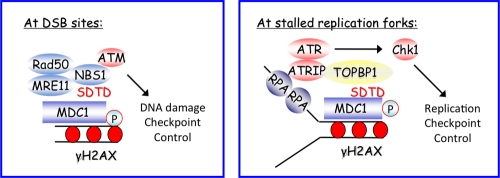
The DNA damage checkpoint protein MDC1 also interacts with TopBP1 to promote DNA replication checkpoint control.
Abstract
Human TopBP1 is a major player in the control of the DNA replication checkpoint. In this study, we identified MDC1, a key checkpoint protein involved in the cellular response to DNA double-strand breaks, as a TopBP1-associated protein. The specific TopBP1–MDC1 interaction is mediated by the fifth BRCT domain of TopBP1 and the Ser-Asp-Thr (SDT) repeats of MDC1. In addition, we demonstrated that TopBP1 accumulation at stalled replication forks is promoted by the H2AX/MDC1 signaling cascade. Moreover, MDC1 is important for ATR-dependent Chk1 activation in response to replication stress. Collectively, our data suggest that MDC1 facilitates several important steps in both cellular DNA damage response and the DNA replication checkpoint.
Introduction
In eukaryotic cells, the DNA damage response helps to maintain genomic integrity. DNA damage induces signaling pathways that activate DNA repair processes and cell cycle checkpoints. The phosphoinositide kinase-related kinases ataxia telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) are involved in DNA damage response and replication checkpoint control, respectively. ATM is activated primarily by DNA double-strand breaks (DSBs), whereas ATR responds principally to replication blockage or replication stress. In response to DNA DSBs, the histone variant H2AX is phosphorylated by ATM, which recruits a downstream checkpoint protein, mediator of DNA damage checkpoint protein 1 (MDC1), to sites of DNA damage. In addition, MDC1 is also phosphorylated on DNA damage and further facilitates the loading of the E3 ubiquitin ligase RNF8 to DSB sites. RNF8 ubiquitinates H2AX, and probably other substrates, and facilitates the accumulation of many DNA damage repair proteins at sites of DSBs (Wood and Chen, 2008; Yan and Jetten, 2008; Messick and Greenberg, 2009). The accumulation of these DNA damage repair proteins at DSB sites via the H2AX/MDC1-dependent pathway is generally believed to facilitate DNA damage repair and checkpoint control in response to DSBs.
A similar signal transduction pathway exists for cellular response to replication stress. We showed recently that both the replication checkpoint protein TopBP1 and a DNA helicase, BACH1 (also known as FANCJ), are recruited to stalled replication forks, facilitating the accumulation of additional replication protein A (RPA)-coated single-stranded DNA (ssDNA) at stalled replication forks (Gong et al., 2010). This efficient accumulation of RPA-coated ssDNA leads to the assembly of multiprotein complexes, including ATR–ATR interacting protein (ATR–ATRIP), TopBP1, and Rad9–Hus1–Rad1 (dubbed as 9-1-1) at stalled replication forks, which is required for the activation of ATR kinase activity and for subsequent Chk1 phosphorylation and activation (Kumagai and Dunphy, 2006; Burrows and Elledge, 2008; Cimprich and Cortez, 2008; Yan and Michael, 2009).
Human TopBP1 and its orthologues in other organisms play important roles in DNA replication and replication checkpoint control (Saka et al., 1994; Wang and Elledge, 1999; Yamamoto et al., 2000; Mäkiniemi et al., 2001; Van Hatten et al., 2002; Yamane et al., 2002; Kim et al., 2005). It has been suggested that TopBP1 has acquired diverse functions by its abilities to interact with many binding partners via its multiple protein–protein interaction domains, including eight BRCA1 C-terminal (BRCT) phospho-peptide recognition motifs. For instance, TopBP1 regulates DNA replication initiation. Early studies in yeast suggested that this function of Dpb11, the yeast orthologue of TopBP1, can interact with Sld3 through BRCT1-2 of Dpb11 and with Sld2 through BRCT3-4 of Dpb11 (Tanaka et al., 2007; Zegerman and Diffley, 2007). More recently, Treslin/Ticrr has been shown to collaborate with TopBP1 in promoting replication initiation (Kumagai et al., 2010; Sansam et al., 2010). Although Treslin/Ticrr does not share any obvious sequence homology with yeast Sld2 or Sld3, the same N-terminal tandem BRCT1-2 domains are involved in this interaction, which suggests that the functions of TopBP1 are evolutionarily conserved.
