PKA RIα subunit is localized to MVBs by the A-kinase–anchoring protein AKAP11 when disassociated from the PKA catalytic subunit.
Abstract
Although RII protein kinase A (PKA) regulatory subunits are constitutively localized to discrete cellular compartments through binding to A-kinase–anchoring proteins (AKAPs), RI subunits are primarily diffuse in the cytoplasm. In this paper, we report a novel AKAP-dependent localization of RIα to distinct organelles, specifically, multivesicular bodies (MVBs). This localization depends on binding to AKAP11, which binds tightly to free RIα or RIα in complex with catalytic subunit (holoenzyme). However, recruitment to MVBs occurs only with the release of PKA catalytic subunit (PKAc). This recruitment is reversed by reassociation with PKAc, and it is disrupted by the presence of AKAP peptides, mutations in the RIα AKAP-binding site, or knockdown of AKAP11. Cyclic adenosine monophosphate binding not only unleashes active PKAc but also leads to the targeting of AKAP11:RIα to MVBs. Therefore, we show that the RIα holoenzyme is part of a signaling complex with AKAP11, in which AKAP11 may direct RIα functionality after disassociation from PKAc. This model defines a new paradigm for PKA signaling.
Introduction
Although PKA has been extensively studied for decades, the focus has mainly been on its catalytic subunit activity or the PKA holoenzyme (heterotetramers of regulatory and catalytic subunits). The role of its regulatory subunits (R-subunits) that is independent of the PKA catalytic subunits (PKAc’s) has not been well characterized. The classically known functions of R-subunits are to inhibit PKAc and facilitate in targeting PKAc to compartmentalized regions of the cell by binding to A-kinase–anchoring proteins (AKAPs) at its N-terminal domain (dimerization/docking [D/D] domain). Through the binding of cAMP at the C-terminal domains of R-subunits, PKAc is released from the holoenzyme complex and activated to phosphorylate downstream substrates.
Two general classes of R-subunits exist, RI and RII (Reimann et al., 1971), which can be further resolved at the genetic level into four separate gene products: RIα, RIβ, RIIα, and RIIβ (Lee et al., 1983; Jahnsen et al., 1986; Scott et al., 1987; Clegg et al., 1988). Although over 50 AKAPs have been identified (Wong and Scott, 2004), the majority of them bind to RII subunits so that RII subunits are typically associated with membranous organelles. Meanwhile, because RI subunits are mainly diffused in the cytoplasm, the localization of the RI holoenzyme has received less attention and is not well appreciated. Several studies have indicated that RIα, probably associated with PKAc, can be recruited to specific sites, for example, to the “cap” site of activated T lymphocytes (Levy et al., 1996). The RI holoenzyme also localized to microtubules during the entire cell cycle (Imaizumi-Scherrer et al., 2001) and to protrusions at the front of migrating cells through its interaction with α4 integrin (Lim et al., 2007).
Although free RI was previously reported to be less stable when not in complex with PKAc (Steinberg and Agard, 1981; Orellana and McKnight, 1990), more recent studies have revealed that free RI subunits localize to subcellular sites beyond the cytoplasm. For example, some studies have shown the association of RI with membranes (Rubin, 1979; Boeshans et al., 1999). Another study reported that, independent of PKAc, the N terminus of RIα, but not RII, was needed to bind RFC40 and transport it into the nucleus (Gupte et al., 2005). In addition, this translocation from the cytoplasm to the nucleus has been correlated with the formation of the interchain disulfide bonds found in the D/D domain of RI but not found in RII (Brennan et al., 2006). Mavrakis et al. (2006) also reported the localization of RIα to late endosomes and autophagosomes.
Because the function of a protein is closely linked to its subcellular location, the different localizations of R-subunit isoforms could explain some of their known functional differences. RIα is the only isoform that acted as a tissue-specific extinguisher (Boshart et al., 1991; Jones et al., 1991) and the only one that has been pursued as an anticancer drug target (Chen et al., 2000; Goel et al., 2006). Its down-regulation or overexpression disrupted normal cell cycle progression (Tortora et al., 1994a,b). It was also the only isoform that resulted in embryonic lethality when deleted in mice (Amieux and McKnight, 2002). On the other hand, deletion of RIβ resulted in defects in hippocampal function (Brandon et al., 1995). Deletion of RIIα showed no gross organ dysfunction (Burton et al., 1999), whereas deletion of RIIβ resulted in several changes, such as a lean phenotype with elevated body temperatures and metabolic rates (McKnight et al., 1998) or a decreased sensitivity to increased ethanol consumption (Thiele et al., 2000). Thus, although four isoforms of the R-subunits exist, they are not functionally redundant.
RIα is also the only isoform that can compensate for excess PKAc activity. When PKAc was overexpressed in NIH 3T3 cells, RIα levels increased with no change in RII subunit levels (Uhler and McKnight, 1987). RIα knockout mice were no longer embryonically lethal when PKAc was also knocked out, but the double knockout mice displayed developmental defects later in life (Amieux and McKnight, 2002). Thus, a tightly coordinated regulation exists between RIα and PKAc that is crucial for normal development.
RIα may also have functions independent of PKAc. Overexpression of RIα is associated with breast cancer (Miller, 2002), whereas underexpression of RIα is linked to systemic lupus erythematosus (Kammer et al., 1996; Laxminarayana et al., 1999) and Carney complex (Kirschner et al., 2000). Mutations in RIα that did not affect RIα expression levels or PKAc activity still resulted in Carney complex (Veugelers et al., 2004). In light of these findings, the localization of RIα in the absence of PKAc is important to characterize.
In this study, we investigate a novel localization of RIα that was observed after expression in mammalian cells. In contrast to the other proteins we expressed, RIα formed puncta. The aims of our study were to characterize these puncta and determine their requirements for formation. By expressing RIα-GFP in HeLa and RIα knockout mouse embryonic fibroblasts (MEFs), we showed that these puncta were RI specific. Furthermore, we established that it was free RI subunit that was required for formation, as overexpressed RIα holoenzyme was cytoplasmic and showed no evidence of puncta formation. Upon stimulation with forskolin/isobutylmethylxanthine (IBMX), however, puncta formation occurred and was reversible for up to 3 h. Using fractionation and correlated light and electron microscopy, we established that RIα puncta localized to multivesicular bodies (MVBs). Through mutagenesis experiments or coexpression with isoform-specific AKAP-disrupting peptides, we demonstrated that AKAP binding was essential for this localization. In addition, our knockdown experiments established that it required a specific AKAP, AKAP11 (also known as AKAP220). Finally, we determined that the endogenous RIα holoenzyme, though diffuse in the cytoplasm, was still tightly associated with AKAP11; however, translocation to MVBs only occurred when PKAc was released. Therefore, in contrast to RII subunits, which appear constitutively localized to discrete regions of the cell via high affinity binding to AKAPs, RIα is assembled with AKAP11 as a soluble and cytoplasmic signaling complex. Then, upon stimulation with cAMP, AKAP11:RIα can be dynamically recruited to specific sites. We thus show that AKAP11 is highly specific for RIα and also define a new paradigm for RIα signaling and targeting through AKAP-mediated mechanisms.
Results
Punctate pattern of overexpressed RIα is RI specific
Type I PKA holoenzyme and free RIα are predominantly thought to be cytoplasmic; however, when we expressed GFP-tagged RIα in HeLa cells, we found a distinct punctate pattern (Fig. 1 A, top right). When we expressed RIα in RIα knockout MEFs (Prkar1a−/− MEFs), we saw a similar punctate pattern (Fig. 1 A, bottom right). In both cell lines, we observed the punctate pattern even when expression levels were low. To further confirm that puncta formation was not an artifact of overexpression, we used quantitative Western blot analysis to compare the levels of transfected RIα-GFP in Prkar1a−/− MEFs to endogenous levels of RIα in Prkar1a+/+ MEFs (Fig. S1). Transfection efficiency of RIα-GFP in Prkar1a−/− MEFs was typically around 15–30%. Based on this efficiency and the difference in band intensities between RIα-GFP versus endogenous RIα (15.4-fold for experiment 1 and 18.1-fold for experiment 2), the expression levels of RIα-GFP with 100% transfection efficiency would range from 2.3- to 5.4-fold lower than the levels of endogenous RIα. Although these values are approximations, the results clearly demonstrate that RIα-GFP was not overexpressed in relation to the levels of endogenous RIα in Prkar1a+/+ MEFs. In the remainder of this paper, the term “overexpression” is still used to indicate exogenous expression but is not indicative of the true levels of expression.
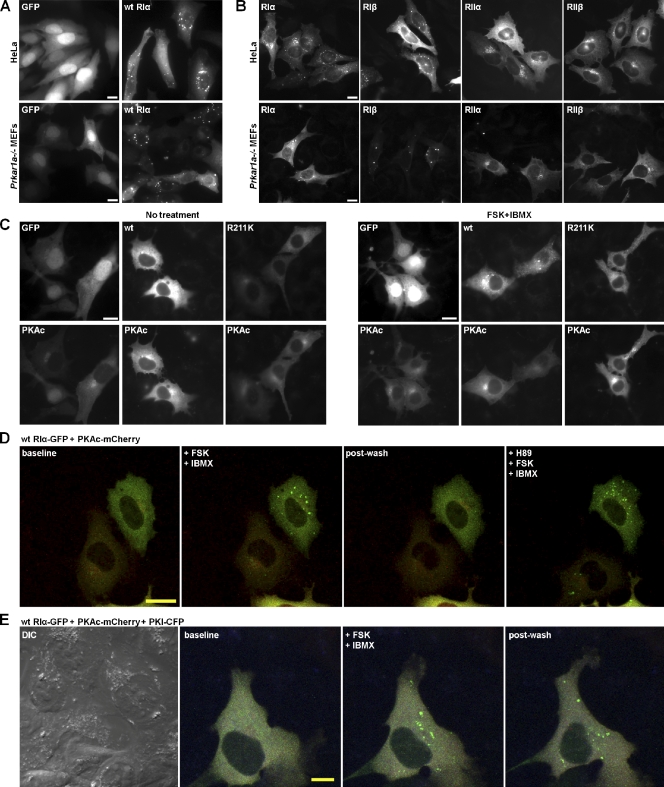
Figure 1.
