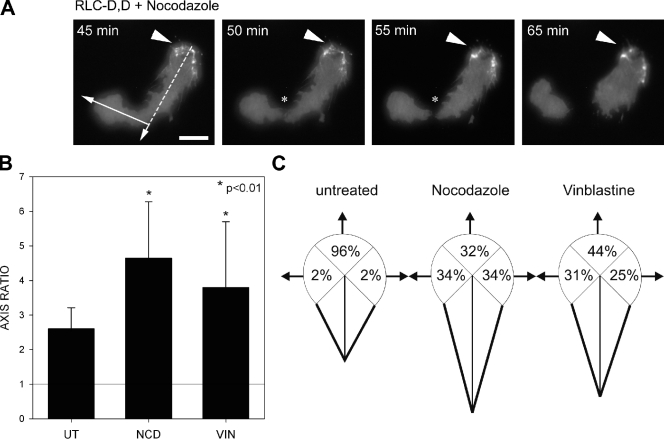
We next determined the relation between the actomyosin bundles and microtubules in establishing front–back polarity. Treating cells with nocodazole, an inhibitor of microtubule polymerization, either at the time of plating or after 45 min (when front–back polarity is already established) did not affect the formation of the proto-bundle in cells adhering to either fibronectin or PLL (Table I and Fig. 2), which indicates that microtubules are not needed for proto-bundle formation and initial polarization. Similar results were obtained using vinblastine (Fig. 2, B and C). However, microtubules do appear to position the protrusion opposite the rear and thereby fine-tune the polarization. In cells expressing the RLC-D,D mutant, proto-bundles form and become part of the rear of the cell during spreading even in the presence of nocodazole or vinblastine. However, the leading protrusion becomes unusually long (Fig. 2 B) and often extends in directions that are not directly opposed to the rear (Fig. 2, A and C). Occasionally, it extended so far that it detached from the main body of the cell (Fig. 2 A and Video 3), an observation described previously by others (Verkhovsky et al., 1999). These data show that MII establishes the rear, and that the microtubules position the protrusion across from the rear and constrain the length of the cell.
Figure 2.
Microtubules position the leading edge across from the actomyosin-enriched rear. (A) TIRF microscopy time-lapse series of a cell expressing RLC-D,D–GFP (localized at the rear, marked by arrowheads) in the presence of the microtubule inhibitor nocodazole (5 µM). Note that the direction of movement of the leading protrusion (arrow) is bent (almost perpendicular to the major polarity axis, indicated by a dashed line), and the leading protrusion has separated from the rear; the asterisks point to the site of ripping. The complete movie is shown in Video 3. Bar, 10 µm. (B) Cells expressing RLC-D,D–GFP and migrating on fibronectin in the presence or absence of 5 µM nocodazole (NCD) or 0.1 µM vinblastine (VIN). Axis ratio is calculated as the ratio between long (migratory) and short (perpendicular, passing through the center of the nucleus) axes; thus, an AR of 1 (indicated with a horizontal line) denotes a round cell. Error bars indicate ±SD. (C) Diagrams showing the relative direction of the leading protrusion with respect to the axis established by the extended rear in control and 5 µM nocodazole– or 0.1 µM vinblastine–treated cells. n > 100 cells were quantified from two independent experiments.
MIIA and MIIB synergize to form actomyosin bundles and regulate adhesion
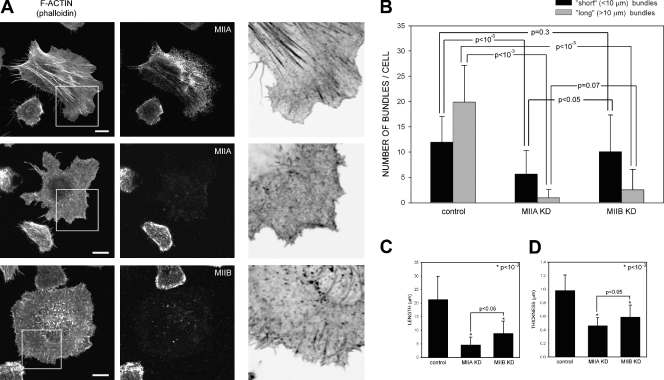
Once fibroblasts spread and acquire a motile morphology, large actin bundles and adhesions are the hallmarks of a well-defined rear (Vicente-Manzanares et al., 2008). To probe the roles of MIIA and MIIB in generating these large actin bundles and adhesions, we investigated the effect of isoform-specific small hairpin RNA (shRNA)-mediated knockdowns of MIIA and MIIB on their formation. In contrast to wild-type cells (Fig. 3 A), MIIA-depleted cells, which still express MIIB, exhibit almost no large, elongated adhesions (Fig. 4 A) and only a few visible actin bundles (Fig. 3 A, B), which are short (Fig. 3 C) and thin (Fig. 3 D). In these cells, MIIB appeared largely disorganized (unpublished data). These effects are similar to blebbistatin treatment (unpublished data). The MIIA knockdown is also dose dependent; knockdowns containing residual MIIA levels show some visible actomyosin bundles, although reduced compared with wild-type cells (Fig. S3). This phenotype indicates that MIIA is required for the formation of actin bundles of sufficient size and organization to be visualized by fluorescence microscopy.
Figure 3.
MIIB enhances MIIA-initiated actomyosin bundling in CHO.K1 cells. (A) Cells were transfected with control, MHCIIA, or MHCIIB shRNA-containing plasmids to inhibit expression of the indicated isoform. The cells were plated on 2 µg/ml fibronectin for 60 min, then fixed and stained with rhodamine-conjugated phalloidin to visualize actin (left) or antibodies against the indicated MII isoform (middle). Representative cells are shown. The right column shows inverse colored, higher magnification of the boxed regions in left column. In the MIIA-depleted cells, there are few organized actomyosin bundles; in the MIIB-depleted cells, the bundles are small and thin. Images were captured using a confocal microscope (FV300; Olympus). Bar, 10 µm. (B–D). Quantification of the actin bundling phenotypes illustrated in A. Parameters evaluated are: (B) number of bundles per cell, divided into “short” (<10 µm) and “long” (>10 µm), n ≥ 35 cells/condition; (C) bundle length, and (D) thickness (n ≥ 150 bundles from >30 cells/condition). (B–D) P is the significance using the nonparametric Mann-Whitney U test (in C and D; asterisks refer to P vs. control cells). Error bars indicate ±SD.
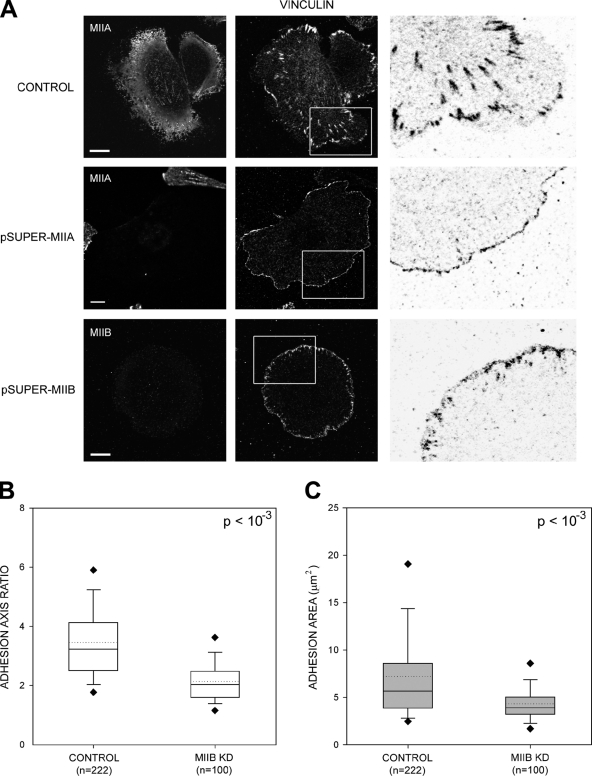
Figure 4.