TopBP1 also plays a key role in replication checkpoint control. An ATR-activating domain within TopBP1 interacts directly with ATR–ATRIP and thus activates ATR kinase activity (Kumagai et al., 2006). In addition, TopBP1 also interacts with the phosphorylated Rad9 tail of the 9-1-1 complex through its N-terminal tandem BRCT1-2 domains (Delacroix et al., 2007; Lee et al., 2007); this interaction is also required for Chk1 activation. The same N-terminal BRCT domains of TopBP1 interact with Rad9, NBS1, and (as recently shown) Treslin/Ticrr (Delacroix et al., 2007; Lee et al., 2007; Yoo et al., 2009), which indicates that the diverse roles of TopBP1 in replication and replication checkpoint control may be mediated by its distinct binding partners. Recently, we reported that TopBP1 associates with BACH1 through the very C-terminal tandem BRCT domains of TopBP1, which are required for early replication checkpoint control (Gong et al., 2010). However, we showed that BACH1 is not required for the accumulation of TopBP1 at stalled replication forks (Gong et al., 2010). Thus, despite all of these advances, we still do not know how TopBP1 accumulates at stalled replication forks. Although we showed that the fifth BRCT domain (BRCT5) of TopBP1 is required for TopBP1 focus formation after DNA damage (Yamane et al., 2002), the identity of an upstream regulator that would bind to TopBP1 BRCT5 and facilitate the recruitment of TopBP1 to DNA damage sites remains elusive.
In this study, we report a functional interaction between TopBP1 and MDC1. MDC1 is a large adaptor protein, best known for its roles in DNA damage response after DNA DSBs (Jungmichel and Stucki, 2010). MDC1 binds to the phosphorylated Ser139 site of H2AX (γ-H2AX) through its tandem BRCT domains, which further amplify DNA damage signals. MDC1 also binds to RNF8 and initiates an ubiquitination-mediated signaling cascade at DSB sites. Recently, we and others have shown that phosphorylation of the conserved Ser-Asp-Thr (SDT) repeats at the N terminus of MDC1 facilitates the recruitment and retention of NBS1 at DNA damage sites, thereby increasing the local concentration of the MRE11–RAD50–NBS1 (MRN) complex, which is required for intra–S phase checkpoint control after DNA DSBs (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008). Here, we describe a physical interaction between MDC1 and TopBP1 and suggest that MDC1 plays a similar, but unexpected, role in replication checkpoint control.