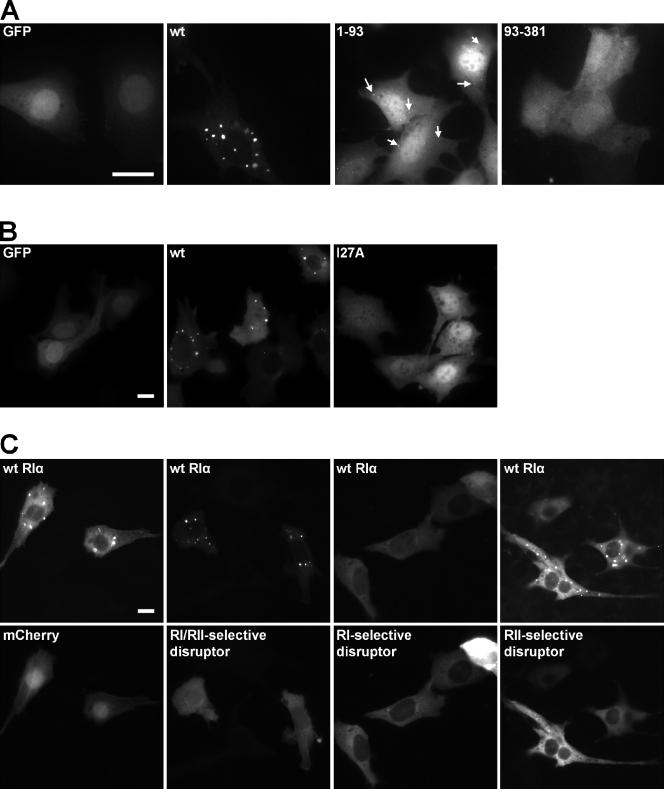
Expression of wild-type RIα in mammalian cells revealed a punctate pattern that is RI specific. (A) Transient transfection of control GFP empty vector or GFP-tagged RIα in HeLa cells (top) and Prkar1a−/− MEFs (bottom). (B) Expression of fluorescently tagged R-subunit isoforms in HeLa cells (top) and Prkar1a−/− MEFs (bottom). (C) Coexpression of control GFP empty vector or GFP-tagged RIα with mCherry-tagged PKAc in Prkar1a−/− MEFs without treatment (left) and with 20 µM forskolin/200 µM IBMX treatment (right). Labels in the top left corner of each image indicate which protein is observed. (D) Representative frames from Video 1 showing Prkar1a−/− MEFs transiently transfected with GFP-tagged RIα (green) and mCherry-tagged PKAc (red). 10 µM H89 was added to inhibit PKAc activity (last representative frame). (E) Representative frames from Video 3 showing Prkar1a−/− MEFs transiently transfected with GFP-tagged RIα (green), mCherry-tagged PKAc (red), and CFP-tagged PKI (blue). (D and E) Baseline represents the cell before any treatment, whereas postwash represents the cell after removal of treatment. 20 µM forskolin/200 µM IBMX was added to activate PKAc. DIC, differential interference contrast. wt, wild type. FSK, forskolin. Bars, 20 µm.
Four isoforms of the PKA R-subunits exist, and they display different localization patterns. We therefore next explored whether puncta formation was common to all R-subunits or unique to the RIα isoform. Constructs of the four R-subunit isoforms were expressed in HeLa cells and Prkar1a−/− MEFs (Fig. 1 B). Overexpression of the RI isoforms showed a punctate pattern, whereas overexpression of the RII isoforms produced perinuclear localization. Therefore, the punctate pattern was an RI isoform-specific phenotype but not unique to RIα.
Free RIα, but not the holoenzyme, is associated with punctate pattern
In normal resting cells, RIα is predominantly present as an inactive heterotetramer. It has been shown that free RIα and the RIα holoenzyme localize to discrete locations. We therefore wondered whether it was free RIα or the type I PKA holoenzyme that was forming puncta. To answer this question, we coexpressed GFP-tagged wild-type RIα with mCherry-tagged PKAc. When both RIα and PKAc were overexpressed in Prkar1a−/− MEFs, RIα localization was diffuse in the cytoplasm (Fig. 1 C, left). These results suggested that type I PKA holoenzyme did not form puncta.
To test whether the overexpressed RIα could be recruited to puncta after its disassociation from PKAc, cells were treated with forskolin/IBMX to elevate cAMP levels. Forskolin, by activating adenylyl cyclase, increases the intracellular levels of cAMP, whereas IBMX, a nonspecific inhibitor of phosphodiesterases, prevents its breakdown. cAMP binds to RIα and releases PKAc from the holoenzyme complex. Under these conditions, the punctate pattern appeared (Fig. 1 C, right). However, when RIα(R211K), which is defective in cAMP binding (Bubis et al., 1988; Herberg et al., 1996), was coexpressed with PKAc, no puncta were observed with or without forskolin/IBMX treatment. Therefore, formation of puncta appears to require free RIα. These results also confirm that overexpressed wild-type RIα did not readily recruit endogenous PKAc, unlike the overexpressed mutant, which binds to endogenous PKAc to form the holoenzyme (Fig. 1 C and see Fig. 5 B). Consistent with this data, overexpression of RIα(R211K) alone did not result in a punctate pattern (unpublished data).
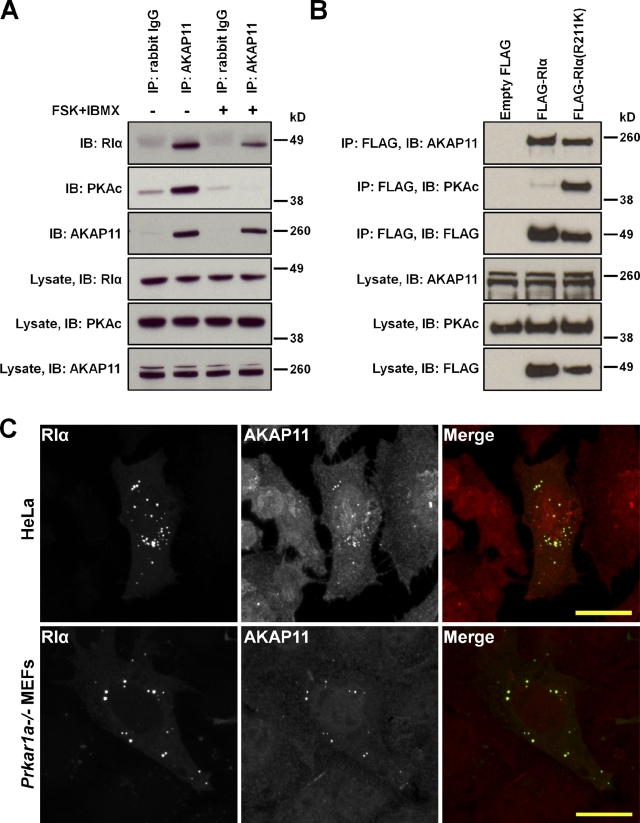
Figure 5.
AKAP11 binds to free RIα and the RIα holoenzyme. (A) Endogenous AKAP11 was immunoprecipitated from HEK 293 cells in the absence and presence of 20 µM forskolin (FSK)/200 µM IBMX. Samples from the lysate and from the elution of the immunoprecipitation (IP) were immunoblotted (IB) with antibodies against RIα, PKAc, or AKAP11. (B) HEK 293 cells were transiently transfected with control FLAG empty vector, FLAG-tagged RIα, or FLAG-tagged RIα(R211K), an RIα mutant that is defective in cAMP binding. Samples from the lysate and from the elution of the IP were immunoblotted with antibodies against AKAP11, PKAc, or FLAG. (A and B) The lanes marked Lysate were loaded with 5% of the total cell lysates. (C) HeLa cells and Prkar1a−/− MEFs were transiently transfected with GFP-tagged RIα (green) and stained for endogenous AKAP11 (red). Bars, 20 µm.
Live-cell imaging demonstrates that puncta formation is reversible
To follow the formation of the puncta, we performed live-cell imaging experiments with Prkar1a−/− MEFs transiently transfected with RIα-GFP and PKAc-mCherry (Fig. 1 D and Video 1). After 18 h of transfection, the cells were equilibrated in Opti-MEM for 1 h and imaged live at 37°C on a confocal microscope. The cells were treated through two rounds of forskolin/IBMX with four washes of Opti-MEM in between treatments. Before treatment (baseline), both RIα and PKAc displayed a diffuse pattern. Within ∼2 min of the first treatment, RIα showed a punctate pattern. This phenotype was reversible as indicated by the diffuse pattern within 4 min after the washout of drugs (postwash). The diffuse pattern suggested a reassociation of RIα with PKAc. When the cells were treated with the second forskolin/IBMX treatment, the punctate pattern reappeared, which was also reversible when the drugs were washed away again (shown in Video 1 but not in Fig. 1 D).
In a separate experiment, when cells were treated with forskolin/IBMX for ∼1.5 h and then washed three times with Opti-MEM and left without drugs for ∼45 min, the punctate pattern diminished but never became fully diffuse. Then, the strong punctate pattern reappeared within ∼3 min of adding a second round of forskolin/IBMX. This result indicated that any diffuse pattern observed during postwash was not simply a result of targeting RIα for rapid degradation (Video 2).
Punctate pattern is independent of PKAc activity and irreversible in the presence of a protein kinase inhibitor (PKI)
Upon activation, PKAc is released to phosphorylate downstream substrates. We therefore wanted to determine whether PKAc activity was required for puncta formation. When the cells from Video 1 displayed a diffuse pattern after undergoing two rounds of forskolin/IBMX treatments and washes, an inhibitor of PKA, the ATP analogue H89, was added for 10 min. Then, a third round of forskolin/IBMX treatment was added. Despite inhibiting PKAc activity, puncta formation still occurred, proving that PKAc activity was not necessary (Fig. 1 D, last image).
Another inhibitor of PKAc is the heat-stable PKI. To further prove that the punctate pattern is independent of PKAc activity, cells were transfected with RIα-GFP, PKAc-mCherry, and PKI-CFP simultaneously (Fig. 1 E and Video 3). These cells were given one round of forskolin/IBMX treatment followed by four washes of Opti-MEM. At the beginning, the cells showed a diffuse pattern, and then upon treatment, they presented a punctate pattern. However, in this case, the punctate pattern was not reversible when the drugs were removed, which indicated the inability of RIα to reassociate with PKAc in the presence of PKI. Thus, although PKAc activity was not required for puncta formation, release of PKAc from the RIα holoenzyme was necessary.
Fractionation experiments of MEFs reveal a migration of free RIα to membranes
To establish whether the puncta corresponded to membranous organelles or to protein aggregates, fractionation experiments were implemented (Fig. 2 A). Prkar1a+/+ MEFs were used to look at the migration of endogenous RIα. At basal levels of cAMP, the majority of RIα was in the cytosolic fraction. However, upon 60 min of forskolin/IBMX treatment, RIα showed a significant shift to the membrane fraction. Meanwhile, no change in PKAc levels was seen. Under these conditions, RIα and PKAc were not seen in the nuclear fraction. RIα was also not present in the cytoskeletal fraction, which included all residual proteins and protein aggregates. Markers for each fraction were used to verify equal sample loading and to confirm minimal cross-contamination during subcellular fractionation. We performed the same experiment with Prkar1a−/− MEFs, overexpressing both GFP-tagged RIα with mCherry-tagged PKAc (unpublished data). Likewise, an increase of RIα levels in the membrane fraction was observed after forskolin/IBMX treatment. These results confirmed that free RIα was migrating to membranes and that free RIα was not forming protein aggregates.
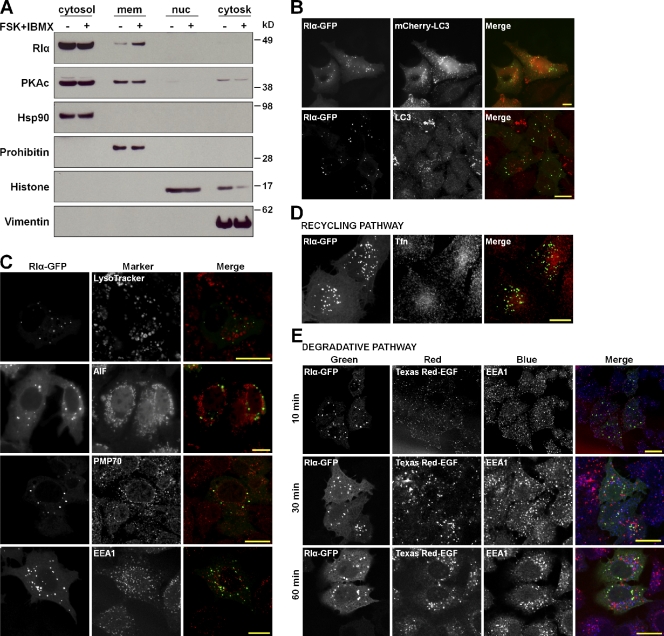
Figure 2.