MIIA and MIIB control different stages of adhesion maturation. (A) Cells were transfected with control, MHCIIA, or MHCIIB shRNA-containing plasmids to inhibit expression of the indicated isoform. The cells were plated on coverslips coated with 2 µg/ml fibronectin for 60 min, fixed, and stained with antibodies against the indicated MII isoform (left) or endogenous vinculin (middle). Images were captured using a confocal microscope (FV300; Olympus). Representative cells are shown. The right column shows inverse colored, higher magnification of the boxed regions in the middle column. In the MIIA-depleted cells, there are no elongated adhesions; in the MIIB-depleted cells, some adhesions around the periphery are somewhat elongated. Bar, 10 µm. (B and C) Quantification of the adhesion phenotypes including adhesion elongation represented as the axial ratio (B) and area (C). MIIA-depleted cells were not included in the analysis as they only display small, nonelongated, nascent adhesions. n is indicated, and P represents significance using the nonparametric Mann-Whitney U test.
MIIB-depleted cells, in contrast, exhibit visible actin bundles throughout the cell; however, they are shorter, thinner, and oriented differently than those in wild-type cells (Fig. 3). MIIA decorates these bundles (unpublished data). Although long bundles are almost completely absent, the number of short bundles is similar to control cells, particularly around the periphery of the cell (Fig. 3, A and B). The adhesions are also smaller (Fig. 4 C) and less elongated (Fig. 4 B) than those in wild-type cells. As described previously, MIIB-depleted cells protrude randomly and do not exhibit extended rears, and their migration is inhibited (Lo et al., 2004; Vicente-Manzanares et al., 2007). Collectively, these data show that both MIIA and MIIB are required for the formation of large actomyosin bundles and lead to the hypothesis that MIIA initiates the formation of visible actin bundles, whereas MIIB increases their size.
To test this hypothesis, we visualized the relative kinetics of MIIA and MIIB entry into filaments using a truncated promoter expression system (Watanabe and Mitchison, 2002) that prevents overexpression artifacts (e.g., increased cell contractility and protein mislocalization; Vicente-Manzanares et al., 2007) and enhances visualization by removing background fluorescence (Choi et al., 2008). Dual-color imaging using low levels of expressed MIIA and MIIB (coupled to mCherry and GFP, respectively) reveals a hierarchical recruitment into actomyosin bundles. MIIA decorates actin bundles in protrusions (Video 4). MIIB, in contrast, decorates bundles localized more centrally. When MIIB appears in protrusions, it accumulates in MIIA-containing bundles and increases their thickness (Fig. 5, A and B). A fraction of small MIIA-only decorated bundles in more anterior parts of the protrusion disassemble as the protrusion evolves (Fig. 5 B, arrows), whereas those decorated with both MIIA and MIIB do not (Fig. 5 B, arrowheads). The stability of the MIIB-containing filaments is consistent with its higher apparent affinity for actomyosin filaments and duty ratio (Kim et al., 2005; Vicente-Manzanares et al., 2008).
Figure 5.
MIIB incorporates into actomyosin bundles preformed by MIIA and inhibits disassembly of actomyosin bundles and adhesions. (A) Cells were transfected with mChe-MIIA (magenta) and GFP-MIIB (green), plated on fibronectin, and filmed using TIRF microscopy. Representative time points are shown. Magenta-to-green arrowheads point to representative sites of MIIB incorporation after MIIA has created the initial bundle. Video 4 shows the entire sequence. (B) Cells were transfected and filmed as in A. Arrows point to MIIA bundles that turn over; arrowheads point to a MIIB bundle that enlarges as MIIB incorporates into the filaments (note the magenta-to-white color transition in the bundle as it thickens, which denotes colocalization of MIIA and MIIB). (C) Cells were transfected with GFP-MIIA (green) and paxillin-mCherry (magenta), plated on fibronectin, and filmed using TIRF microscopy. Arrows in magenta point to elongated adhesions bound to MIIA-decorated actomyosin bundles that disassemble over the course of the experiment; red arrowheads point to adhesions that do not disassemble. Video 5 shows the entire sequence. (D) Cells were transfected with GFP-MIIB (green) and paxillin-mCherry (magenta), plated on fibronectin, and filmed using TIRF microscopy. Arrowheads point to representative elongated adhesions bound to MIIB-decorated actomyosin bundles that enlarge or do not disassemble over the course of the experiment. Video 6 shows the entire sequence. Bars, 5 µm. (E) Quantification of the adhesion turnover and maturation as shown in C and D. n = 222 adhesions/12 cells (MIIA); n = 153 adhesions/10 cells (MIIB).
We next investigated the dynamics of the adhesions associated with MIIA- and MIIB-containing actomyosin filaments. Dual-color imaging of cells expressing low levels of GFP-MIIA or -MIIB and paxillin-mCherry shows that small MIIA-containing actin bundles associate with adhesions in protrusions that are growing and elongating centripetally (maturing). These adhesions turn over in parallel with the disappearance of the MIIA-associated actomyosin bundle (Fig. 5, C and E; and Video 5). In contrast, adhesions associated with GFP-MIIB–containing actomyosin bundles did not turn over; instead, they enlarged (Fig. 5, D and E; and Video 6). These data show that actomyosin bundles containing only MIIA mediate initial adhesion maturation, whereas the incorporation of MIIB enlarges and stabilizes them.
MII-dependent actin bundling regulates adhesive signaling to inhibit Rac activation
The absence of protrusions and presence of large, stable adhesions and actin bundles in the rear suggests that either the actin bundles that comprise the rear and the sides of the cell do not support the robust actin polymerization typical of protrusions, or (2) the large, stable adhesions associated with the rear region do not signal to Rac.
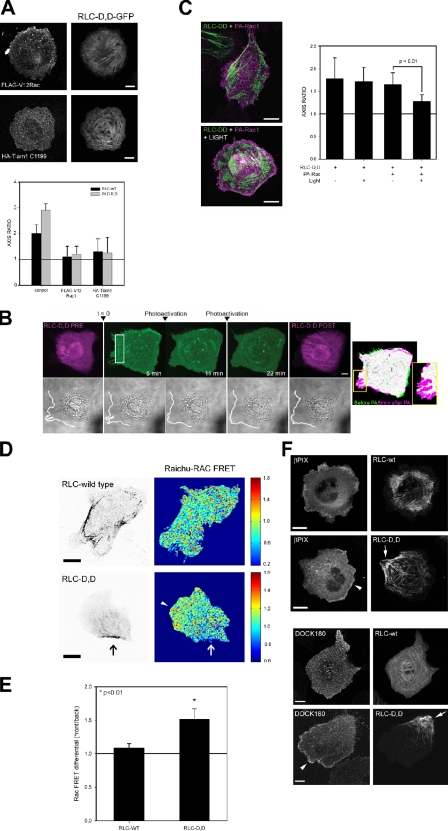
To test the first hypothesis, we activated Rac in cells coexpressing RLC-D,D to produce well-defined rears. When activated V12-Rac (or L61, not depicted) or an activated mutant of the Rac GEF Tiam1 (Tiam1 C1199) were coexpressed with RLC-D,D, they produced a depolarized cell with protrusions emanating around the cell, despite the continued presence of large actomyosin bundles (Fig. 6 A) and adhesions (not depicted). We also coexpressed a photoactivatable Rac (mVenus-tagged PA-Rac1; Wu et al., 2009) with RLC-D,D and then photoactivated Rac locally in the vicinity of the RLC-D,D–decorated bundles that go on to mark the rear. Protrusions emerged at the photoactivation site, i.e., in the vicinity of the RLC-D,D–decorated actomyosin bundles (Fig. 6 B). Furthermore, global photoactivation of mVenus-tagged PA-Rac in RLC-D,D–expressing cells led to round, largely depolarized cells (Fig. 6 C), similar to the effect of overexpressed activated Tiam1 or constitutively activated Rac (Fig. 6 A). Collectively, these observations show that Rac activation supersedes the polarizing effect of the RLC-D,D mutant. They also show that the downstream effectors of Rac that mediate the actin polymerization are available throughout the cell, including the rear, and point to inhibition of Rac itself as a major mechanism by which protrusion is inhibited at the rear.
Figure 6.