Results and discussion
TopBP1 accumulation at stalled replication forks requires TopBP1 BRCT5 domain
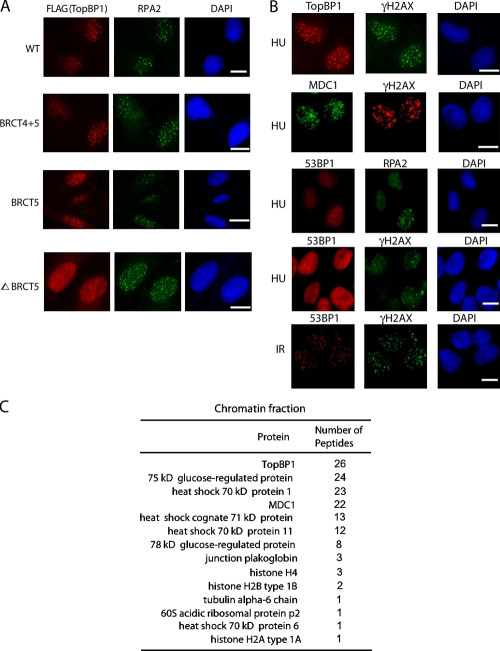
Previous work by our group documented that TopBP1 BRCT5 domain is important for TopBP1 focus formation in response to DNA damage (Yamane et al., 2002). To finely map the focus localization region of TopBP1, we generated several TopBP1 constructs. Similar to our previous results (Yamane et al., 2002), we found that deletion of TopBP1 BRCT5 domain abolished TopBP1 focus formation after hydroxyurea (HU) treatment, whereas normal focus localization was observed when a construct containing both the BRCT4 and BRCT5 domains (BRCT4+5) of TopBP1 was used (Fig. 1 A). We found that a region containing BRCT5 domain of TopBP1 (residues 545–722) is sufficient for TopBP1 focus formation after HU treatment (Fig. 1 A). HU treatment should lead to replication stress only in S phase cells. Indeed, we found that HU-induced TopBP1 focus formation was restricted to S phase cells, which were also positive for cyclin A staining (Fig. S1 A). To distinguish whether HU-induced TopBP1 focus formation represents stalled replication forks or fork-derived DNA DSBs, we used 53BP1 as a marker of DNA DSBs and found that under our experimental condition (after 2 h of treatment with 2 mM HU), HU treatment did not induce a significant amount of DNA DSBs (Fig. 1 B). This result was further confirmed by a time-course experiment after HU treatment (Fig. S1 B).
Figure 1.
TopBP1 accumulation at stalled replication forks requires TopBP1 BRCT5 domain. (A) U2OS cells were transfected with plasmids encoding SFB-tagged WT or deletion mutants of TopBP1. Immunostaining was performed with the indicated antibodies in cells treated with 2 mM HU. (B) U2OS cells were treated with HU (top) or IR (bottom). After 2 h, cells were fixed and immunostaining was performed with the indicated antibodies. Bars, 10 µm. (C) TopBP1 BRCT4+5-associated proteins in the chromatin fraction identified by mass spectrometric analysis.
Next, we wanted to identify the upstream signaling molecules that facilitate TopBP1 accumulation at stalled replication forks. Thus, we performed tandem affinity purification using lysates derived from 293T cells stably expressing triple-tagged (S protein, FLAG, and streptavidin-binding peptide [SFB]-tagged) BRCT4+5 domain of TopBP1. Surprisingly, mass spectrometric analysis showed that MDC1 was the major TopBP1-associated protein in the chromatin fraction (Fig. 1 C), which suggests that MDC1 may be involved in TopBP1 accumulation at stalled replication forks.
TopBP1 focus localization after replication stress requires a H2AX/MDC1-dependent signaling pathway
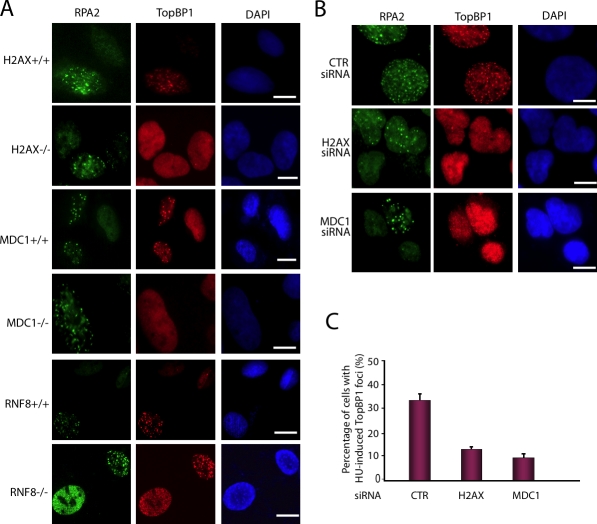
We next explored whether MDC1 might be essential for the focus accumulation of TopBP1 at stalled replication forks after replication stress. We used a panel of mouse embryonic fibroblast (MEF) cell lines deficient in MDC1 or MDC1-associated molecules. We found that TopBP1 focus formation was greatly reduced in H2AX−/− and MDC1−/− MEFs compared with their wild-type (WT) counterparts (Fig. 2 A), which indicates that HU-induced focus formation of TopBP1 requires both H2AX and MDC1. In contrast, normal TopBP1 focus formation was observed in both RNF8−/− MEFs and their WT counterparts (Fig. 2 A), which suggests that the RNF8-dependent ubiquitination cascade is not involved in TopBP1 accumulation after replication stress.