Disassociated RIα localized to membranous organelles, and some organelles were ruled out from costaining with organelle markers. (A) Prkar1a+/+ MEFs were treated with 20 µM forskolin (FSK)/200 µM IBMX and fractionated. Cytosol refers to the cytosolic fraction, mem refers to the membrane fraction, nuc refers to the nuclear fraction, and cytosk refers to the cytoskeletal fraction. (B, top) HeLa cells were transiently cotransfected with GFP-tagged RIα (green) and mCherry-tagged LC3 (red). (bottom) Autophagy was induced in Prkar1a−/− MEFs, which were transiently transfected with GFP-tagged RIα (green), and then autophagosomes (LC3) were stained (red). (C) Cells were transfected with GFP-tagged RIα (green). Prkar1a−/− MEFs were stained for lysosomes (LysoTracker), mitochondria (AIF), and peroxisomes (PMP70), whereas HeLa cells were stained for early endosomes (EEA1). (D) Rhodamine-transferrin (Tfn) was used to follow the recycling pathway. (E) Texas red–EGF (EGF) was used to follow the degradative pathway, and cells were fixed at three different time points. EEA1 was stained so that a merge with EGF indicated early endosomes. Bars, 20 µm.
Colocalization experiments of RIα
To determine which possible membranous organelles were associated with free RIα, commercially available antibodies were used to stain various organelles in Prkar1a−/− MEFs and HeLa cells (Fig. 2, B–E). Attempts to see the localization of free endogenous RIα were unsuccessful because of the lack of suitable antibodies for immunostaining RIα, thus we used RIα-GFP. Because it was previously reported that RIα colocalized with autophagosomes (Mavrakis et al., 2006), these were the first organelles we checked. However, when we coexpressed RIα-GFP with light chain 3 (LC3)–mCherry, we saw no colocalization (Fig. 2 B, top). Because transiently transfected LC3 can be incorporated into protein aggregates without actually labeling autophagosomes (Kuma et al., 2007), we also checked for colocalization by staining for LC3 after autophagy induction. Likewise, we saw no evidence of colocalization between RIα and endogenous LC3 (Fig. 2 B, bottom). Colocalization with lysosomes (LysoTracker) or mitochondria (apoptosis-inducing factor [AIF]) was also not observed (Fig. 2 C). When staining for peroxisomes using 70-kD peroxisomal membrane protein (PMP70), we found a partial overlap with GFP-tagged RIα in both Prkar1a−/− MEFs and HeLa cells (unpublished data) but not enough to suggest a localization limited to peroxisomes.
We also transiently transfected HeLa cells with GFP-tagged RIα and stained for early endosomes (early endosome antigen 1 [EEA1]; Fig. 2 C, bottom). No overlap was detected, though the puncta were similar in size to EEA1 speckles. They were also very close and almost touching but not overlapping. Because the puncta were in such close proximity to EEA1, we looked for colocalization along the endosomal pathway. The recycling pathway was marked using rhodamine-transferrin but no overlap was found (Fig. 2 D). A search for localization along the degradative pathway was achieved using Texas red–labeled EGF (Fig. 2 E). Cells were fixed at different time points after the uptake of EGF to follow the different stages of the endosomal pathway. A great amount of overlap between Texas red–EGF and EEA1 at 10 min indicated that EGF localized to the early endosomes at that time point. However, no overlap between RIα and EGF/EEA1 was seen, although close proximity was consistently observed.
Electron microscopy experiments reveal localization to MVBs
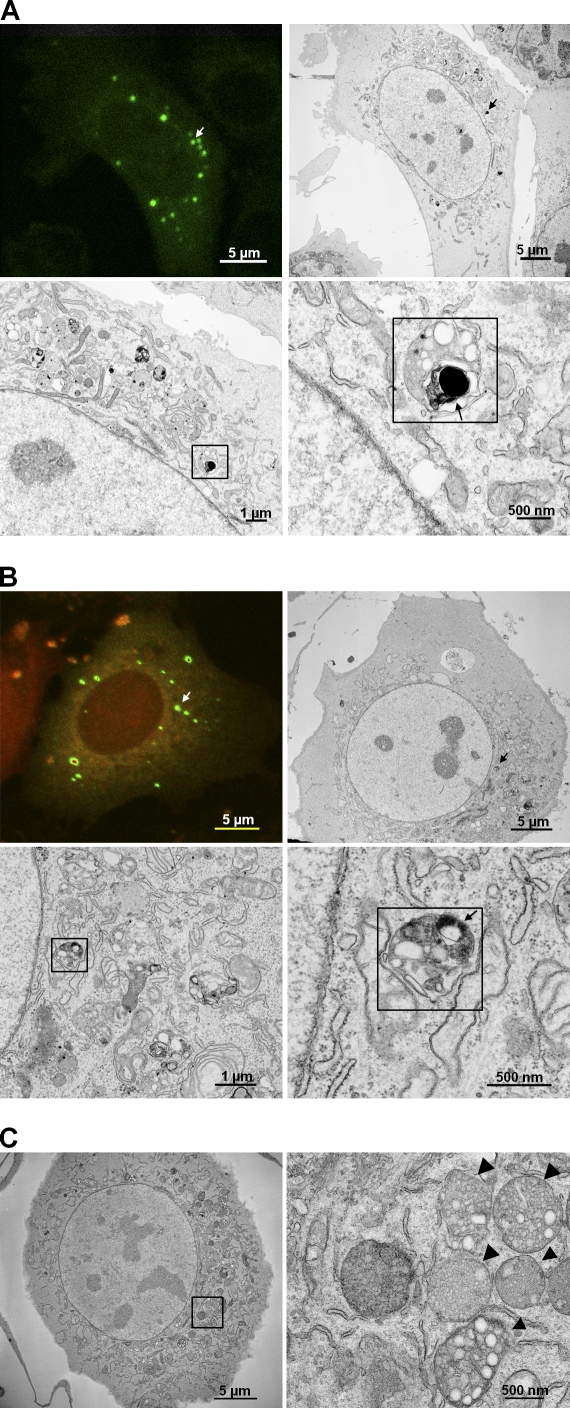
Because our immunostaining was unsuccessful in identifying the organelles to which RIα-GFP localized, we used correlated light and electron microscopy for an ultrastructural analysis. We transiently transfected Prkar1a−/− MEFs with either GFP-tagged RIα alone or GFP-tagged RIα and mCherry-tagged PKAc (Fig. 3, A and B, respectively). A poststain with lead was performed before imaging to enhance the visualization of cell membranes. With the transfection of RIα-GFP, we observed electron-dense particles in organelles, making antibody staining unnecessary. In both cases, we discovered a localization of RIα-GFP to membranous organelles that resembled MVBs. As a control, we also looked at untransfected Prkar1a−/− MEFs that were treated with forskolin/IBMX (Fig. 3 C). No electron-dense product was observed in the MVBs from these cells, further verifying that the aforementioned dark electron-dense material was correlated with RIα-GFP. Thus, we found the localization of RIα-GFP puncta to MVBs. However, this localization was not a mechanism for its rapid degradation because we observed puncta that remained despite reintroducing holoenzyme formation conditions in live-cell imaging (Video 2).
Figure 3.
Ultrastructural analysis revealed localization of RIα-GFP in MVBs. Light and electron microscopy pictures were correlated to look for the localization of GFP-tagged RIα. (A and B, top) White/black arrows point to an MVB that was correlated. (bottom) The MVB is indicated by black squares. (bottom right) RIα-GFP is the dark black area within the MVB indicated by a black arrow. (A) Prkar1a−/− MEFs were transfected with RIα-GFP (green). (B) Prkar1a−/− MEFs were cotransfected with RIα-GFP (green) and PKAc-mCherry (red) and treated with 20 µM forskolin/200 µM IBMX. (C) Untransfected Prkar1a−/− MEFs were treated with 20 µM forskolin/200 µM IBMX. An enlarged image of MVBs from within the black square is shown on the right. The MVBs are indicated by black arrowheads.
Localization to MVBs is not caused by ubiquitination
Because ubiquitination of proteins is used as a sorting signal to MVBs (Katzmann et al., 2001; Reggiori and Pelham, 2001), we wondered whether the RIα targeted to MVBs was ubiquitinated. The PEST sequence is one signal that is recognized by the ubiquitin machinery, and RIα has a potential PEST sequence in its linker region (Bergold et al., 1992) that has been correlated with its ubiquitination upon cAMP binding (Hegde et al., 1993). To test the involvement of this PEST sequence, we used a PEST sequence predictor to determine that mutating the proline-rich region would remove it. If the PEST sequence were involved in the localization of RIα to MVBs, removing it would disrupt puncta formation. However, when we mutated the PEST sequence, the punctate pattern was still observed, indicating that the PEST sequence was not essential for targeting to MVBs (Fig. S2). In addition, mass spectrometry analysis of human embryonic kidney (HEK) 293 cells overexpressing wild-type RIα did not detect ubiquitination of RIα (unpublished data). We also saw no evidence of higher molecular weight ubiquitinated forms of RIα in any of our Western blot analyses.
Localization of puncta is through AKAP binding
To deduce which region of RIα was essential for puncta formation, we expressed two GFP-tagged truncation mutants, RIα(1–93) and RIα(93–381). RIα(1–93) includes the D/D domain, whereas RIα(93–381) includes the two cAMP-binding domains. A faint punctate pattern was observed with the RIα(1–93) construct but not with the RIα(93–381) construct (Fig. 4 A). Thus, the N-terminal part of RIα was necessary for the punctate pattern. Because RII anchoring to membranous organelles occurs via AKAP binding at the D/D domain, we next tested whether an AKAP was involved in the recruitment of RIα to MVBs.
Figure 4.
Disruption of AKAP binding removed the punctate pattern. (A) Control GFP empty vector, GFP-tagged wild-type (wt) RIα, and GFP-tagged deletion mutants, RIα(1–93) and RIα(93–381), were expressed in Prkar1a−/− MEFs. White arrows indicate a few of the faint puncta that were seen with the expression of RIα(1–93)-GFP. (B) A mutation that has been reported to disrupt D-AKAP1 binding, RIα(I27A), was GFP-tagged and expressed in Prkar1a−/− MEFs. GFP empty vector and GFP-tagged wild-type RIα were included as controls. (C) mCherry empty vector or mCherry-tagged AKB domain from D-AKAP2 (RI/RII selective, RI selective, or RII selective) was coexpressed with GFP-tagged RIα in Prkar1a−/− MEFs. Bars, 20 µm.
Previously, it has been shown that several mutations in RIα reduced its binding to a dual-specific AKAP1, D-AKAP1, in vitro (Banky et al., 1998). We introduced one of these mutations (I27A) into Prkar1a−/− MEFs, and the punctate pattern disappeared (Fig. 4 B). This result provided evidence that formation of the RIα puncta required AKAP binding.
To test this theory further, GFP-tagged RIα was coexpressed in Prkar1a−/− MEFs with mCherry-tagged peptide disruptors of AKAP binding (Fig. 4 C). These disruptors were derived from the C terminus of a dual-specific AKAP, D-AKAP2(623–662), which contains the PKA-binding motif. Residues identified for isoform specificity in a previous study (Burns-Hamuro et al., 2003) were mutated to make RI or RII selectivity. Coexpressing wild-type RIα with the RI/RII-selective AKAP disruptor resulted in no change to the punctate pattern. On the other hand, coexpressing wild-type RIα with an RI-selective AKAP disruptor caused the punctate pattern to become diffuse. However, the punctate pattern remained in the presence of the RII-selective AKAP disruptor. Although it might have been expected that the punctate pattern would become diffuse in the presence of the RI/RII-selective AKAP disruptor, the lack of change is not surprising. Burns-Hamuro et al. (2003) reported that D-AKAP2 binds with greater affinity to RIIα versus RIα (Kd = 2.2 nM vs. 48 nM, respectively); therefore, the nonspecific disruptor could naturally be more selective for RIIα because of its higher affinity to RIIα. Collectively, these results supported the notion that an RIαspecific AKAP is involved in localizing RIα to MVBs.