RLC-D,D restricts Rac activation and GEF localization to the front of polarized cells. (A) CHO.K1 cells were transfected with constitutively active Rac1 (myc-V12Rac1, top) or Tiam1 (HA-Tiam1 C1199, bottom) and RLC-D,D–GFP, plated onto fibronectin-coated coverslips (2 µg/ml, 60 min), stained for myc or HA, and imaged using a confocal microscope (FV300; Olympus). Representative morphologies are shown. (A, bottom) Axis ratio as defined in Fig. 2. Data are the mean ± SD of three independent experiments (error bars; n > 200). (B) CHO.K1 cells were transfected with RLC-D,D–mCherry (magenta) and mVenus-tagged PA-Rac (green), plated onto 5 µg/ml fibronectin for 15–30 min (initial phases of spreading and polarization), and photoactivated in the rear; e.g., region of RLC bundles (marked with a box). Photoactivation and imaging were done with a confocal microscope (FV1000; Olympus). Protrusion was observed by differential interference contrast to define the edge of the cell (bottom, white line). Image on the right depicts a color overlay before (green) and after (magenta) photoactivation (5 min) using mVenus fluorescence. Inset shows the detail of a fragment of the photoactivated area that displays the most robust protrusion. A representative experiment is shown. (C) CHO.K1 cells were transfected with RLC-D,D–mCherry (green) and PA-Rac (magenta), plated onto fibronectin as in Fig. 6 B, and photoactivated globally. Representative examples are on the left. (C, right) Data are the mean ± SEM (error bars) of the polarity axis scored in three independent experiments. P represents significance using a Student’s t test. (D) CHO.K1 cells were cotransfected with RLC wild type (top left) or RLC-D,D (bottom left) coupled to mCherry and the Raichu-Rac FRET sensor (right). Localized activation of Rac was visualized in a confocal microscope (FV1000) by imaging the intensity ratio image of YFP and CFP, which represents FRET efficiency. Note the lower FRET index at the rear as defined by the RLC-D,D bundles (arrows). FRET values outside of the cell contour were set to zero for representation. Note that the scales are different because of differences in expression levels and imaging parameters, resulting in differences in the absolute values of the fluorescence intensity. The arrowhead points to the region of higher Rac activity at the leading edge of the RLC-D,D cell, across from the rear. Representative cells are shown. Bar, 20 µm. (E) FRET quantification. Data are the mean ± SD (error bars) of the difference between FRET indices at the front and rear (RLC-D,D) and rear/sides (RLC-WT) from three independent experiments (RLC-D,D, n = 40; RLC-WT, n = 34). P represents significance using a Student’s t test. (F) CHO.K1 cells were transfected with wild-type RLC or RLC-D,D coupled to GFP (top) or mCherry (bottom) and mCherry-βPIX (top) or FLAG-DOCK180 (bottom), plated on fibronectin, fixed, stained with an anti-FLAG antibody (bottom), and imaged using confocal microscopy (FV300). Arrowheads point to clusters of βPIX and DOCK180, respectively, whereas arrows denote the RLC-D,D–decorated rears. Representative morphologies are shown. Bars: (A–C) 10 µm; (D and F) 20 µm.
To test this hypothesis, we localized Rac activation and the GEFs that activate Rac in control or RLC-D,D–expressing cells. A fluorescence resonance energy transfer (FRET)-based Rac biosensor revealed that Rac activation is lowest in the vicinity of the RLC-D,D–decorated actomyosin bundles that define the rear of the cell (Fig. 6, D and E). In contrast, cells expressing wild-type RLC, which were not as polarized, displayed a more modest difference in Rac activity between the leading protrusion and other regions of the cell (Fig. 6, D and E). Finally, pull-down assays of endogenous Rac revealed that the RLC-D,D mutant did not produce a major effect on the level of total active Rac (unpublished data).
Two GEFs—βPIX and DOCK180—are implicated in Rac activation at the front of migrating cells (Kiyokawa et al., 1998; Kunisaki et al., 2006; Nayal et al., 2006). Both βPIX and DOCK180 were highly polarized and localized primarily in adhesions near the leading edge of protrusions (Fig. 6 F, arrowheads) across from the RLC-D,D–decorated rears (Fig. 6 F, arrows). In addition, the small fraction (∼10–20%, Fig. S1) of wild-type RLC-expressing cells that displayed extended rears similar to those seen in RLC-D,D–expressing cells also showed an asymmetric distribution of the GEFs away from the actomyosin-enriched rears (unpublished data).
Adhesion protein phosphorylation and adaptor recruitment is reduced in adhesions associated with actomyosin bundles containing RLC-D,D
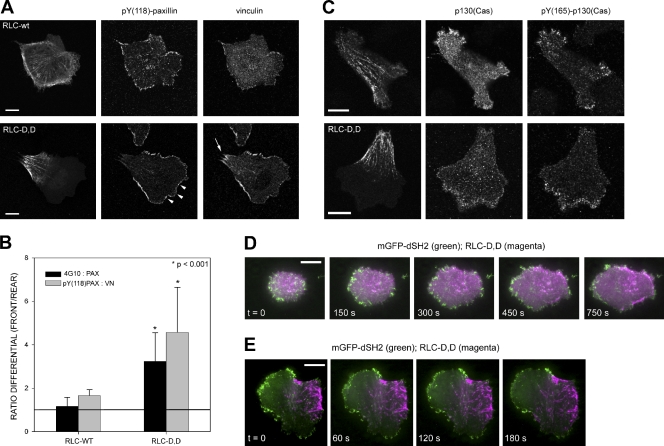
DOCK180 and βPIX are recruited to the plasma membrane through their interaction with the paxillin–p130(Cas)–CrkII and paxillin–GIT–PIX–PAK complexes, respectively (Matsuda et al., 1996; Gumienny et al., 2001; Nayal et al., 2006). Kinases like Src or focal adhesion kinase (FAK) phosphorylate adhesion adaptors, e.g., paxillin (Schaller and Parsons, 1995) and create docking sites that recruit these complexes to adhesions (Brown and Turner, 2004; Tomar and Schlaepfer, 2009). To determine whether RLC-D,D polarizes the phosphorylation of adhesion proteins, we assayed for tyrosine phosphorylation using the 4G10 mAb, a reagent with a broad specificity for phosphotyrosine. Like the GEFs, the phosphorylation was polarized toward the front, protruding region of the cell, opposite the RLC-D,D–induced rear (Fig. 7 B). Antibodies specific for phosphorylation on pY(397)-FAK, pY(31)-paxillin (not depicted), and pY(118)-paxillin (Fig. 7, A and B) also localized preferentially in the protruding front, opposite the RLC-D,D–containing rear.
Figure 7.
RLC-D,D–polarized cells create a region containing large adhesions with low Tyr phosphorylation at the rear and small, highly Tyr-phosphorylated, p130(Cas)-containing dynamic adhesions at the front. (A) CHO.K1 cells were transfected with wild-type RLC or RLC-D,D (fused to GFP), stained for phosphorylated Tyr118 paxillin and endogenous vinculin, and imaged with a confocal microscope (FV1000; Olympus). Arrowheads point to several clusters of pY(118)-paxillin at the leading edge; the arrow points to larger accumulations of vinculin adjacent to the RLC-D,D–decorated bundles. Representative morphologies are shown. (B) Quantification of the phosphorylation ratio using the endogenous staining pairs indicated. Data are the mean ± SD (error bars) of the ratio of the relative intensities at the front and the back (back and sides in wild-type cells). 12 cells/condition (adhesion, n > 50) were analyzed from three independent experiments. P represents significance using a Student’s t test. (C) CHO.K1 cells were transfected with wild-type RLC (top) or RLC-D,D (fused to mCherry, bottom) and GFP-p130(Cas), stained for phosphorylated Tyr(165)-p130(Cas), and imaged as in A. (D) TIRF microscopy time-lapse series of a CHO.K1 cell transfected with mGFP-dSH2 and RLC-D,D–mCherry and allowed to spread on fibronectin. Images were collected starting 5 min after plating. Video 7 shows the entire sequence. (E) TIRF microscopy time-lapse series of a CHO.K1 cell transfected with mGFP-dSH2 and RLC-D,D–mCherry migrating on fibronectin. The cell was allowed to spread and polarize for 35 min before image collection began. Video 8 shows the entire sequence. Bars, 10 µm.
We also determined whether the asymmetric distribution of these phosphorylations reflected the presence of the protein or their localized phosphorylation. Previous studies have shown large FAK and paxillin-containing adhesions at the ends of RLC-D,D–decorated actomyosin bundles at the rear (Vicente-Manzanares et al., 2008). These adhesions are only weakly phosphorylated, despite their size (Fig. 7, A and B). Thus, the phosphorylation, rather than the molecule itself, localizes preferentially at the leading edge and away from adhesions in which RLC-D,D actomyosin bundles terminate.