Figure 2.
TopBP1 focus formation depends on H2AX/MDC1 but not on RNF8 after replication stress. (A) Cells deficient in H2AX, MDC1, and RNF8, and their WT counterparts were treated with HU, and immunostaining experiments were performed with anti-TopBP1 and anti-RPA2 antibodies. (B) U2OS cells were transfected with control siRNA (CTR), H2AX-specific, or MDC1-specific siRNA. Cells were treated with HU, fixed, and immunostained with anti-TopBP1 and anti-RPA2 antibodies. Bars, 10 µm. (C) Percentages of cells stained positive for TopBP1 foci were determined in cells transfected with the indicated siRNAs. Data are presented as mean ± SD (error bars) from three different experiments.
Similarly, we observed that HU-induced TopBP1 focus formation was reduced in U2OS cells with H2AX or MDC1 knockdown (Fig. 2, B and C; and Fig. S2 C). Collectively, these data demonstrate that TopBP1 acts downstream of H2AX and MDC1, but is independent of RNF8, in response to replication stress.
MDC1 interacts with TopBP1
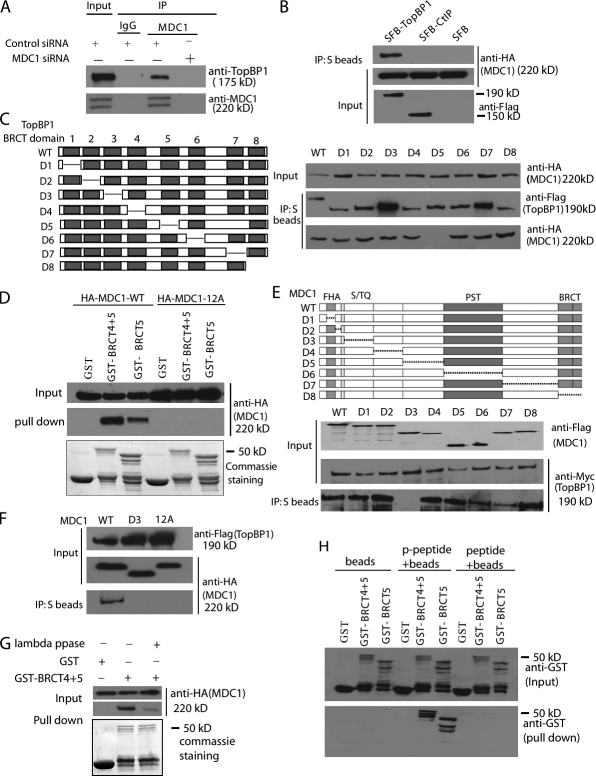
In addition to identifying MDC1 as a major TopBP1-associated protein, we also repeatedly identified TopBP1 as a major MDC1-associated protein in the chromatin fraction by tandem affinity purification using lysates derived from 293T cell lines stably expressing SFB-tagged human MDC1 (unpublished data). Collectively, these results provide support for a physical interaction between TopBP1 and MDC1. We performed coimmunoprecipitation (co-IP) experiments and confirmed an interaction not only between endogenous proteins (Fig. 3 A), but also between overexpressed MDC1 and TopBP1 (Fig. 3 B), which suggests that TopBP1 is a bona fide MDC1-interacting protein. Moreover, knocking down the other MDC1 binding partner, NBS1, did not affect the binding of MDC1 with TopBP1 (Fig. S3 A).
Figure 3.