Mass spectrometry analysis reveals AKAP11 as a binding partner to disassociated RIα
Because >50 AKAPs exist (Wong and Scott, 2004), we developed affinity methods and mass spectrometry techniques for an unbiased screen of RIα-interacting partners, including AKAPs. We used untransfected HEK 293 cells and HEK 293 cells that were stably expressing tandem affinity purification (TAP)–tagged RIα.
First, to validate the accuracy of our methods, cAMP resin was used on untransfected HEK 293 cells to find proteins that bind to all four R-subunit isoforms (Table I, first method). The cAMP resin was directly subjected to trypsin digestion instead of buffer elution because it proved difficult to elute RIα from the cAMP resin. The digested peptides were then analyzed by mass spectrometry, and we considered a result as a true positive if it had at least two unique peptides. Three R-subunit isoforms were identified: RIα, RIIα, and RIIβ. Two AKAPs were also identified: AKAP9 and AKAP1.
Table I.
cAMP-binding proteins and AKAPs identified from mass spectrometry using different purification methods on HEK 293 cell lysates
| Method/cell type | Protein description | IPI | Number of hits | Unique peptides |
| cAMP resin/untransfected | PKA type I-α R-subunit | IPI00021831 | 64 | 19 |
| PKA type II-α R-subunit | IPI00219774 | 104 | 31 | |
| PKA type II-β R-subunit | IPI00554752 | 6 | 4 | |
| AKAP9 | IPI00019223 | 148 | 70 | |
| AKAP1 | IPI00022585 | 9 | 5 | |
| cAMP resin/stably transfected TAP-tagged wild-type RIα | PKA type I-α R-subunit | IPI00021831 | 57 | 19 |
| PKA type II-α R-subunit | IPI00219774 | 15 | 12 | |
| PKA type II-β R-subunit | IPI00554752 | 9 | 6 | |
| AKAP11 | IPI00007411 | 9 | 8 | |
| AKAP9 | IPI00220628 | 5 | 4 | |
| AKAP5 | IPI00307794 | 2 | 2 | |
| Streptavidin and calmodulin resins/stably transfected TAP-tagged wild-type RIα | PKA type I-α R-subunit | IPI00021831 | 434 | 29 |
| PKA α–catalytic subunit | IPI00217960 | 2 | 2 | |
| PKA α–catalytic subunit | IPI00396630 | 6 | 3 | |
| AKAP11 | IPI00007411 | 13 | 9 | |
| Streptavidin and cAMP resins/stably transfected TAP-tagged wild-type RIα | PKA type I-α R-subunit | IPI00021831 | 721 | 63 |
| PKA type I-β R-subunit | IPI00787996 | 21 | 14 | |
| AKAP11 | IPI00007411 | 113 | 59 |
IPI, International Protein Index.
We also used cAMP to find proteins that bind to all four R-subunit isoforms in the presence of stably transfected wild-type RIα (Table I, second method). The proteins that were identified included the same three R-subunit isoforms: RIα, RIIα, and RIIβ. In this case, though, three AKAPs were identified: AKAP11, AKAP9, and AKAP5. However, it was not clear which R-subunit isoforms bind to these proteins.
Therefore, to detect proteins associated with RIα instead of the other isoforms, we used the TAP tag system, in which stably transfected TAP-tagged RIα was purified with streptavidin and calmodulin resins (Table I, third method). In this case, we expected to find RIα, PKAc, and any protein that would bind to either RIα or to the RIα holoenzyme. Cell lysates were first bound to streptavidin resin, and then binding proteins were eluted and bound to calmodulin resin. As with the cAMP resin, proteins were not eluted from the calmodulin resin because of difficulty in disassociating TAP-tagged proteins from these beads. Instead, we ran the protein-bound calmodulin resin on an SDS-PAGE gel to separate any calmodulin that was attached to the beads. The gel pieces were digested and analyzed by mass spectrometry to identify proteins. Out of the three AKAPs that were identified when using cAMP alone, only one was detected using this technique, AKAP11.
To mimic the scenario from our microscopy experiments, we used the TAP tag system followed by cAMP to disassociate any PKAc (Table I, fourth method). From this method, we pulled out proteins that bound directly to free TAP-tagged RIα. AKAP11 was the only AKAP detected, and the unique peptides covered almost the entire AKAP11 sequence (unpublished data). Therefore, our mass spectrometry results suggested that AKAP11 is the best AKAP candidate responsible for the targeting of RIα to MVBs. These results also demonstrated that binding of AKAP11 to RIα is very robust, as it survived multiple stringent washes. Complete mass spectrometry data for all proteins that were pulled out under the aforementioned conditions are included as supplemental tables (Table S1, Table S2, Table S3, and Table S4).
AKAP11 binds to RIα in its free or holoenzyme form
To independently validate the interaction between AKAP11 and RIα, we immunoprecipitated endogenous AKAP11 from untransfected HEK 293 cells using the AKAP11 antibody or rabbit IgG as a negative control. Samples were analyzed by Western blotting using antibodies for RIα, PKAc, and AKAP11 (Fig. 5 A). AKAP11 bound to both the RIα holoenzyme and free RIα. Inversely, we immunoprecipitated FLAG-tagged constructs of RIα that were transiently transfected into HEK 293 cells. As mentioned earlier, one mutation, RIα(R211K), is defective in cAMP binding. Samples were analyzed by Western blotting using antibodies for AKAP11, PKAc, and FLAG (Fig. 5 B). Both wild-type and mutant RIα interacted with AKAP11. As expected, a lower amount of PKAc was purified with wild-type RIα versus RIα(R211K) because RIα(R211K) would remain in complex with PKAc, despite the basal levels of cAMP. Together with the mass spectrometry data, this information led us to deduce that AKAP11 binds tightly to both free RIα and RIα in complex with PKAc.
We also demonstrated the colocalization of AKAP11 with RIα through immunofluorescence. HeLa cells and Prkar1a−/− MEFs were transiently transfected with RIα-GFP and stained for endogenous AKAP11. As observed from Fig. 5 C, the punctate pattern of AKAP11 overlapped with a majority of the punctate pattern from RIα-GFP, further suggesting their interaction at MVBs.
AKAP11 is responsible for the targeting of free RIα
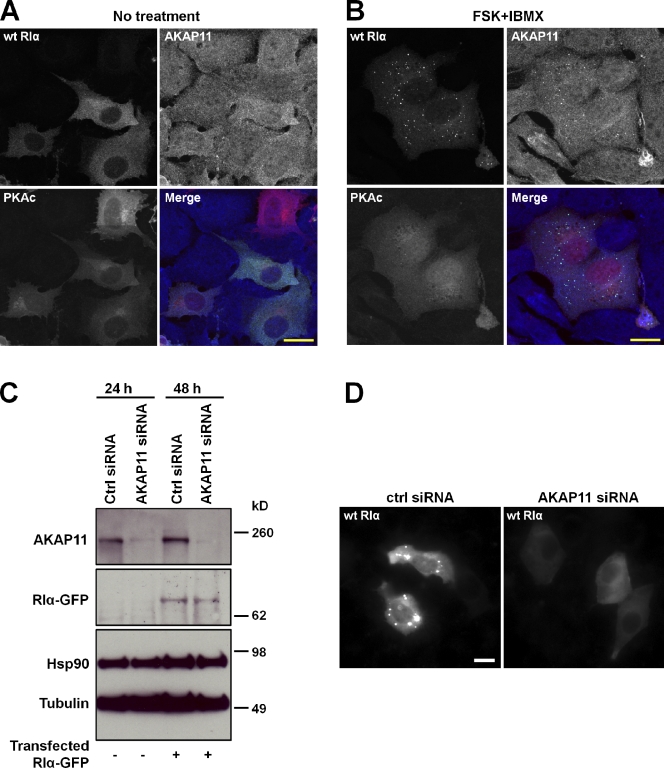
Because we ascertained that AKAP11 binds to RIα and that free RIα, but not the holoenzyme, is targeted to MVBs, we next confirmed the colocalization of AKAP11 with RIα that was disassociated from PKAc. Prkar1a−/− MEFs were transiently transfected with RIα-GFP and PKAc-mCherry. One set of cells was left untreated (Fig. 6 A), whereas the other set was treated with forskolin/IBMX (Fig. 6 B). Then, both sets were stained for endogenous AKAP11. Before treatment, AKAP11 showed a diffuse pattern. After treatment, AKAP11 displayed a punctate pattern that overlapped with a majority of the RIα-GFP puncta.
Figure 6.
AKAP11 targets free RIα. (A) Prkar1a−/− MEFs were transiently cotransfected with GFP-tagged RIα (green) and mCherry-tagged PKAc (red) and stained for endogenous AKAP11 (blue). (B) Prkar1a−/− MEFs were transiently cotransfected with GFP-tagged RIα (green) and mCherry-tagged PKAc (red), treated with 20 µM forskolin (FSK)/200 µM IBMX, and then stained for endogenous AKAP11 (blue). (C) siRNA of AKAP11 effectively knocked down the levels of endogenous AKAP11 protein in Prkar1a−/− MEFs after 1–2 d. No effect on the expression level of transiently transfected RIα-GFP was observed, as judged by immunoblotting of whole-cell extracts with antibodies against AKAP11, RIα, Hsp90, or tubulin. (D) Knockdown of endogenous AKAP11 disrupted the formation of RIα-GFP puncta in Prkar1a−/− MEFs. Ctrl, control. wt, wild type. Bars, 20 µm.
Because AKAP11 displayed a punctate pattern only if RIα were punctate, we next addressed the question of whether AKAP11 is responsible for the targeting of RIα to form puncta. If this were the case, knocking down AKAP11 by RNA interference should have an effect on the targeting of RIα to MVBs. We thus treated Prkar1a−/− MEFs for 1–2 d with siRNA directed against AKAP11 and observed that siRNA effectively knocked down nearly all of the endogenous protein (Fig. 6 C). After 1 d of siRNA treatment, cells were transfected with wild-type RIα-GFP and analyzed through Western blotting and fluorescence microscopy analysis. We observed that the levels of RIα-GFP were unaffected by the knockdown of AKAP11 (Fig. 6 C); however, cells knocked down for AKAP11 no longer displayed a punctate pattern of RIα-GFP (Fig. 6 D). Collectively, these data suggest that AKAP11 is responsible for the targeting of free RIα to MVBs.
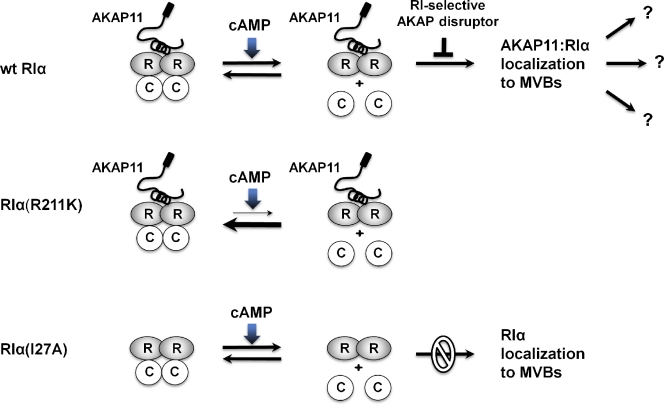
We thus propose a model for the targeting of free RIα that is blocked by the formation of the holoenzyme or by the disruption of AKAP binding and is independent of PKAc activity (Fig. 7). According to this model, AKAP11 is associated with the RIα holoenzyme when it is diffuse in the cytoplasm, and release of PKAc is required for recruitment of RIα to MVBs. Thus, holoenzyme formation not only inhibits the catalytic activity of PKAc but also prevents the targeting of free RIα to MVBs.