The CrkII–p130(Cas) complex requires phosphorylation of several adhesion proteins, e.g., paxillin on Tyr31 and Tyr118, for its recruitment to adhesions (Schaller and Parsons, 1995). In control, wild-type RLC-expressing cells, p130(Cas) localizes in small adhesions distributed around the periphery. In RLC-D,D–expressing cells, it was largely absent at the rear and instead localized primarily in the small adhesions that populate the leading edge (Fig. 7 C). Furthermore, staining with an antibody that recognizes p130(Cas) phosphorylated on Tyr165, which is a site that becomes available in response to mechanical stretch (Sawada et al., 2006), revealed a highly polarized distribution toward the leading edge and away from the large adhesions that terminate in RLC-D,D–decorated bundles (Fig. 7 C).
We then determined the relative kinetics of tyrosine phosphorylation and the acquisition of polarity using an SH2 domain sensor, comprised of tandem SH2 domains from Src coupled to mGFP (Kirchner et al., 2003) and driven by a truncated promoter (Watanabe and Mitchison, 2002), to minimize dominant interfering effects and ensure proper localization. This sensor binds to phosphorylated tyrosines with broad specificity (Kirchner et al., 2003) and reveals the dynamics of these phosphorylations. In spreading cells coexpressing the SH2 domain sensor and RLC-D,D–mCherry, the sensor localized along the entire edge of the cell; however, it disappeared from the vicinity of the proto-bundles as they began to form (Fig. 7 D and Video 7). In fully polarized cells, the SH2 domain sensor remained asymmetrically distributed, with the majority of the signal in the adhesions in protrusions (Fig. 7 E and Video 8); however, the signal persisted even when the protrusion stopped. Conversely, in cells depleted of MIIB, Tyr phosphorylation was not polarized, as revealed by the SH2 domain sensor or 4G10 staining, even in the presence of the RLC-D,D mutant (Fig. S4, A and B; and not depicted). An analogous, polarized distribution was also seen in U2OS cells, which have large, elongated adhesions in protrusions; interestingly the SH2 domain sensor was also present in regions where protrusion had paused, which suggests that phosphorylation is necessary but not sufficient for protrusion and showed robust retrograde flux away from the protrusion (unpublished data).
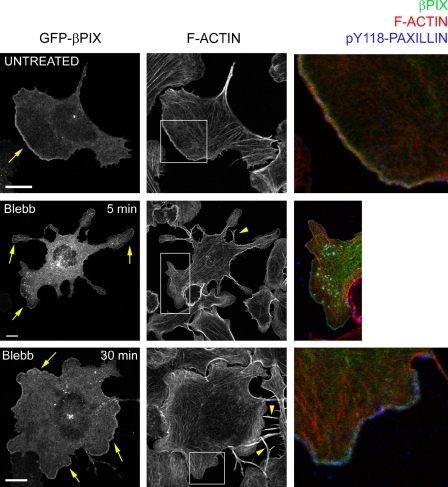
We also studied the localization of the signaling adaptor p130(Cas) and the Rac GEFs βPIX and DOCK180 in MIIB-deficient cells, in the presence or absence of RLC-D,D. Like the SH2 domain, both GEFs were evenly distributed around the cell periphery regardless of expression of the RLC-D,D mutant (Fig. S4 C), which indicates that MIIB activation by diphosphorylation of the RLC leads to regional exclusion of the Rac GEFs and the signaling adaptors. Similar results were obtained by treating cells plated for longer periods of time (2–5 h), when they reach maximal polarization, with the MII ATPase inhibitor blebbistatin (Fig. 8). 5 min after treatment, retraction defects appear (Fig. 8, arrowheads), and a fraction of the cells extend multidirectional, abnormally thin protrusions decorated with βPIX (and also DOCK180, not depicted) at their tips (arrows). After longer times (30–120 min), retraction defects become more apparent and most cell bodies are round and depolarized except for sparse actin bundles that remain. In these cells, the GEFs are evenly distributed around the cell periphery. Interestingly, co-staining with fluorescent phalloidin and an anti-phospho (Y118) paxillin antibody reveals that the GEFs concentrate near the cell edge where most of the phospho-paxillin is also localized (Fig. 8, color images).
Figure 8.
MII inhibition alters GEF localization. Cells expressing GFP-βPIX (green, right) and plated on fibronectin for 2 h were treated with 50 µM blebbistatin (Blebb) for the indicated time points, stained for actin (red) and pY118-paxillin (blue), and imaged using a confocal microscope (FV1000; Olympus). Arrowheads point to retraction defects; arrows point to the GEFs. Note the relatively even distribution of βPIX in the Blebb-treated cells. Color insets: detail (enlarged from the boxed regions) of the thin actin-rich band near the leading edge, where βPIX and paxillin phosphorylation (Y118) are more prominent. Bar, 10 µm.
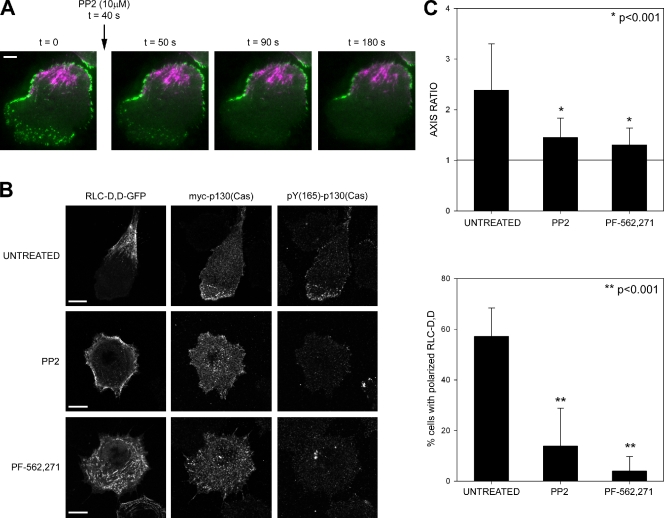
FAK and Src phosphorylate paxillin on Y(31),Y(118) and promote binding of the CrkII–p130(Cas) complex (Schaller and Parsons, 1995). To study their involvement in the polarized phosphorylation of paxillin induced by RLC-D,D, we treated RLC-D,D–expressing cells with the Src inhibitor PP2 or the FAK/Pyk2 inhibitor PF-562,271 (Roberts et al., 2008). Inhibition of either kinase abrogated Tyr phosphorylation at nascent adhesions, as revealed by disappearance of the GFP-SH2 sensor from the protrusions immediately after treatment (Fig. 9 A). They both also induced a global decrease of Tyr phosphorylation (4G10) and pY(118)-paxillin (unpublished data). Consistently, p130(Cas) was no longer polarized and phosphorylated on Tyr165 (Fig. 9 B). Surprisingly, long-term (>1 h) treatment with the drugs led to generalized adhesion maturation and enlargement (not depicted), and also inhibited the formation of localized, RLC-D,D–containing rears (Fig. 9 B). Actomyosin bundling was not impaired by the drug treatments; instead, the bundles distributed around the cell periphery (Fig. 9 C). Similar results were obtained using FAK knockdown cells (unpublished data). This suggests that FAK/Src activation is also required for maintaining the polarized rear.
Figure 9.
FAK and Src control the asymmetric distribution of adhesive signaling and actomyosin bundles to the front and rear, respectively. (A) TIRF microscopy time-lapse movie of a CHO.K1 cell expressing RLC-D,D–mCherry and mGFP-dSH2 migrating on fibronectin before and after treatment with the Src inhibitor PP2. Note the rapid disappearance of the GFP signal from the front. (B) Localization of p130(Cas) and phospho-Tyr(165) p130(Cas) in RLC-D,D–expressing cells treated with 10 µM PP2 (Src inhibitor) or 0.1 µM PF-562,271 (FAK inhibitor) imaged using a confocal microscope (FV1000; Olympus). Note the almost complete disappearance of phospho-Tyr(165) p130(Cas) in inhibited cells. Representative cells are shown. Bars, 10 µm. (C) Quantification of the axis ratio (top) and accumulation of RLC-D,D–containing bundles to the rear (bottom) in cells treated with 10 µM PP2 or 0.1 µM PF-562,271. n > 200 cells scored from three independent experiments. P represents significance using the nonparametric Mann-Whitney U test.