TopBP1 interacts with MDC1. (A) Endogenous interaction between TopBP1 and MDC1. HeLa cells were transfected with control siRNA or MDC1-specific siRNA. Control or anti-MDC1 immunoprecipitates were immunoblotted with the indicated antibodies. (B) TopBP1 binds specifically to MDC1. Constructs encoding SFB-tagged TopBP1, CtIP, or vector alone were cotransfected with plasmids encoding HA-tagged MDC1. Immunoprecipitation (IP) and immunoblotting were performed as indicated. (C) Schematic presentation of WT and deletion mutants of TopBP1 used in this study (left). 293T cells were transfected with plasmids encoding HA-tagged MDC1 together with plasmids encoding WT or deletion mutants of SFB-tagged TopBP1. IP reactions were conducted using S protein beads and then subjected to Western blotting using the indicated antibodies (right). (D) Beads coated with bacterially expressed GST, GST fusion of TopBP1 BRCT4+5 domains, or TopBP1 BRCT5 domain were incubated with cell lysates containing exogenously expressed HA-tagged WT MDC1. Immunoblotting experiments were performed using the indicated antibodies (top). (E) Schematic diagram of WT and deletion mutants of MDC1 used in this study (top). 293T cells were transfected with plasmids encoding Myc-tagged TopBP1 together with plasmids encoding WT or deletion mutants of SFB-tagged MDC1. IP reactions were conducted using S protein beads and then subjected to Western blot analyses using antibodies as indicated. (F) TopBP1 binds to the phosphorylated SDT repeats of MDC1. 293T cells were transfected with plasmids encoding HA-tagged WT, D3 mutant, or 12A mutant of MDC1 together with plasmids encoding SFB-tagged TopBP1. IP reactions were conducted using S protein beads and then subjected to Western blotting using the indicated antibodies. (G) Extracts prepared from 293T cells expressing HA-tagged MDC1 were mock-treated or treated with λ phosphatase. Extracts were then incubated with 10 µg of bacterially expressed and purified GST or GST-BRCT4+5 fusion proteins immobilized on glutathione agarose beads for 2 h at 4°C. The complex was separated by SDS-PAGE, and the amount of MDC1 that bound specifically to TopBP1 BRCT5 domain was evaluated by immunoblotting. (H) Phosphorylated or control MDC1 peptides were incubated with purified GST, or GST-BRCT4+5 or GST-BRCT5 fusion proteins. Input (top) and GST fusion proteins associated with peptides (bottom) were assessed by immunoblotting using anti-GST antibodies.
To determine the regions on TopBP1 required for its interaction with MDC1, we subjected SFB-tagged WT TopBP1 and a series of TopBP1 internal-deletion mutants (Fig. 3 C, left) to co-IP experiments with full-length HA-tagged MDC1. Only deletion of TopBP1 BRCT5 domain led to a dramatic decrease in the TopBP1–MDC1 interaction (Fig. 3 C, right), confirming that TopBP1 BRCT5 domain is responsible for the binding of TopBP1 to MDC1. Furthermore, using bacterially expressed and purified proteins, we found that both TopBP1 BRCT4+5 and TopBP1 BRCT5 domains bound to MDC1 (Fig. 3 D). To determine which residues in the BRCT5 domain of TopBP1 are required for the association of TopBP1 with MDC1, we mutated two highly conserved Trp residues in BRCT5 (W711R and W720R). These mutants disrupted the TopBP1–MDC1 interaction and accordingly abolished TopBP1 focus formation (Fig. S3, B and C).
Next, we sought to define the TopBP1 binding regions on MDC1. Again, a series of MDC1 internal-deletion mutants were coexpressed with Myc-tagged TopBP1 in 293T cells. The interaction between MDC1 and TopBP1 was dramatically decreased by D3, a mutant with deletion of a region of MDC1 that is enriched for SDT repeats (Fig. 3 E). This result indicates that the SDT repeats of MDC1 may be involved in its interaction with TopBP1, just as they are involved in its interaction with NBS1 (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008). These SDT repeats are phosphorylated by CK2 kinase (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008). If TopBP1 binds to these phosphorylated repeats of MDC1, we would expect that a 12A mutant of MDC1, in which the Ser/Thr residues in all six SDT repeats were changed to Ala, would abolish the MDC1–TopBP1 interaction. Indeed, we found this to be the case (Fig. 3, D and F). In addition, the binding between MDC1 and TopBP1 was dramatically decreased when extracts were pretreated with λ phosphatase (Fig. 3 G). Moreover, only biotinylated phosphopeptides containing the consensus (p)SD(p)TDXE motif of MDC1, but not unphosphorylated peptides with the identical sequence, could pull down TopBP1 BRCT4+5 or TopBP1 BRCT5 fusion proteins (Fig. 3 H). Together, these data indicate that TopBP1 associates only with MDC1 that is phosphorylated at these conserved SDTD motifs.