Figure 7.
Free RIα is recruited to MVBs by AKAP11. A model for the mechanism of targeting RIα is depicted. AKAP11 binds to the RIα holoenzyme. Upon activation of PKA, AKAP11 stays bound to RIα, which is recruited to multivesicular bodies (MVBs). If the RIα holoenzyme is not activated, as in the case of RIα(R211K):PKAc, RIα does not get targeted. Likewise, free RIα is not targeted properly when AKAP11 binding is disrupted, as in the case with the RIα(I27A) mutation or in the presence of RI-selective AKAP disruptors. R, RIa; C, PKAc; wt, wild type.
Discussion
Although the initial driving force for this study was to characterize the distinct puncta that formed when the free RIα subunit was expressed in mammalian cells, the finding that both free RIα and the RIα holoenzyme were associated with AKAP11 was another important discovery. Whereas RII subunits are constitutively localized to discrete areas in the cell through their tight binding to AKAPs, the RIα subunits are typically diffuse in the cytoplasm and only recruited to specific sites in response to stimuli, for example, through the activation of T lymphocytes (Levy et al., 1996) or addition of hydrogen peroxide (Brennan et al., 2006). Here, we show that the association of RIα with AKAP11 defines a novel signaling mechanism for the RIα subunit that is distinct from the RII subunits and separate from its function as a regulator of PKAc.
We disproved several of the conclusions from an earlier study that analyzed RIα-GFP in cells (Mavrakis et al., 2006). Our data refuted their study of RIα localization to autophagosomes. Additionally, Mavrakis et al. (2006) claimed a statistically significant difference in the number of autophagosomes observed between Prkar1a+/+ MEFs and Prkar1a−/− MEFs, which is contradictory to our data. Analysis of the number of autophagosomes as correlated by their volume density in the cytoplasm at the electron microscopy level produced no statistically significant difference in the number of autophagosomes between Prkar1a+/+ MEFs and Prkar1a−/− MEFs (volume density is 1.5 and 1.4%, respectively). From looking at the number of autophagosomes in Prkar1a−/− MEFs at the light microscopy level, we discovered that cells with RIα-GFP puncta displayed lower induction of autophagy compared with cells without RIα-GFP puncta (representative picture in Fig. 2 B, bottom merge). Therefore, we conclusively proved that RIα was not colocalizing with autophagosomes, and the loss of RIα did not affect the number of autophagosomes in cells.
Instead, our electron microscopy results revealed the localization of RIα to MVBs. MVBs can only be definitively identified at the ultrastructural level (Clague and Urbé, 2008), which could be the reason for our difficulty in detecting colocalization through immunostaining, although we consistently saw close proximity to endosomes. We also determined that localization to MVBs was unique to the free RIα subunit, independent of PKAc activity, and reversible by lowering intracellular levels of cAMP.
To elucidate the molecular basis for RIα targeting to MVBs, we used several strategies to pinpoint that it was the AKAP-binding site within the N terminus that was required and that the AKAP needed was AKAP11. Our analysis also identified AKAP11 as a highly selective AKAP for RIα. Surprisingly, only the free RIα subunit goes to MVBs, even though AKAP11 appears to be associated with both the holoenzyme and the disassociated RIα subunit.
AKAP11, first discovered as an anchoring protein almost 15 yr ago (Lester et al., 1996), is a 220-kD protein that has been reported to bind to both RIα and RIIα in testis (Reinton et al., 2000); however, the interaction to RIα has not been studied in other cell lines, and its significance has never been addressed. In our experiments, we deduced that RIα(I27) was essential for binding with AKAP11 based on our mutagenesis experiments coupled with the recent structure of a D-AKAP2 peptide bound to RIα. In this structure, a mutation of RIα(I27) would affect binding to an AKAP because it disrupts the packing of RIα(I35), a direct interactor to D-AKAP2 (Sarma et al., 2010). Most AKAPs with dual specificity have a 25–100-fold greater affinity for the RII versus RI isoforms (Herberg et al., 2000; Burns-Hamuro et al., 2003). Furthermore, the off rate of AKAPs with RIα is typically fast, making it difficult to analyze protein binding through RI overlay assays. In contrast, we find the binding of AKAP11 to RIα with our most stringent washes during purification (Fig. 5, A and B; and Table I), which led us to deduce the presence of a high affinity RIα-specific binding site in AKAP11. Three putative A-kinase binding (AKB) motifs exist in AKAP11 (Reinton et al., 2000). Based on the nature of the amphipathic helix that is formed from each of these putative AKB motifs and the recently predicted requirements for AKAP binding to RI versus RII subunits (Sarma et al., 2010), we predict that one of the AKB motifs has the potential to bind exclusively to RIα: AKAP11(611–628).
Although the precise role of targeting RIα to MVBs remains to be established, it does not appear to simply be a mechanism for degradation. Although MVBs were initially identified as part of the degradative pathway for proteins (Felder et al., 1990), it has now been discovered that MVBs also give rise to endosomal compartments that do not fuse with lysosomes, and subsequently, their cargo are not destined for degradation. For example, the major histocompatibility complex class II proteins accumulate in MVBs, but instead of being degraded, they are secreted from the cell upon fusion of the MVB with the plasma membrane (van Niel et al., 2006). Thus, MVBs can house proteins in temporary storage compartments without eventual degradation. We believe this is the case for the targeting of RIα to MVBs. Because we saw a reversibility of puncta formation, we suspect that the functionality of targeting RIα to MVBs is to keep RIα in a holding pattern until the holoenzyme reforms or its presence is needed somewhere else in the cell. A closer examination of our live-cell videos revealed that the puncta were merging with each other. The electron microscopy pictures showed that some of the electron-dense RIα-GFP appeared to be clinging to the MVB outer membrane, so it is possible that RIα is associated with the membrane from the cytosolic side, and not all were enclosed in the lumen. Furthermore, we do not believe that RIα is moving along the degradative pathway because of its lack of colocalization with lysosomes (Fig. 2 C, LysoTracker).
To our knowledge, this is the first study of RIα localizing to MVBs, and AKAP11 has never been shown to target there. In rats, AKAP11 potentially targets to peroxisomes because of the final three amino acids CRL (Lester et al., 1996), but human AKAP11 does not contain these final residues. We ruled out the targeting of human AKAP11 to peroxisomes because partial overlap of a couple of puncta, possibly caused by chance, was seen in only a few cells either stained for PMP70 or stained for proteins with another peroxisomal targeting sequence—SKL (unpublished data). Another study suggested the possibility of AKAP11 as an endosomal AKAP because of its colocalization with aquaporin 2 (AQP2) near apical membranes (Okutsu et al., 2008). AQP2 binds to AKAP11 (Okutsu et al., 2008) and is known to target to MVBs in kidney cells (Takata et al., 2008). Therefore, in HEK 293 or HeLa cells, the binding of AKAP11 to AQP2 may be the method by which the AKAP11:RIα complex is targeted to MVBs. AQP2 is not known to be expressed in MEFs, but in this cell line, another protein may be acting in a similar fashion to target AKAP11:RIα to MVBs. Elucidating the molecular basis for targeting AKAP11:RIα to MVBs is our next challenge.
Although other researchers have reported the association of RIα with proteins, such as RSK1 (p90 ribosomal S6 kinase; Chaturvedi et al., 2006), RFC40 (Gupte et al., 2005), and cytochrome c oxidase (Yang et al., 1998), we describe a unique signaling paradigm for the RIα subunit. Our model involves its dynamic and reversible recruitment to a specific organelle once it is disassociated from PKAc. We have known that the formation of the holoenzyme leads to the inactivation of PKAc, but we now discover that the holoenzyme also retains the RI subunit in a soluble signaling complex that prevents it from localizing to MVBs. The signaling complex, comprised of AKAP11 and RIα, is constitutively present, but the dynamic recruitment of RIα to MVBs requires the release of PKAc. AKAP11 has been suggested to regulate the cell cycle and play a role in driving oral carcinogenesis (Garnis et al., 2005), whereas RIα is involved in several different signaling pathways, cellular functions, and diseases. Therefore, it is likely that AKAP11 directs some of the RIα functions. Through our characterization of RIα puncta, we begin to have a handle on examining the fate of the disassociated RIα subunit, whether it is to compensate for excess PKAc levels, bind to other proteins to necessitate their translocation, or participate in the control of cell cycle and cell growth.
Materials and methods
Antibodies
The following antibodies were used for Western blot analysis: anti–RIα (Transduction Laboratories), anti-PKAc (Transduction Laboratories), anti-Hsp90 (Transduction Laboratories), antiprohibitin (Abcam), antihistone (Cell Signaling Technology), antivimentin (Abcam), anti-AKAP11 (generated by the laboratory of J. Scott, University of Washington, Seattle, WA), anti-AKAP11 (Abnova), anti–FLAG M2 (Sigma-Aldrich), and antitubulin (Sigma-Aldrich).
Cloning and expression of RIα and other proteins
Human RIα was subcloned from pCMV-SPORT6 (Invitrogen) into the HindIII–KpnI site of the pEGFP C2 vector (Takara Bio Inc.) and into the BamHI–HindIII site of the pNTAP vector (Agilent Technologies). Single site mutations were made in the full-length sequence by QuikChange mutagenesis (Agilent Technologies). Truncated RIα(1–93 and 93–381) was isolated by PCR, and the amplified cDNA was subcloned. Vectors for expression of mCherry- and FLAG-tagged proteins were generated as previously described (Eggers et al., 2009). RIα was subcloned into the HindIII–KpnI site of a FLAG vector. D-AKAP2 constructs were subcloned into the EcoRI–XhoI site of the mCherry vector. All constructs were verified by sequencing. R-subunit isoforms cloned into A-kinase activity reporter vectors were provided by F. Ma (University of California, San Diego, La Jolla, CA). PKAc tagged with mCherry and PKI tagged with CFP were provided by R. Tsien (University of California, San Diego, La Jolla, CA). LC3 tagged with mCherry was provided by A. Gustafsson (San Diego State University, San Diego, CA).
Cell lines and culture conditions
HeLa cells were obtained from the American Type Culture Collection. MEFs, immortalized by transformation with pBRSV encoding the SV40 virus, were provided by G.S. McKnight (University of Washington, Seattle, WA) and M. Ginsberg (University of California, San Diego, La Jolla, CA). Cells were maintained in DME supplemented with 10% fetal bovine serum (HyClone), 2 mM GlutaMax (Invitrogen), and 100 U/ml penicillin and streptomycin (Invitrogen) under 8% CO2. Transfections were performed with transfection reagent (Fugene 6; Roche) 1 d before imaging or with Lipofectamine 2000 (Invitrogen) 1 d before immunoprecipitation (IP). Stable cell lines of HEK 293 cells were generated by transfection with Lipofectamine 2000, selected accordingly for geneticin resistance, and maintained as described for HeLa cells and MEFs with the addition of 500 µg/ml geneticin (Invitrogen) to the media.