Together, these data provide direct support for the hypothesis that the large actomyosin filaments containing diphosphorylated RLC locally regulate Rac activation by inhibiting signals that lead to Rac GEF localization in the adhesions associated with the bundles. The mechanism by which the bundles inhibit the adhesive signaling, GEF localization, and Rac activation may not be direct, and is an important, unresolved challenge. The cell front, in contrast, has activated Rac and moves away from the Rac-inhibited rear, thereby polarizing the cell.
Discussion
Front–back polarization of migrating cells results in two defined regions: a protrusive area in the direction of migration and a retracting rear (Ridley et al., 2003). These regions reflect an asymmetry in cytoskeletal components. In protrusions, actin is primarily dendritic or in small bundles (Pollard and Borisy, 2003; Small et al., 2008), whereas it appears in larger, highly bundled filaments at the center and along the sides that form the rear of the cell (Cramer et al., 1997). The microtubules and nucleus are also polarized with the microtubule organizing center (MTOC) positioned in front of the nucleus (Etienne-Manneville and Hall, 2001; Gomes et al., 2005). Emerging evidence points to a key role for MII in front–back polarization. It resides preferentially in the rear of migrating keratocyte fragments, whereas nonmotile cells show an isotropic distribution (Verkhovsky et al., 1999). Local application of calyculin A, which is a general phosphatase inhibitor with some preference for MII phosphatase (Ishihara et al., 1989), at one side of migrating keratocytes induces protrusion at the opposite side, thereby producing a rear (Yam et al., 2007). Finally, ectopic expression of RLC-D,D, a phosphomimetic mutant of RLC, induces the formation of a well-defined, extended rear and increased directional migration in fibroblasts (Vicente-Manzanares et al., 2008).
The studies presented here and other recently published data converge on a mechanism for front–back polarity in migrating cells in which both MII isoforms cooperate to polarize the cell. Our working model proposes that: MIIA breaks the initial symmetry of the cell by assembling dynamic actomyosin proto-bundles and the adhesions with which they associate; MIIB stabilizes and enlarges the initial actomyosin proto-bundles and adhesions to create a stable rear; and the rear, and the sides that it defines, are maintained by large stable actin bundles and adhesions that do not activate Rac because of the absence of Rac GEFs and the activated scaffolds that recruit them.
MIIA and IIB synergize to initiate and stabilize the formation of the cell rear.
Recent evidence shows that the MII generates front–back polarity upstream of microtubules. MHCIIB−/− mouse embryo fibroblasts and shRNA-mediated MIIB knockdown cells do not exhibit a discernable front or back; instead, multiple protrusions emanate from around the periphery (Lo et al., 2004; Vicente-Manzanares et al., 2007). In addition, MIIB also positions the MTOC and microtubules, the Golgi apparatus, and the nucleus. In migrating fibroblasts, the MTOC and Golgi reside anterior to the nucleus. In MIIB-deficient cells, the Golgi is distributed around the nucleus, the MTOC is positioned randomly, and the nucleus rotates (Vicente-Manzanares et al., 2007). MIIA also contributes to polarity through its role in the retraction of protrusions and cell edges and the accompanying disassembly of adhesions (Even-Ram et al., 2007; Vicente-Manzanares et al., 2007).
The data presented here show that a well-defined rear is formed by a localized cluster, or proto-bundle, of actomyosin filaments away from which the cell protrudes. MIIA and MIIB cooperate to form this rear. MIIA alone forms unstable, dynamic actomyosin bundles and adhesions, resulting in transient polarity. MIIB does not form actomyosin bundles alone, but serves to enlarge and stabilize MIIA-containing bundles and associated adhesions. Diphosphorylation of RLC serves to produce a single, large stable bundle; in less polarized cells, the proto-bundles are more distributed and less focal. The formation of these primary actomyosin bundles is microtubule independent, which instead serves to position the front and determine the length of the cell. The different properties of MIIA and MIIB reside in the tail, rather than the actin-binding head domain. This region mediates myosin bundling and controls the differences in the biological properties of the isoforms, which appear to arise from the higher apparent affinity of MIIB for actomyosin bundles (Vicente-Manzanares et al., 2009). Although our studies have focused on MIIB, in some cells (e.g,, B16 melanomas), MIIC performs a similar, symmetry breaking function, but it generates a different morphology (Vicente-Manzanares et al., 2008).
Several studies support these observations. In keratocytes, the localized application of calyculin A, which activates MII, generates actin flow toward the region of application, presumably to create local actomyosin bundles as that region becomes a rear (Yam et al., 2007). Likewise, local treatment of endothelial cells with blebbistatin, which inhibits MII and disassembles myosin filaments, induces localized protrusion (Fischer et al., 2009). Finally, keratocyte fragments are motile only when MII is localized at the rear and are immobile when MII is isotropic (Verkhovsky et al., 1999).
MII regulates signaling by adhesions to generate front–back polarity by restricting Rac activation.
On migrating fibroblasts, the rear and the sides that form it are characterized by the absence of protrusions, low levels of Rac activity, and large actin bundles that terminate in large stable adhesions (Cramer et al., 1997; Kraynov et al., 2000). In these cells, Rac activation is regulated by GEFs, e.g., DOCK180 or βPIX (Rossman et al., 2005; García-Mata and Burridge, 2007). GEF-activated Rac, in turn, promotes actin polymerization (Joneson et al., 1996). The GEFs are recruited to adhesions by the phosphorylation of scaffold proteins like FAK, paxillin, and p130(Cas)/Crk-II (Brown and Turner, 2004; Defilippi et al., 2006; Tomar and Schlaepfer, 2009). Consequently, inhibiting Src, FAK, paxillin, or mutating their key phosphorylation sites affects protrusion and adhesion (Arthur et al., 2000; Webb et al., 2004; Zaidel-Bar et al., 2007). We have now implicated the inhibition of these pathways to Rac activation as an important mechanism by which phosphorylated RLC and MIIB create a stable rear. However, these pathways can also control MII activation in a reciprocal signaling loop that links MII, adhesive signaling, and Rac activation.
Much evidence points to the mechanosensitive activation of adhesion molecules, including integrin (Friedland et al., 2009), talin (del Rio et al., 2009), vinculin (Pasapera et al., 2010), or p130(Cas) (Sawada et al., 2006), as major mechanisms regulating signaling by adhesions. Force mapping of cells migrating on flexible substrates shows that mechanical forces are highest in protrusions at the cell front (Beningo et al., 2001; Munevar et al., 2001). In addition, more central adhesions do not appear to be under similar tension (Beningo et al., 2006; Gardel et al., 2008). Thus, forces at the front could activate adhesion molecules, which in turn would recruit GEFs to activate Rac and promote actin polymerization; conversely, lower tension in other regions would not recruit Rac GEFs. p130(Cas) phosphorylation is mechanosensitive, and it is part of a signaling complex that recruits DOCK180 to the plasma membrane (Kiyokawa et al., 1998). However, it remains possible that other mechanosensitive pathways are involved, as Src activation appears to be a key regulator acting upstream of DOCK180.
Although it is generally assumed that RLC activation produces high levels of tension, the high duty ratio, low ATPase-based contractility, and other features of MIIB might result in a state that produces large adhesions and actomyosin bundles without the higher contractility associated with MIIA. In support of this, expression of a noncontractile, cross-linking mutant of MIIB (R709C), which locks MIIB in an actin-bound state and thus cross-links actin, produces a phenotype very similar to that of RLC-D,D; e.g., large actomyosin bundles that form a polarized rear, and the absence of phosphorylation in their vicinity (unpublished data).
Conclusions.
It appears that migrating, fibroblast-like cells polarize by the local formation of stable actomyosin bundles that inhibit the adhesive signals that lead to Rac activation. The cell then protrudes away from these stable structures. Rac activation and the formation of a leading protrusion are regulated by adhesion-associated signaling complexes that assemble on signaling scaffolds like paxillin and FAK, which show greatly reduced tyrosine phosphorylation in the adhesions that define the rear.