Both TopBP1 and MDC1 are required for ATR activation in response to replication stress
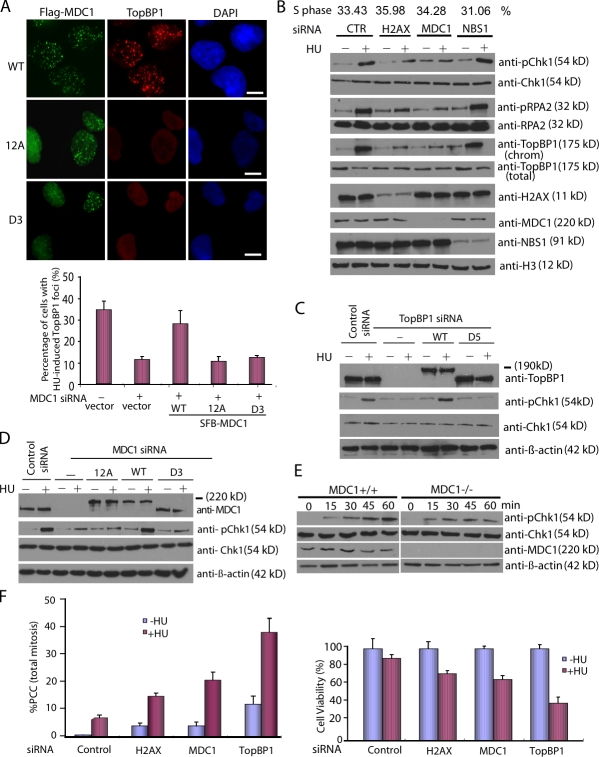
Given that TopBP1 focus formation requires MDC1, we next determined whether the TopBP1-binding region on MDC1 is also required for TopBP1 accumulation in response to replication stress. We transfected HeLa cells with FLAG-tagged siRNA-resistant WT MDC1, D3 mutant, or 12A mutant of MDC1. After siRNA-mediated depletion of endogenous MDC1, HU-induced TopBP1focus formation was observed only in the cells reconstituted with WT MDC1 and not in the cells reconstituted with D3 mutant or 12A mutant of MDC1 (Fig. 4 A), which indicates that these SDT repeats of MDC1 are required for the TopBP1 focus formation after replication stress.
Figure 4.