Live-cell imaging
Prkar1a−/− MEFs were grown overnight on poly-d-lysine–coated glass-bottom dishes (MatTek) and cotransfected with GFP-tagged RIα and mCherry-tagged PKAc (Fig. 1 D, Video 1, and Video 2) or cotransfected with GFP-tagged RIα, mCherry-tagged PKAc, and CFP-tagged PKI (Fig. 1 E and Video 3). 18 h after transfection, DME growth media were replaced with Opti-MEM (Invitrogen). cAMP levels were elevated by the addition of 20 µM forskolin (Sigma-Aldrich) and 200 µM IBMX (Sigma-Aldrich) simultaneously at 37°C. PKA activity was inhibited by the addition of 10 µM H89 (Millipore). Live-cell imaging sessions were conducted in a heated (37°C), sealed, humidified, and CO2-equilibrated environmental chamber (WeatherStation; Precision Control) paired with a confocal laser-scanning microscope (FluoView1000; Olympus). Images were collected using an oil immersion 60× objective with an NA of 1.42 (Olympus). The data in Fig. 1 D, Video 1, and Video 2 were collected using 488- and 561-nm laser excitations, whereas the data in Fig. 1 E and Video 3 were collected using a 405-, 488-, and 561-nm laser excitations. Stacks of 25–30 slices (300-nm thick) were acquired using sequential scanning to prevent bleed through between channels. The collected volumes were processed using a customized ImageJ macro (written by O. Kwon, National Center for Microscopy and Imaging Research/Center for Research in Biological Systems, University of California, San Diego, La Jolla, CA) to generate a maximum intensity projection for each channel and time point and then merged into an AVI video.
Fractionation
Prkar1a+/+ MEFs were grown on 15-cm Nunc dishes to ∼80% confluency. Prkar1a−/− MEFs were transfected using Lipofectamine 2000 and also grown to ∼80% confluency. One set was left untreated, whereas the other set was treated with 20 µM forskolin simultaneously with 200 µM IBMX for 1 h at 37°C. Cells were harvested in 2 ml of cold PBS and fractionated with a cell compartment kit (Qproteome; QIAGEN). Western blots were run on samples using equal volume loads. All primary antibodies were used at a dilution of 1:1,000 except for anti-Hsp90 (1:2,000 dilution) and antiprohibitin (1:100 dilution).
Immunofluorescence
HeLa cells and Prkar1a−/− MEFs were grown overnight in poly-d-lysine–coated glass-bottom dishes or on glass coverslips (Thermo Fisher Scientific), transfected with GFP-tagged RIα for 18–24 h, and fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min at room temperature. Prkar1a−/− MEFs, which were cotransfected with GFP-tagged RIα and mCherry-tagged PKAc, were left untreated or treated with 20 µM forskolin simultaneously with 200 µM IBMX for 1 h at 37°C and fixed through the same method as cells transfected with GFP-tagged RIα alone. All solutions used during the immunostaining were prepared in Dulbecco’s PBS (Mediatech). The following antibodies were used at 1:100 dilutions: anti-LC3 (Cell Signaling Technology), anti-AIF (Cell Signaling Technology), anti-PMP70 (Sigma-Aldrich), anti-EEA1 (Cell Signaling Technology), anti-AKAP11 (generated by J. Scott, University of Washington, Seattle, WA), Cy5-labeled donkey anti–rabbit IgG antibodies (Jackson ImmunoResearch Laboratories, Inc.), and DyLight 649 (Jackson ImmunoResearch Laboratories, Inc.). LysoTracker (Invitrogen) was used to visualize lysosomes. Cells were permeabilized in 0.2% Triton X-100 (Sigma-Aldrich) for 5 min at room temperature and blocked in 2% BSA (Sigma-Aldrich) followed by sequential incubations in primary and secondary antibodies. The method for staining LC3 was slightly altered: cells were permeabilized in 0.05% Triton X-100 for 15 min and blocked in 3% BSA. When using LysoTracker, cells were incubated with a probe diluted in prewarmed DME (final concentration of 50 nM) for 1 h at 37°C before fixation. Fixed cells were viewed with either a microscope (Axiovert 200M; Carl Zeiss) equipped with an oil immersion 40× objective with an NA of 1.3 (Carl Zeiss) and an electron microscopy charge-coupled device camera (Cascade II:512; Photometrics) or with a confocal laser-scanning microscope (FluoView1000) equipped with an oil immersion 60× objective with an NA of 1.42. Epifluorescent images were acquired with MetaFluor software (Molecular Devices), whereas confocal images were acquired with FluoView ASW 1.7c software (Olympus). The collected confocal images were processed using ImageJ (Abramoff et al., 2004) to generate maximum intensity projection images (z = 30–40 slices; z step = 0.3 µm) and Photoshop (Adobe) to generate composite pictures.
Induction of autophagy
Autophagy was induced in cells by amino acid and glucose starvation using Earle’s balanced salt solution (Invitrogen) for 4 h at 37°C. Two lysosomal inhibitors, 1 µM pepstatin A (EMD) and 10 µM EST (EMD), were used to prevent the degradation of LC3-II.
Endosomal pathway
Recycling pathway.
HeLa cells transfected with RIα-GFP were serum starved in DME supplemented with 0.5% BSA for 2 h. Alexa Fluor 568 transferrin receptor (Invitrogen) was added to the cells at a final concentration of 50 µg/ml for 1 h at 37°C. Cells were washed once with cold PBS, once with cold stripping buffer (500 mM NaCl and 0.5% acetic acid, pH 3.0) for 45 s, and two more times with cold PBS. Cells were fixed with 4% paraformaldehyde for 15 min at room temperature.
Degradative pathway.
HeLa cells transfected with RIα-GFP were serum starved in DME supplemented with 0.5% BSA for 6 h. Texas red–labeled EGF (Invitrogen), diluted in cold DME to a final concentration of 1 µg/ml, was added to the cells. Unbound EGF was removed by washing in cold PBS. Prewarmed DME was added to the cells, and different time points were taken by fixing cells with 4% paraformaldehyde for 15 min at room teseccmperature.
Electron microscopy
Prkar1a−/− MEFs were grown on glass-bottom grid dishes. After 18 h of transfection (using Fugene 6), cells were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, solution for 5 min at room temperature and then moved to ice for a duration of 30 min for optimal cell ultrastructural preservation. The cells were washed five times in ice-cold 0.1 M sodium cacodylate buffer, pH 7.4, and 3 mM calcium chloride, each taking 2 min, to remove excess aldehydes. Cells were viewed with a confocal microscope (MRC-1024; Bio-Rad Laboratories) using 488- and 568-nm laser excitations and a 40× water objective lens with an NA of 1.2 (confocal stacks of z = 37 slices; z step = 0.36 µm). A transmitted image was viewed with either a 10 or 20× objective lens to determine the grid location. Images were acquired using LaserSharp 2000 software (Bio-Rad Laboratories/Carl Zeiss). The cultured cells were postfixed in ice-cold 1% osmium tetroxide with 0.8% potassium ferrocyanide and 3 mM calcium chloride for 30 min, washed five times in double-distilled water for 2 min each to stop the osmium tetroxide reaction, and en bloc stained in ice-cold 2% uranyl acetate overnight to help increase membrane contrast. Afterward, the cultured cells were dehydrated in an ethanol series for 3 min each, starting with 20, 50, 70, and 90% ethanol on ice and ending with four changes of 100% ethanol at room temperature. The dehydrated cells were infiltrated in a 1:1 ratio of absolute ethanol to epoxy resin (Durcupan ACM; Electron Microscopy Sciences) for 30 min and then with three changes of epoxy resin (Durcupan ACM) for 1 h each. Finally, a fourth epoxy resin (Durcupan ACM) change was performed, and immediately, the dish was placed in a vacuum oven at 60°C to be polymerized for 48–72 h. After polymerization, the coverslip was removed from the dish, and plastic blocks were sawed out and glued onto dummy blocks. 80-nm ultrathin serial sections were prepared using an ultramicrotome (Reichert–Jung Ultracut E; American Optical Instrument Company) with a diamond knife (Diatome), and the sections were supported onto 200 mesh copper grids. The 80-nm sections were poststained in Sato’s lead for 1 min, and the stained sections were imaged using an electron microscope (JEM-1200EX II; JEOL) at 80 kV.
Pull-down experiments
Untransfected and stable HEK 293 cells were grown on Nunc dishes and harvested at 80% confluency in PBS. Cells were lysed in 50 mM Tris, pH 7.0, 150 mM NaCl, 1 mM EDTA, and 1% NP-40 by three freeze-thaw cycles. Lysates were bound to 8-AEA ([2-aminoethylamino]adenosine)–cAMP resin, streptavidin and calmodulin resins, or streptavidin and 8-AEA–cAMP resin. 8-AEA–cAMP was purchased from BioLog. Proteins bound to streptavidin resin were eluted with streptavidin buffer, which contains biotin (Agilent Technologies). Mass spectrometry analyses of proteins bound to cAMP or calmodulin resins are described in the next section.
Proteomics analysis
Proteins bound to cAMP resin.
Proteins pulled down with cAMP beads were resuspended in 0.1 M ammonium bicarbonate. Reduction and alkylation of the sample was performed by adding DTT to a final concentration of 2 mM, incubating at room temperature for 30 min, and then adding iodoacetamide to a final concentration of 4 mM and incubating at room temperature for 30 min in the dark. 0.5 µg trypsin was added to the beads and incubated at 37°C overnight. The sample was acidified with formic acid (final concentration of 5%) and then separated from the cAMP beads by microfuge centrifugation and dried in a speed vacuum. The dried peptides were resuspended in 10 µl of buffer A (5% acetonitrile and 0.1% formic acid) and analyzed by automated microcapillary liquid chromatography–tandem mass spectrometry. Fused-silica capillaries (100-µm inner diameter) were pulled using a CO2 laser puller (P-2000; Sutter Instrument) to a 5-µm inner diameter tip and packed with 10 cm of 5-µm Magic C18 material (Agilent Technologies) using a pressure bomb. This column was then placed in line with a quaternary HPLC pump (1100 series; Agilent Technologies) equipped with an autosampler (1100 series). The column was equilibrated in buffer A, and the peptide mixture was loaded onto the column using the autosampler. The HPLC pump flowed at 100 µl/min, and the flow rate to the electrospray tip was reduced to ∼200–300 nL/min by a split. The HPLC separation was provided by a gradient between buffer A and buffer B (90% acetonitrile and 0.1% formic acid). The HPLC gradient was held constant at 100% buffer A for 5 min after peptide loading followed by a 30-min gradient from 5% buffer B to 40% buffer B. The gradient was then switched from 40 to 80% buffer B over 5 min and held constant for 3 min. Finally, the gradient was changed from 80% buffer B to 100% buffer A over 1 min and then held constant at 100% buffer A for another 15 min. The application of a 1.8-kV distal voltage electrosprayed the eluted peptides directly into the ion trap mass spectrometer (LTQ; ThermoFinnigan) equipped with an electrospray ionization source (nanoLC; ThermoFinnigan). Full mass (tandem mass spectrometry) spectra were recorded on the peptides over a 400–2,000-mass/charge range followed by five tandem mass events sequentially generated in a data-dependent manner on the first-, second-, third-, fourth-, and fifth-most intense ions selected from the full mass spectrometry spectrum (at 35% collision energy). Mass spectrometer scan functions and HPLC solvent gradients were controlled by a data system (Xcalibur; ThermoFinnigan).
Isolation of proteins bound to calmodulin resin.
Proteins pulled down with calmodulin resin were run on a 4–12% Bis-Tris SDS-PAGE gel for 5 min. Gel bands were incubated with 20% acetonitrile in 0.1 M ammonium bicarbonate for 20 min and dehydrated with 100% acetonitrile. After drying, the gel pieces were resuspended in 1 mM DTT in 0.1 M ammonium bicarbonate and incubated for 30 min at room temperature. Next, 2 mM iodoacetamide was added, and samples were incubated for 30 min at room temperature. The supernatant was aspirated, and gel pieces were dehydrated with 100% acetonitrile. After drying, 250 ng trypsin was added in 0.1 M ammonium bicarbonate and incubated overnight. After incubation, the supernatant with the peptides was removed. The gel pieces were washed with 10% acetonitrile and 0.1% formic acid, and the washes were combined with the supernatant. This solution was dried in a speed vacuum, and the pellet was resuspended in 0.1% formic acid in 5% acetonitrile and loaded into the ion trap mass spectrometer (LTQ) as described in the methods for analyzing proteins bound to cAMP resin.