This model suggests general principles that may pertain to other cell types. In epithelial sheets, for example, contact with adjacent cells might produce a contact region without adhesions that signal to Rac (Yamada and Nelson, 2007), thereby creating a functional “rear” and localizing Rac activation to protrusions in cell free regions. Similarly, contact inhibition of movement, in which a transient association of two cells inhibits local protrusive activity (Mayor and Carmona-Fontaine, 2010), might create a transient “rear” at the contact site. Finally, weakly adhesive, highly motile cells like leukocytes could polarize from the front through their response to a chemotactic gradient that locally activates Rac and forms a protrusion that moves away from the rest of the cell. In these cells, which express only MIIA (Jacobelli et al., 2004), the rear is made only of dynamic actomyosin bundles and forms a uropod that extends upward, rather than along the plane of the substratum.
Materials and methods
Plasmids
shRNA knockdown vectors for MHCII-A and MHCII-B as well as siRNA-insensitive GFP-MHCII-B have been described previously (Vicente-Manzanares et al., 2007). The plasmid containing the cytomegalovirus (CMV)-truncated (“speckle”) promoter was a gift from T. Mitchison (Harvard Medical School, Boston, MA; Watanabe and Mitchison, 2002). Where indicated, the CMV promoter was swapped with the truncated version. GFP-MHCII-A and GFP-MHCII-B were gifts from R.S. Adelstein (National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD; Wei and Adelstein, 2000). RLC-GFP (wild-type RLC) and RLC 18,19D-GFP (RLC-D,D–GFP) were provided by K. Kelly (National Cancer Institute, Bethesda, MD). Paxillin-GFP has been described previously (Laukaitis et al., 2001). Where indicated, GFP was replaced by mCherry, from R. Tsien (University of San Diego, San Diego, CA; Shaner et al., 2004). GFP-p130(Cas) was generated from myc-p130(Cas), which was a gift from A. Bouton (University of Virginia, Charlottesville, VA). FLAG-DOCK180 was obtained from K. Ravichandran (University of Virginia, Charlottesville, VA) and has been described previously (Brugnera et al., 2002). GFP- and mCherry-βPIX were generated from FLAG-βPIX (Mayhew et al., 2007). HA-Tiam1 C1199 was provided by J. Collard (Het Nederlands Kanker Instituut, Amsterdam, Netherlands; Michiels et al., 1995). FLAG-V12Rac was a gift from A. Hall (Memorial Sloan-Kettering Cancer Center, New York, NY; Ridley et al., 1992). mVenus-tagged PA-Rac1 and the photoactivation protocol used in this study have been described elsewhere (Wu et al., 2009). The Raichu-Rac probe was from M. Matsuda (Osaka University, Osaka, Japan; Itoh et al., 2002), and the mGFP-dSH2 sensor was generated from YFP-dSH2 (Kirchner et al., 2003), a gift from B. Geiger (Weizmann Institute of Science, Rehovot, Israel).
Antibodies and reagents
The following antibodies were used: MHCII-A and MHCII-B (rabbit, pAb) from Covance; p165Tyr-p130(Cas) (rabbit, pAb) from Cell Signaling Technology; myc and HA epitopes (9E10 and 12C5 mAbs, respectively) and paxillin (rabbit, pAb) from Santa Cruz Biotechnology, Inc.; FLAG (M2 mAb) epitope from Agilent Technologies; vinculin (hVin-1, mAb) from Sigma-Aldrich; 4G10 pan-phosphorylated Tyr (mAb) from Millipore; and pY(118)-paxillin (rabbit, pAb) from Invitrogen. Rhodamine-phalloidin was from Cytoskeleton. PP2 (used at 10 µM), blebbistatin (50 µM), nocodazole (5 µM), and vinblastine (0.1 µM) were from EMD; PF-562,271 (FAK/Pyk2 inhibitor, used at 0.1 µM) was from Pfizer and a gift of J.T. Parsons (University of Virginia, Charlottesville, VA).
Cell culture and transfection
CHO-K1 cells (American Type Culture Collection) were cultured in low-glucose DME supplemented with 10% FBS, 4 mM l-glutamine, 1 mM sodium pyruvate, 1% (vol/vol) nonessential amino acids, and penicillin/streptomycin, and transfected with 0.25–1 µg DNA using Lipofectamine (Invitrogen). For knockdown experiments, plasmids containing the shRNA sequences were used in a 10:1 (1 µg:0.1 µg) excess to GFP- or mCherry-containing plasmids to ensure knockdown in fluorescence-positive cells, which was verified by immunofluorescence in parallel samples. For imaging assays, cells were plated on glass-bottomed dishes, pre-incubated overnight with 2 µg/ml fibronectin in CCM1 for the indicated times, and maintained at 37°C at pH 7.4 (migration-promoting conditions).
Immunofluorescence
Cells were allowed to adhere to 2 µg/ml fibronectin-coated coverslips or 100 µg/ml PLL for the following times: analysis of proto-bundle formation, 10 min on fibronectin or 10–180 min on PLL (shown at 45 min); rest of conditions, 60 min on fibronectin, except as indicated. Samples were fixed using 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 for 5 min. The coverslips were blocked using IgG-free BSA (Sigma-Aldrich), and incubated with primary antibodies (see the Antibodies section for dilutions) and a species-appropriate secondary antibody coupled to Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 647 (Invitrogen).
Microscopy and image processing
Confocal images were collected on a microscope (Fluoview 300; Olympus) using a 60×/1.45 NA (oil) Plan-Apochromat 60× OTIRFM objective lens (Olympus). GFP and mCherry were excited using the 488 nm laser line of an Ar ion laser and the 543 nm laser line of a He-Ne laser (CVI Melles Griot), respectively. Fluorescence emission was collected using SDM560 and BA505-525 (EGFP) and BA560-660 (mCherry) filters. Some of the two-color (EGFP or Alexa Fluor 488/Alexa Fluor 568 or mCherry or rhodamine-phalloidin) as well as the three-color fluorescence images of EGFP/Alexa Fluor 568 (or mCherry)/Alexa Fluor 647 were collected in sequential mode in a microscope (Fluoview 1000; Olympus) using a 60×/1.35 NA (oil) UPlan-Sapo objective lens. Alexa Fluor 647 was excited with a 633 nm He-Ne laser set, and emission was collected using a variable long-pass filter set at >650 nm. The use of one or the other confocal microscope is indicated under each individual figure. Fixed and stained samples were acquired at room temperature using Vectashield (Vector) as imaging medium. Live samples were acquired at 37°C using CCM1 medium. Confocal fluorescence and differential interference contrast images were acquired using Fluoview software (Olympus) and displayed using ImageJ in agreement with the JCB guidelines.
ImageJ was also used to measure length, thickness, and intensity of adhesions and actin bundles. To quantify cell polarity (axis ratio), cells were fixed and stained, and images were acquired in a confocal microscope as described previously. For each individual cell, the axis ratio was calculated by dividing the length of the long, migration-defined axis by the perpendicular axis passing by the nucleus of the cell. Cells with proto-bundles were quantified by presence of a cluster of pRLC (untransfected cells) or localized RLC WT/D,D-GFP in knockdown cells.
Total internal reflection fluorescence (TIRF) images were acquired on an inverted microscope (IX70; Olympus) with a 1.45 NA (oil) Plan-Apochromat 60× TIRFM objective lens, fitted with a Ludl modular automation controller (Ludl Electronic Products), and controlled by MetaMorph software (Invitrogen). The excitation laser lines used were as described for confocal microscopy. A dichroic mirror (HQ485/30) was used for GFP-labeled cells. For dual GFP-mCherry/mOrange, a dual-emission filter (z488/543) was used. Images were acquired with a charge-coupled device (CCD) camera (Retiga Exi; Qimaging) and analyzed using MetaMorph. Adhesion turnover quantification was performed as described previously (Webb et al., 2004).
FRET
CHO-K1 cells were cotransfected with the Raichu-Rac probe (Itoh et al., 2002) and RLC-D,D–mCherry, and plated on 2 µg/ml fibronectin-coated glass bottom dishes in CCM1 for 20–30 min at 37°C. A confocal image time series was acquired with a microscope (Fluoview 1000) using a 60× 1.35 NA oil UPlan-Sapo objective lens. For RLC-D,D–mCherry excitation, we used a He-Ne laser with a 543 nm line. Fluorescence emission was collected using a DM BP 560–660-nm filter. For dual-emission ratio imaging of Raichu-Rac, we used the 458 nm line of an Ar ion laser for CFP excitation and DM510, BA480-495 nm, and BA535-565 nm filters for the collection of CFP and YFP fluorescence emission, respectively. 256 × 256 pixel images were collected at 2× optical zoom (0.414 µm/pixel). YFP/CFP intensity ratio images were calculated as described previously (Itoh et al., 2002), and FRET values outside of the cell contour were set to zero for representation purposes.