The TopBP1–MDC1 interaction is required for replication checkpoint control. (A) The SDT repeats of MDC1 are required for TopBP1 focus formation in response to HU. HeLa cells were transfected with FLAG-tagged siRNA-resistant WT, D3 mutant, or 12A mutant of MDC1, and with MDC1 siRNA twice with a 24-h interval. Cells were selected with puromycin for 48 h and then treated with HU. Immunostaining experiments were performed with anti-FLAG and anti-TopBP1 antibodies. Percentages of cells stained positive for TopBP1 foci were determined. Data are presented as mean ± SD from three different experiments. Bars, 10 µm. (B) Both H2AX and MDC1 are required for Chk1 activation in response to replication stress. U2OS cells transfected with control siRNA, H2AX siRNA, MDC1 siRNA, or NBS1 siRNA were mock-treated or treated with HU and harvested 1 h later. Cell lysates were prepared and immunoblotted with antibodies as indicated. (C) The interaction between TopBP1 and MDC1 is required for Chk1 activation. U2OS cells or U2OS cells stably expressing siRNA-resistant WT or D5 deletion mutant of TopBP1 were transfected with TopBP1 siRNA. 72 h after initial siRNA transfection, cells were treated with HU and collected 1 h later. Cell lysates were immunoblotted with the indicated antibodies. (D) HeLa cells were transfected with FLAG-tagged siRNA-resistant WT, D3 mutant, or 12A mutant of MDC1, and with MDC1 siRNA twice with a 24-h interval. Cells were selected with puromycin for 48 h and then treated with HU and collected 1 h later. Cell lysates were immunoblotted with the indicated antibodies. (E) MDC1−/− MEFs and WT control MEFs treated with HU for different time periods and p-Chk1 levels were determined by Western blotting. (F, left) MDC1 prevents PCC after replication stress. HeLa cells were transfected with control siRNA, TopBP1 siRNA, or MDC1 siRNA and then treated with 2 mM HU and 200 ng/ml nocodazole for 20 h. Mitotic spreads were prepared, and percentages of cells containing PCC were evaluated under the microscope. Data are presented as mean ± SD (error bars) from three independent experiments. (F, right) HeLa cells were arrested by HU for 12 h and released by changing with fresh medium. Cell survival after HU treatment was measured with clonogenic assay. Data are presented as mean ± SD (error bars) from three independent experiments.
TopBP1 is required for Chk1 activation after replication stress (Burrows and Elledge, 2008). Although MDC1 is clearly involved in DNA damage response, its function in the replication stress pathway remains to be determined. We first examined the role of MDC1 in Chk1 phosphorylation after replication stress. Consistent with previous results (Kim et al., 2005), we found that depletion of TopBP1 inhibited HU-induced Chk1 phosphorylation (Fig. 4 C). MDC1-depleted cells also exhibited obvious reductions in HU-induced Chk1 and RPA2 phosphorylation, whereas NBS1-depleted cells displayed normal Chk1 phosphorylation in response to HU (Fig. 4 B).
Although the expression of siRNA-resistant WT TopBP1 completely restored Chk1 activation in cells depleted of endogenous TopBP1, reconstitution with a TopBP1 mutant with deletion of BRCT5 domain failed to rescue HU-induced Chk1 phosphorylation (Fig. 4 C). Furthermore, the expression of siRNA-resistant WT MDC1 fully rescued Chk1 activation in MDC1-depleted cells, whereas the expression of siRNA-resistant D3 mutant or 12A mutant of MDC1 failed to rescue the Chk1 phosphorylation defect after HU treatment (Fig. 4 D). These data indicate that the TopBP1–MDC1 interaction plays an important role in Chk1 activation after replication stress.
To further explore whether MDC1 is required for initial ATR activation or for signal amplification, we performed a detailed time-course experiment. MDC1−/− MEF cells and WT control MEF cells were treated with HU for different time periods, and Chk1 phosphorylation levels were determined by Western blotting. Chk1 phosphorylation was increased after 15 min of HU treatment in both MDC1−/− MEF cells and WT control MEF cells (Fig. 4 E). However, the Chk1 phosphorylation level kept on increasing continuously in WT cells but not in MDC1−/− MEF cells. These results indicate that MDC1 is not required for initial ATR activation, but is involved in the amplification of ATR signaling after replication stress.
It is well established that replication checkpoint defects that abrogate the ATR–Chk1 pathway would lead to premature chromosome condensation (PCC; Nghiem et al., 2001). TopBP1- or MDC1-depleted HeLa cells displayed a substantial increase of PCC after HU treatment compared with cells transfected with control siRNA (Fig. 4 F, left). Moreover, TopBP1 or MDC1 depletion also reduced cell survival after HU treatment (Fig. 4 F, right). These results confirm that MDC1 is involved in replication checkpoint control.