Data analysis.
Tandem mass spectrometry spectra were extracted from the RAW file with ReADW.exe. The resulting mzXML file contained all the data for all tandem mass spectrometry spectra and was read by the subsequent analysis software. The tandem mass spectrometry data were searched with InsPecT (Tanner et al., 2005) against the human International Protein Index database v3.31 (Kersey et al., 2004) with optional modifications: +16 on methionine, +57 on cysteine, and +80 on threonine, serine, and tyrosine. Only peptides with a P ≥ 0.01 were analyzed furthseccer.
Co-IPs
For endogenous AKAP11 co-IPs, HEK 293 cells were grown on 10-cm Nunc dishes to ∼80% confluency. One set was left untreated, whereas the other set was treated with 20 µM forskolin simultaneously with 200 µM IBMX for 1 h at 37°C. A co-IP kit (Universal Magnetic; Active Motif) was used to perform whole-cell extractions and then to perform the co-IPs. Proteins were analyzed by Western blotting. For FLAG-RIα co-IPs, HEK 293 cells were grown in 10-cm dishes to ∼80% confluency and transfected with FLAG plasmids. After 18–20 h, cells were harvested and lysed in a buffer (50 mM Tris, pH 7.0, 150 mM NaCl, 1 mM EDTA, and 1% NP-40) and bound to anti–Flag M2 affinity resin (Sigma-Aldrich) for 1 h at 4°C. Proteins that bound to the beads were eluted with SDS sample buffer and analyzed by Western blotting.
RNA interference
To knock down levels of AKAP11 with siRNA, Prkar1a−/− MEFs were transfected with 10 nM AKAP11 siRNA (siGENOME SMARTpool; Thermo Fisher Scientific) using Lipofectamine RNAiMAX (Invitrogen). The siGENOME nontargeting siRNA Pool #2 was used as a negative control. Cells were analyzed 1–2 d after siRNA treatment. Any transfection of plasmid DNA was performed 24 h after siRNA treatment.
Quantitative Western blot
Prkar1a+/+ MEFs and Prkar1a−/− MEFs were grown to ∼80% confluency on 10-cm Nunc dishes and transfected with empty GFP or GFP-tagged RIα, respectively. After 16–18 h, cells were harvested with cold CE1 buffer (10 mM Hepes, 60 mM KCl, 1 mM EDTA, 1 mM DTT, 1% BSA, and 0.5% NP-40) and lysed with sonication. Lysates were spun down at 14,000 rpm for 5 min. Proteins from the supernatant were analyzed by Western blotting. Band intensity levels were measured using the AlphaView Q software on an imaging system (FluorChem Q; Cell Biosciences, Inc.).
Stereology
The volume density of autophagosomes was determined using stereological observations similar to those previously described (Yuan et al., 2007) on electron microscopy images as follows. Point counting was used to determine the volume densities by overlaying a 100-point square grid on each digitized image (4,033 × 6,010 pixels with a pixel size of 2.8 nm) in Photoshop. Observations were made at the points of intersection (15 × 21 points) of the grid. First, the volume available for autophagosomes to occupy was counted on each image. This was accomplished by excluding those points lying on top of any nucleus present and any empty space, that is, space not occupied by the cytoplasm. Next, the points lying directly on top of autophagosomes were counted. The volume density of mitochondria was calculated by dividing the number of autophagosome points by the cytoplasm points.
Online supplemental material
Fig. S1 shows that puncta formation is not an artifact of RIα-GFP overexpression. Fig. S2 shows that ubiquitination of RIα is not the signal for targeting to MVBs. Video 1 shows that RIα puncta formation is reversible and independent of PKAc activity. Video 2 shows that RIα puncta is not rapidly degraded, and formation is reversible for up to 3 h. Video 3 shows that RIα puncta formation is not reversible in the presence of PKI. Table S1 lists the proteins identified to bind to all four R-subunit isoforms in untransfected HEK 293 cells. Table S2 lists the proteins identified to bind to all four R-subunit isoforms in HEK 293 cells that are stably expressing TAP-tagged RIα. Table S3 lists the proteins identified to bind to TAP-tagged RIα, which were free or in complex with PKAc, in HEK 293 cells that are stably expressing TAP-tagged RIα. Table S4 lists the proteins identified to bind to free TAP-tagged RIα in HEK 293 cells that are stably expressing TAP-tagged RIα. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201010034/DC1.
Acknowledgments
We thank Christopher Eggers for useful discussions and valuable assistance with colocalization experiments along the endosomal pathway. We wish to thank Ohkyung Kwon for help with image processing and analysis and both Ohkyung and Hiroyuki Hakozaki for exceptional assistance with microscope setup. We also wish to thank Philip Chang for assistance with the analysis of RIαGFP expression levels. We greatly appreciate discussions with Hilde Abrahamsen regarding autophagy. We also thank Christopher Benner for careful reviews of manuscript drafts.
This work was supported by National Institutes of Health grants DK54441 to S.S. Taylor and J.D. Scott, P41-RR004050 to M.H. Ellisman, and a Ruth L. Kirschstein National Research Service Award NIH/NCI T32 CA009523 to M.E. Day.
Footnotes
Abbreviations used in this paper:
- AIF
- apoptosis-inducing factor
- AKAP
- A-kinase–anchoring protein
- AKB
- A-kinase binding
- AQP2
- aquaporin 2
- D/D
- dimerization/docking
- EEA1
- early endosome antigen 1
- HEK
- human embryonic kidney
- IBMX
- isobutylmethylxanthine
- IP
- immunoprecipitation
- LC3
- light chain 3
- MEF
- mouse embryonic fibroblast
- MVB
- multivesicular body
- PKAc
- PKA catalytic subunit
- PKI
- protein kinase inhibitor
- PMP70
- 70-kD peroxisomal membrane protein
- R-subunit
- regulatory subunit
- TAP
- tandem affinity purification
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. 2004. Image processing with ImageJ. Biophoton. Int. 11:36–42 [Google Scholar]
- Amieux P.S., McKnight G.S. 2002. The essential role of RI alpha in the maintenance of regulated PKA activity. Ann. NY Acad. Sci. 968:75–95 10.1111/j.1749-6632.2002.tb04328.x [DOI] [PubMed] [Google Scholar]
- Banky P., Huang L.J., Taylor S.S. 1998. Dimerization/docking domain of the type Ialpha regulatory subunit of cAMP-dependent protein kinase. Requirements for dimerization and docking are distinct but overlapping. J. Biol. Chem. 273:35048–35055 10.1074/jbc.273.52.35048 [DOI] [PubMed] [Google Scholar]
- Bergold P.J., Beushausen S.A., Sacktor T.C., Cheley S., Bayley H., Schwartz J.H. 1992. A regulatory subunit of the cAMP-dependent protein kinase down-regulated in aplysia sensory neurons during long-term sensitization. Neuron. 8:387–397 10.1016/0896-6273(92)90304-V [DOI] [PubMed] [Google Scholar]
- Boeshans K.M., Resing K.A., Hunt J.B., Ahn N.G., Shabb J.B. 1999. Structural characterization of the membrane-associated regulatory subunit of type I cAMP-dependent protein kinase by mass spectrometry: identification of Ser81 as the in vivo phosphorylation site of RIalpha. Protein Sci. 8:1515–1522 10.1110/ps.8.7.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M., Weih F., Nichols M., Schütz G. 1991. The tissue-specific extinguisher locus TSE1 encodes a regulatory subunit of cAMP-dependent protein kinase. Cell. 66:849–859 10.1016/0092-8674(91)90432-X [DOI] [PubMed] [Google Scholar]
- Brandon E.P., Zhuo M., Huang Y.Y., Qi M., Gerhold K.A., Burton K.A., Kandel E.R., McKnight G.S., Idzerda R.L. 1995. Hippocampal long-term depression and depotentiation are defective in mice carrying a targeted disruption of the gene encoding the RI beta subunit of cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 92:8851–8855 10.1073/pnas.92.19.8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J.P., Bardswell S.C., Burgoyne J.R., Fuller W., Schröder E., Wait R., Begum S., Kentish J.C., Eaton P. 2006. Oxidant-induced activation of type I protein kinase A is mediated by RI subunit interprotein disulfide bond formation. J. Biol. Chem. 281:21827–21836 10.1074/jbc.M603952200 [DOI] [PubMed] [Google Scholar]
- Bubis J., Neitzel J.J., Saraswat L.D., Taylor S.S. 1988. A point mutation abolishes binding of cAMP to site A in the regulatory subunit of cAMP-dependent protein kinase. J. Biol. Chem. 263:9668–9673 [PubMed] [Google Scholar]
- Burns-Hamuro L.L., Ma Y., Kammerer S., Reineke U., Self C., Cook C., Olson G.L., Cantor C.R., Braun A., Taylor S.S. 2003. Designing isoform-specific peptide disruptors of protein kinase A localization. Proc. Natl. Acad. Sci. USA. 100:4072–4077 10.1073/pnas.2628038100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton K.A., Treash-Osio B., Muller C.H., Dunphy E.L., McKnight G.S. 1999. Deletion of type IIalpha regulatory subunit delocalizes protein kinase A in mouse sperm without affecting motility or fertilization. J. Biol. Chem. 274:24131–24136 10.1074/jbc.274.34.24131 [DOI] [PubMed] [Google Scholar]
- Chaturvedi D., Poppleton H.M., Stringfield T., Barbier A., Patel T.B. 2006. Subcellular localization and biological actions of activated RSK1 are determined by its interactions with subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 26:4586–4600 10.1128/MCB.01422-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.X., Marshall J.L., Ness E., Martin R.R., Dvorchik B., Rizvi N., Marquis J., McKinlay M., Dahut W., Hawkins M.J. 2000. A safety and pharmacokinetic study of a mixed-backbone oligonucleotide (GEM231) targeting the type I protein kinase A by two-hour infusions in patients with refractory solid tumors. Clin. Cancer Res. 6:1259–1266 [PubMed] [Google Scholar]
- Clague M.J., Urbé S. 2008. Multivesicular bodies. Curr. Biol. 18:R402–R404 10.1016/j.cub.2008.02.068 [DOI] [PubMed] [Google Scholar]
- Clegg C.H., Cadd G.G., McKnight G.S. 1988. Genetic characterization of a brain-specific form of the type I regulatory subunit of cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 85:3703–3707 10.1073/pnas.85.11.3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers C.T., Schafer J.C., Goldenring J.R., Taylor S.S. 2009. D-AKAP2 interacts with Rab4 and Rab11 through its RGS domains and regulates transferrin receptor recycling. J. Biol. Chem. 284:32869–32880 10.1074/jbc.M109.022582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder S., Miller K., Moehren G., Ullrich A., Schlessinger J., Hopkins C.R. 1990. Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell. 61:623–634 10.1016/0092-8674(90)90474-S [DOI] [PubMed] [Google Scholar]
- Garnis C., Rosin M.P., Zhang L., Lam W.L. 2005. Alteration of AKAP220, an upstream component of the Rb pathway, in oral carcinogenesis. Int. J. Cancer. 116:813–819 10.1002/ijc.21065 [DOI] [PubMed] [Google Scholar]