Online supplemental material
Fig. S1 shows that proto-bundles containing wild-type RLC are more dynamic and evolve to form extended rears less frequently than those containing RLC-D,D. Fig. S2 shows that actomyosin-containing proto-bundles on cells adhering to PLL do not show prominent accumulations of adhesion proteins or fibronectin. Fig. S3 show that MIIA knockdowns show a dose-dependent decrease in visible actomyosin bundles. Fig. S4 shows that tyrosine phosphorylation and Rac GEFs are evenly distributed in MIIB-deficient cells. Video 1 shows that RLC-D,D localizes and determines the rear of spreading cells. Video 2 shows that RLC-D,D cannot determine the rear in MIIB-depleted cells. Video 3 show that MT inhibition alters the location of the cell front. Video 4 shows that MIIB incorporates into actomyosin bundles after MIIA. Video 5 shows adhesions associated with MIIA turnover. Video 6 shows that MIIB inhibits adhesion turnover. Video 7 shows that tyrosine phosphorylation, as revealed by GFP-SH2, becomes asymmetric during spreading concomitant with RLC-D,D assembly. Video 8 shows that tyrosine phosphorylation, as revealed by GFP-SH2, is asymmetrically distributed in RLC-D,D–polarized cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201012159/DC1.
Acknowledgments
We thank Robert Adelstein, Tim Mitchison, Kathleen Kelly, Kodi Ravichandran, Roger Tsien, Michiyuki Matsuda, Amy Bouton, Yi Wu, Klaus Hahn, Alan Hall, John Collard, and Benjamin Geiger for generously sharing plasmids (see Materials and methods for a complete list), and J. T. Parsons for the FAK/Pyk2 inhibitor PF-562,271, from Pfizer. We also thank Emilio Tejera for helpful discussions.
This work was supported by National Institutes of Health grant GM23244 and the Cell Migration Consortium (U54 GM064346).
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- FAK
- focal adhesion kinase
- FRET
- fluorescence resonance energy transfer
- MHC
- myosin heavy chain
- MTOC
- microtubule organizing center
- PLL
- poly-l-lysine
- RLC
- regulatory light chain
- shRNA
- small hairpin RNA
- TIRF
- total internal reflection fluorescence
References
- Arthur W.T., Petch L.A., Burridge K. 2000. Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr. Biol. 10:719–722 10.1016/S0960-9822(00)00537-6 [DOI] [PubMed] [Google Scholar]
- Beningo K.A., Dembo M., Kaverina I., Small J.V., Wang Y.L. 2001. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153:881–888 10.1083/jcb.153.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beningo K.A., Hamao K., Dembo M., Wang Y.L., Hosoya H. 2006. Traction forces of fibroblasts are regulated by the Rho-dependent kinase but not by the myosin light chain kinase. Arch. Biochem. Biophys. 456:224–231 10.1016/j.abb.2006.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.C., Turner C.E. 2004. Paxillin: adapting to change. Physiol. Rev. 84:1315–1339 10.1152/physrev.00002.2004 [DOI] [PubMed] [Google Scholar]
- Brugnera E., Haney L., Grimsley C., Lu M., Walk S.F., Tosello-Trampont A.C., Macara I.G., Madhani H., Fink G.R., Ravichandran K.S. 2002. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4:574–582 [DOI] [PubMed] [Google Scholar]
- Choi C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., Horwitz A.R. 2008. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10:1039–1050 10.1038/ncb1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M., Burridge K. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133:1403–1415 10.1083/jcb.133.6.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer L.P. 2010. Forming the cell rear first: breaking cell symmetry to trigger directed cell migration. Nat. Cell Biol. 12:628–632 10.1038/ncb0710-628 [DOI] [PubMed] [Google Scholar]
- Cramer L.P., Siebert M., Mitchison T.J. 1997. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 136:1287–1305 10.1083/jcb.136.6.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defilippi P., Di Stefano P., Cabodi S. 2006. p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol. 16:257–263 10.1016/j.tcb.2006.03.003 [DOI] [PubMed] [Google Scholar]
- del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J.M., Sheetz M.P. 2009. Stretching single talin rod molecules activates vinculin binding. Science. 323:638–641 10.1126/science.1162912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 106:489–498 10.1016/S0092-8674(01)00471-8 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. 2003. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr. Opin. Cell Biol. 15:67–72 10.1016/S0955-0674(02)00005-4 [DOI] [PubMed] [Google Scholar]
- Even-Ram S., Doyle A.D., Conti M.A., Matsumoto K., Adelstein R.S., Yamada K.M. 2007. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 9:299–309 10.1038/ncb1540 [DOI] [PubMed] [Google Scholar]
- Fischer R.S., Gardel M., Ma X., Adelstein R.S., Waterman C.M. 2009. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr. Biol. 19:260–265 10.1016/j.cub.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland J.C., Lee M.H., Boettiger D. 2009. Mechanically activated integrin switch controls alpha5beta1 function. Science. 323:642–644 10.1126/science.1168441 [DOI] [PubMed] [Google Scholar]
- García-Mata R., Burridge K. 2007. Catching a GEF by its tail. Trends Cell Biol. 17:36–43 10.1016/j.tcb.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Gardel M.L., Sabass B., Ji L., Danuser G., Schwarz U.S., Waterman C.M. 2008. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 183:999–1005 10.1083/jcb.200810060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., Gundersen G.G. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Gumienny T.L., Brugnera E., Tosello-Trampont A.C., Kinchen J.M., Haney L.B., Nishiwaki K., Walk S.F., Nemergut M.E., Macara I.G., Francis R., et al. 2001. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 107:27–41 10.1016/S0092-8674(01)00520-7 [DOI] [PubMed] [Google Scholar]
- Ishihara H., Martin B.L., Brautigan D.L., Karaki H., Ozaki H., Kato Y., Fusetani N., Watabe S., Hashimoto K., Uemura D., et al. 1989. Calyculin A and okadaic acid: inhibitors of protein phosphatase activity. Biochem. Biophys. Res. Commun. 159:871–877 10.1016/0006-291X(89)92189-X [DOI] [PubMed] [Google Scholar]
- Itoh R.E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., Matsuda M. 2002. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22:6582–6591 10.1128/MCB.22.18.6582-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanicki M.P., Vomastek T., Tilghman R.W., Martin K.H., Banerjee J., Wedegaertner P.B., Parsons J.T. 2008. FAK, PDZ-RhoGEF and ROCKII cooperate to regulate adhesion movement and trailing-edge retraction in fibroblasts. J. Cell Sci. 121:895–905 10.1242/jcs.020941 [DOI] [PubMed] [Google Scholar]
- Jacobelli J., Chmura S.A., Buxton D.B., Davis M.M., Krummel M.F. 2004. A single class II myosin modulates T cell motility and stopping, but not synapse formation. Nat. Immunol. 5:531–538 10.1038/ni1065 [DOI] [PubMed] [Google Scholar]
- Joneson T., McDonough M., Bar-Sagi D., Van Aelst L. 1996. RAC regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 274:1374–1376 10.1126/science.274.5291.1374 [DOI] [PubMed] [Google Scholar]
- Kim K.Y., Kovács M., Kawamoto S., Sellers J.R., Adelstein R.S. 2005. Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J. Biol. Chem. 280:22769–22775 10.1074/jbc.M503488200 [DOI] [PubMed] [Google Scholar]
- Kirchner J., Kam Z., Tzur G., Bershadsky A.D., Geiger B. 2003. Live-cell monitoring of tyrosine phosphorylation in focal adhesions following microtubule disruption. J. Cell Sci. 116:975–986 10.1242/jcs.00284 [DOI] [PubMed] [Google Scholar]
- Kiyokawa E., Hashimoto Y., Kobayashi S., Sugimura H., Kurata T., Matsuda M. 1998. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 12:3331–3336 10.1101/gad.12.21.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. 1998. Cytoplasmic dynamics of myosin IIA and IIB: spatial ‘sorting’ of isoforms in locomoting cells. J. Cell Sci. 111:2085–2095 [DOI] [PubMed] [Google Scholar]
- Kolega J. 2003. Asymmetric distribution of myosin IIB in migrating endothelial cells is regulated by a rho-dependent kinase and contributes to tail retraction. Mol. Biol. Cell. 14:4745–4757 10.1091/mbc.E03-04-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov V.S., Chamberlain C., Bokoch G.M., Schwartz M.A., Slabaugh S., Hahn K.M. 2000. Localized Rac activation dynamics visualized in living cells. Science. 290:333–337 10.1126/science.290.5490.333 [DOI] [PubMed] [Google Scholar]
- Kunisaki Y., Nishikimi A., Tanaka Y., Takii R., Noda M., Inayoshi A., Watanabe K., Sanematsu F., Sasazuki T., Sasaki T., Fukui Y. 2006. DOCK2 is a Rac activator that regulates motility and polarity during neutrophil chemotaxis. J. Cell Biol. 174:647–652 10.1083/jcb.200602142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukaitis C.M., Webb D.J., Donais K., Horwitz A.F. 2001. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 153:1427–1440 10.1083/jcb.153.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C.M., Buxton D.B., Chua G.C., Dembo M., Adelstein R.S., Wang Y.L. 2004. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol. Biol. Cell. 15:982–989 10.1091/mbc.E03-06-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Ota S., Tanimura R., Nakamura H., Matuoka K., Takenawa T., Nagashima K., Kurata T. 1996. Interaction between the amino-terminal SH3 domain of CRK and its natural target proteins. J. Biol. Chem. 271:14468–14472 10.1074/jbc.271.24.14468 [DOI] [PubMed] [Google Scholar]
- Maupin P., Phillips C.L., Adelstein R.S., Pollard T.D. 1994. Differential localization of myosin-II isozymes in human cultured cells and blood cells. J. Cell Sci. 107:3077–3090 [DOI] [PubMed] [Google Scholar]
- Mayhew M.W., Jeffery E.D., Sherman N.E., Nelson K., Polefrone J.M., Pratt S.J., Shabanowitz J., Parsons J.T., Fox J.W., Hunt D.F., Horwitz A.F. 2007. Identification of phosphorylation sites in betaPIX and PAK1. J. Cell Sci. 120:3911–3918 10.1242/jcs.008177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R., Carmona-Fontaine C. 2010. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 20:319–328 10.1016/j.tcb.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels F., Habets G.G., Stam J.C., van der Kammen R.A., Collard J.G. 1995. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 375:338–340 10.1038/375338a0 [DOI] [PubMed] [Google Scholar]
- Montell D.J. 2008. Morphogenetic cell movements: diversity from modular mechanical properties. Science. 322:1502–1505 10.1126/science.1164073 [DOI] [PubMed] [Google Scholar]
- Mseka T., Coughlin M., Cramer L.P. 2009. Graded actin filament polarity is the organization of oriented actomyosin II filament bundles required for fibroblast polarization. Cell Motil. Cytoskeleton. 66:743–753 10.1002/cm.20403 [DOI] [PubMed] [Google Scholar]
- Munevar S., Wang Y.L., Dembo M. 2001. Distinct roles of frontal and rear cell-substrate adhesions in fibroblast migration. Mol. Biol. Cell. 12:3947–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayal A., Webb D.J., Brown C.M., Schaefer E.M., Vicente-Manzanares M., Horwitz A.R. 2006. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 173:587–589 10.1083/jcb.200509075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent C.A., Devreotes P.N. 1999. A cell’s sense of direction. Science. 284:765–770 10.1126/science.284.5415.765 [DOI] [PubMed] [Google Scholar]
- Pasapera A.M., Schneider I.C., Rericha E., Schlaepfer D.D., Waterman C.M. 2010. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188:877–890 10.1083/jcb.200906012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Paterson H.F., Johnston C.L., Diekmann D., Hall A. 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 70:401–410 10.1016/0092-8674(92)90164-8 [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Roberts W.G., Ung E., Whalen P., Cooper B., Hulford C., Autry C., Richter D., Emerson E., Lin J., Kath J., et al. 2008. Antitumor activity and pharmacology of a selective focal adhesion kinase inhibitor, PF-562,271. Cancer Res. 68:1935–1944 10.1158/0008-5472.CAN-07-5155 [DOI] [PubMed] [Google Scholar]
- Rossman K.L., Der C.J., Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- Sawada Y., Tamada M., Dubin-Thaler B.J., Cherniavskaya O., Sakai R., Tanaka S., Sheetz M.P. 2006. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 127:1015–1026 10.1016/j.cell.2006.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M.D., Parsons J.T. 1995. pp125FAK-dependent tyrosine phosphorylation of paxillin creates a high-affinity binding site for Crk. Mol. Cell. Biol. 15:2635–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Small J.V., Resch G.P. 2005. The comings and goings of actin: coupling protrusion and retraction in cell motility. Curr. Opin. Cell Biol. 17:517–523 10.1016/j.ceb.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Small J.V., Auinger S., Nemethova M., Koestler S., Goldie K.N., Hoenger A., Resch G.P. 2008. Unravelling the structure of the lamellipodium. J. Microsc. 231:479–485 10.1111/j.1365-2818.2008.02060.x [DOI] [PubMed] [Google Scholar]
- Tomar A., Schlaepfer D.D. 2009. Focal adhesion kinase: switching between GAPs and GEFs in the regulation of cell motility. Curr. Opin. Cell Biol. 21:676–683 10.1016/j.ceb.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G., Wu Y., Sasaki Y., Hartshorne D.J., Yamakita Y., Yamashiro S., Matsumura F. 2004. Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J. Cell Biol. 164:427–439 10.1083/jcb.200306172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P.J., Devreotes P.N. 2004. Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 5:626–634 10.1038/nrm1435 [DOI] [PubMed] [Google Scholar]
- Verkhovsky A.B., Svitkina T.M., Borisy G.G. 1999. Self-polarization and directional motility of cytoplasm. Curr. Biol. 9:11–20 10.1016/S0960-9822(99)80042-6 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Zareno J., Whitmore L., Choi C.K., Horwitz A.F. 2007. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 176:573–580 10.1083/jcb.200612043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Koach M.A., Whitmore L., Lamers M.L., Horwitz A.F. 2008. Segregation and activation of myosin IIB creates a rear in migrating cells. J. Cell Biol. 183:543–554 10.1083/jcb.200806030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R.S., Horwitz A.R. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Mitchison T.J. 2002. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 295:1083–1086 10.1126/science.1067470 [DOI] [PubMed] [Google Scholar]
- Webb D.J., Donais K., Whitmore L.A., Thomas S.M., Turner C.E., Parsons J.T., Horwitz A.F. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154–161 10.1038/ncb1094 [DOI] [PubMed] [Google Scholar]
- Wei Q., Adelstein R.S. 2000. Conditional expression of a truncated fragment of nonmuscle myosin II-A alters cell shape but not cytokinesis in HeLa cells. Mol. Biol. Cell. 11:3617–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.I., Frey D., Lungu O.I., Jaehrig A., Schlichting I., Kuhlman B., Hahn K.M. 2009. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 461:104–108 10.1038/nature08241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wang F., Van Keymeulen A., Herzmark P., Straight A., Kelly K., Takuwa Y., Sugimoto N., Mitchison T., Bourne H.R. 2003. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 114:201–214 10.1016/S0092-8674(03)00555-5 [DOI] [PubMed] [Google Scholar]
- Yam P.T., Wilson C.A., Ji L., Hebert B., Barnhart E.L., Dye N.A., Wiseman P.W., Danuser G., Theriot J.A. 2007. Actin-myosin network reorganization breaks symmetry at the cell rear to spontaneously initiate polarized cell motility. J. Cell Biol. 178:1207–1221 10.1083/jcb.200706012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Nelson W.J. 2007. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J. Cell Biol. 178:517–527 10.1083/jcb.200701058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R., Milo R., Kam Z., Geiger B. 2007. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 120:137–148 10.1242/jcs.03314 [DOI] [PubMed] [Google Scholar]