- Goel S., Desai K., Macapinlac M., Wadler S., Goldberg G., Fields A., Einstein M., Volterra F., Wong B., Martin R., Mani S. 2006. A phase I safety and dose escalation trial of docetaxel combined with GEM231, a second generation antisense oligonucleotide targeting protein kinase A R1alpha in patients with advanced solid cancers. Invest. New Drugs. 24:125–134 10.1007/s10637-006-2378-x [DOI] [PubMed] [Google Scholar]
- Gupte R.S., Weng Y., Liu L., Lee M.Y. 2005. The second subunit of the replication factor C complex (RFC40) and the regulatory subunit (RIalpha) of protein kinase A form a protein complex promoting cell survival. Cell Cycle. 4:322–329 10.4161/cc.4.2.1470 [DOI] [PubMed] [Google Scholar]
- Hegde A.N., Goldberg A.L., Schwartz J.H. 1993. Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: a molecular mechanism underlying long-term synaptic plasticity. Proc. Natl. Acad. Sci. USA. 90:7436–7440 10.1073/pnas.90.16.7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg F.W., Taylor S.S., Dostmann W.R. 1996. Active site mutations define the pathway for the cooperative activation of cAMP-dependent protein kinase. Biochemistry. 35:2934–2942 10.1021/bi951647c [DOI] [PubMed] [Google Scholar]
- Herberg F.W., Maleszka A., Eide T., Vossebein L., Tasken K. 2000. Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J. Mol. Biol. 298:329–339 10.1006/jmbi.2000.3662 [DOI] [PubMed] [Google Scholar]
- Imaizumi-Scherrer T., Faust D.M., Barradeau S., Hellio R., Weiss M.C. 2001. Type I protein kinase a is localized to interphase microtubules and strongly associated with the mitotic spindle. Exp. Cell Res. 264:250–265 10.1006/excr.2001.5164 [DOI] [PubMed] [Google Scholar]
- Jahnsen T., Hedin L., Kidd V.J., Beattie W.G., Lohmann S.M., Walter U., Durica J., Schulz T.Z., Schiltz E., Browner M., et al. 1986. Molecular cloning, cDNA structure, and regulation of the regulatory subunit of type II cAMP-dependent protein kinase from rat ovarian granulosa cells. J. Biol. Chem. 261:12352–12361 [PubMed] [Google Scholar]
- Jones K.W., Shapero M.H., Chevrette M., Fournier R.E. 1991. Subtractive hybridization cloning of a tissue-specific extinguisher: TSE1 encodes a regulatory subunit of protein kinase A. Cell. 66:861–872 10.1016/0092-8674(91)90433-Y [DOI] [PubMed] [Google Scholar]
- Kammer G.M., Khan I.U., Kammer J.A., Olorenshaw I., Mathis D. 1996. Deficient type I protein kinase A isozyme activity in systemic lupus erythematosus T lymphocytes: II. Abnormal isozyme kinetics. J. Immunol. 157:2690–2698 [PubMed] [Google Scholar]
- Katzmann D.J., Babst M., Emr S.D. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- Kersey P.J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. 2004. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 4:1985–1988 10.1002/pmic.200300721 [DOI] [PubMed] [Google Scholar]
- Kirschner L.S., Carney J.A., Pack S.D., Taymans S.E., Giatzakis C., Cho Y.S., Cho-Chung Y.S., Stratakis C.A. 2000. Mutations of the gene encoding the protein kinase A type I-alpha regulatory subunit in patients with the Carney complex. Nat. Genet. 26:89–92 10.1038/79238 [DOI] [PubMed] [Google Scholar]
- Kuma A., Matsui M., Mizushima N. 2007. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 3:323–328 [DOI] [PubMed] [Google Scholar]
- Laxminarayana D., Khan I.U., Mishra N., Olorenshaw I., Taskén K., Kammer G.M. 1999. Diminished levels of protein kinase A RI alpha and RI beta transcripts and proteins in systemic lupus erythematosus T lymphocytes. J. Immunol. 162:5639–5648 [PubMed] [Google Scholar]
- Lee D.C., Carmichael D.F., Krebs E.G., McKnight G.S. 1983. Isolation of a cDNA clone for the type I regulatory subunit of bovine cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 80:3608–3612 10.1073/pnas.80.12.3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester L.B., Coghlan V.M., Nauert B., Scott J.D. 1996. Cloning and characterization of a novel A-kinase anchoring protein. AKAP 220, association with testicular peroxisomes. J. Biol. Chem. 271:9460–9465 10.1074/jbc.271.16.9460 [DOI] [PubMed] [Google Scholar]
- Levy F.O., Rasmussen A.M., Taskén K., Skålhegg B.S., Huitfeldt H.S., Funderud S., Smeland E.B., Hansson V. 1996. Cyclic AMP-dependent protein kinase (cAK) in human B cells: co-localization of type I cAK (RI alpha 2 C2) with the antigen receptor during anti-immunoglobulin-induced B cell activation. Eur. J. Immunol. 26:1290–1296 10.1002/eji.1830260617 [DOI] [PubMed] [Google Scholar]
- Lim C.J., Han J., Yousefi N., Ma Y., Amieux P.S., McKnight G.S., Taylor S.S., Ginsberg M.H. 2007. Alpha4 integrins are type I cAMP-dependent protein kinase-anchoring proteins. Nat. Cell Biol. 9:415–421 10.1038/ncb1561 [DOI] [PubMed] [Google Scholar]
- Mavrakis M., Lippincott-Schwartz J., Stratakis C.A., Bossis I. 2006. Depletion of type IA regulatory subunit (RIalpha) of protein kinase A (PKA) in mammalian cells and tissues activates mTOR and causes autophagic deficiency. Hum. Mol. Genet. 15:2962–2971 10.1093/hmg/ddl239 [DOI] [PubMed] [Google Scholar]
- McKnight G.S., Cummings D.E., Amieux P.S., Sikorski M.A., Brandon E.P., Planas J.V., Motamed K., Idzerda R.L. 1998. Cyclic AMP, PKA, and the physiological regulation of adiposity. Recent Prog. Horm. Res. 53:139–159; discussion 160–161 [PubMed] [Google Scholar]
- Miller W.R. 2002. Regulatory subunits of PKA and breast cancer. Ann. NY Acad. Sci. 968:37–48 10.1111/j.1749-6632.2002.tb04325.x [DOI] [PubMed] [Google Scholar]
- Okutsu R., Rai T., Kikuchi A., Ohno M., Uchida K., Sasaki S., Uchida S. 2008. AKAP220 colocalizes with AQP2 in the inner medullary collecting ducts. Kidney Int. 74:1429–1433 10.1038/ki.2008.402 [DOI] [PubMed] [Google Scholar]
- Orellana S.A., McKnight G.S. 1990. The S49 Kin- cell line transcribes and translates a functional mRNA coding for the catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 265:3048–3053 [PubMed] [Google Scholar]
- Reggiori F., Pelham H.R. 2001. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20:5176–5186 10.1093/emboj/20.18.5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann E.M., Walsh D.A., Krebs E.G. 1971. Purification and properties of rabbit skeletal muscle adenosine 3′,5′-monophosphate-dependent protein kinases. J. Biol. Chem. 246:1986–1995 [PubMed] [Google Scholar]
- Reinton N., Collas P., Haugen T.B., Skâlhegg B.S., Hansson V., Jahnsen T., Taskén K. 2000. Localization of a novel human A-kinase-anchoring protein, hAKAP220, during spermatogenesis. Dev. Biol. 223:194–204 10.1006/dbio.2000.9725 [DOI] [PubMed] [Google Scholar]
- Rubin C.S. 1979. Characterization and comparison of membrane-associated and cytosolic cAMP-dependent protein kinases. Studies on human erythrocyte protein kinases. J. Biol. Chem. 254:12439–12449 [PubMed] [Google Scholar]
- Sarma G.N., Kinderman F.S., Kim C., von Daake S., Chen L., Wang B.C., Taylor S.S. 2010. Structure of D-AKAP2:PKA RI complex: insights into AKAP specificity and selectivity. Structure. 18:155–166 10.1016/j.str.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.D., Glaccum M.B., Zoller M.J., Uhler M.D., Helfman D.M., McKnight G.S., Krebs E.G. 1987. The molecular cloning of a type II regulatory subunit of the cAMP-dependent protein kinase from rat skeletal muscle and mouse brain. Proc. Natl. Acad. Sci. USA. 84:5192–5196 10.1073/pnas.84.15.5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg R.A., Agard D.A. 1981. Turnover of regulatory subunit of cyclic AMP-dependent protein kinase in S49 mouse lymphoma cells. Regulation by catalytic subunit and analogs of cyclic AMP. J. Biol. Chem. 256:10731–10734 [PubMed] [Google Scholar]
- Takata K., Matsuzaki T., Tajika Y., Ablimit A., Hasegawa T. 2008. Localization and trafficking of aquaporin 2 in the kidney. Histochem. Cell Biol. 130:197–209 10.1007/s00418-008-0457-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S., Shu H., Frank A., Wang L.C., Zandi E., Mumby M., Pevzner P.A., Bafna V. 2005. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77:4626–4639 10.1021/ac050102d [DOI] [PubMed] [Google Scholar]
- Thiele T.E., Willis B., Stadler J., Reynolds J.G., Bernstein I.L., McKnight G.S. 2000. High ethanol consumption and low sensitivity to ethanol-induced sedation in protein kinase A-mutant mice. J. Neurosci. 20:RC75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortora G., Pepe S., Bianco C., Baldassarre G., Budillon A., Clair T., Cho-Chung Y.S., Bianco A.R., Ciardiello F. 1994a. The RI alpha subunit of protein kinase A controls serum dependency and entry into cell cycle of human mammary epithelial cells. Oncogene. 9:3233–3240 [PubMed] [Google Scholar]
- Tortora G., Pepe S., Bianco C., Damiano V., Ruggiero A., Baldassarre G., Corbo C., Cho-Chung Y.S., Bianco A.R., Ciardiello F. 1994b. Differential effects of protein kinase A sub-units on Chinese-hamster-ovary cell cycle and proliferation. Int. J. Cancer. 59:712–716 10.1002/ijc.2910590521 [DOI] [PubMed] [Google Scholar]
- Uhler M.D., McKnight G.S. 1987. Expression of cDNAs for two isoforms of the catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 262:15202–15207 [PubMed] [Google Scholar]
- van Niel G., Porto-Carreiro I., Simoes S., Raposo G. 2006. Exosomes: a common pathway for a specialized function. J. Biochem. 140:13–21 10.1093/jb/mvj128 [DOI] [PubMed] [Google Scholar]
- Veugelers M., Wilkes D., Burton K., McDermott D.A., Song Y., Goldstein M.M., La Perle K., Vaughan C.J., O’Hagan A., Bennett K.R., et al. 2004. Comparative PRKAR1A genotype-phenotype analyses in humans with Carney complex and prkar1a haploinsufficient mice. Proc. Natl. Acad. Sci. USA. 101:14222–14227 10.1073/pnas.0405535101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W., Scott J.D. 2004. AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5:959–970 10.1038/nrm1527 [DOI] [PubMed] [Google Scholar]
- Yang W.L., Iacono L., Tang W.M., Chin K.V. 1998. Novel function of the regulatory subunit of protein kinase A: regulation of cytochrome c oxidase activity and cytochrome c release. Biochemistry. 37:14175–14180 10.1021/bi981402a [DOI] [PubMed] [Google Scholar]
- Yuan H., Gerencser A.A., Liot G., Lipton S.A., Ellisman M., Perkins G.A., Bossy-Wetzel E. 2007. Mitochondrial fission is an upstream and required event for bax foci formation in response to nitric oxide in cortical neurons. Cell Death Differ. 14:462–471 10.1038/sj.cdd.4402046 [DOI] [PubMed] [Google Scholar]