Our data presented here are different from some of the observations recently described by another group (Cescutti et al., 2010). As we demonstrated in a previous study, recruitment of TopBP1 to sites of replication stress does not require the very C-terminal tandem BRCT domains (Gong et al., 2010). Moreover, as presented here, we took an unbiased approach and identified MDC1 as a TopBP1-associated protein. Our follow-up studies fully supported our initial finding and established that a physical interaction between TopBP1 and MDC1 is required for the stable accumulation of TopBP1 at sites of replication stress. We did not recover any 53BP1 peptides when we performed mass spectrometric analysis of TopBP1 BRCT5-associated proteins (Fig. 1 C), raising the possibility that the 53BP1–TopBP1 interaction may be relatively weak and thus insufficient to recruit TopBP1 to DNA damage sites. Indeed, we showed that ionizing radiation (IR) or HU-induced TopBP1 focus formation was easily detected in 53BP1−/− MEFs (Fig. S3 D), which suggests that 53BP1 does not play a major role in recruiting TopBP1 after IR or HU treatment. A possible explanation for some of the conflicts between our data and those of Cescutti et al. (2010) is that the experiments were conducted in different ways (i.e., our experimental condition is after 2 h of HU treatment) and/or how the conclusions were deduced.
MDC1 is best known for its role in cellular response to DNA DSBs. In this role, MDC1 binds to phosphorylated H2AX to amplify DNA damage signals. In addition, MDC1 interacts with NBS1 and is required for the retention of NBS1 at sites of DNA breaks (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008). The findings we report here agree with these results of earlier studies. In addition, this study has also clarified a role of H2AX in replication checkpoint control. Although we reported several years ago that H2AX is phosphorylated by ATR after replication stress (Ward and Chen, 2001), the exact role of H2AX phosphorylation in replication checkpoint control was not known before the current study. We propose that γ-H2AX plays an indirect role in the recruitment of TopBP1 via its direct binding to MDC1. We propose that, similar to their roles at DSB sites, γ-H2AX and MDC1 are also involved in the amplification of replication stress signals (our working model is presented in Fig. 5). In essence, ssDNA region coated by RPA at stalled replication forks is right next to the double-stranded DNA region coated by H2AX and MDC1. These two molecules are involved in the amplification of replication stress signals. At stalled replication forks, initial phosphorylation of H2AX by ATR or other related kinases triggers the recruitment of MDC1, which then leads to the accumulation of TopBP1 at stalled replication forks via a direct protein–protein interaction. The role of H2AX and MDC1 is to increase the local concentration of TopBP1 at and near stalled replication forks, and therefore facilitate the efficient activation of ATR kinase activity and, subsequently, Chk1 phosphorylation at stalled replication forks. Further analyses of these key molecules involved in DNA damage and replication checkpoint controls will provide insights into the interplay between these two major checkpoint pathways, which are critical for the maintenance of genomic integrity and for tumor suppression.
Figure 5.
A proposed model of replication checkpoint control. This model involves a signal amplification step mediated by H2AX and MDC1 (see text for details).
Materials and methods
Cell culture and plasmids
293T, U2OS, and HeLa cells were maintained in RPMI 1640 medium. MEFs cells were cultivated in DME medium. All media were supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C in a humidified incubator with 5% CO2 (vol/vol). H2AX−/−, MDC1−/−, RNF8−/−, and their respective WT MEFs have been described previously (Lou et al., 2006; Huen et al., 2007). TopBP1 or MDC1 cDNA was cloned or subcloned using Gateway technology (Invitrogen). All internal-deletion mutants were generated by site-directed mutagenesis and verified by sequencing. We used a BRCT5 domain of TopBP1 (residues 545–722) that contained both BRCT5 domain and the region between BRCT4 domain and BRCT5 domain. The construct containing HA-tagged 12A mutant of MDC1 was a gift from J. Lukas (Institute of Cancer Biology, Danish Cancer Society, Copenhagen, Denmark).
Antibodies
Rabbit polyclonal anti-TopBP1 and anti-MDC1 antibodies have been described previously (Yamane et al., 2002; Kim et al., 2005; Lou et al., 2006). Monoclonal anti-Flag M2, anti-HA, and anti-β-actin antibodies were obtained from Sigma-Aldrich. The anti-Myc (9E10) antibody was obtained from Covance. p-DNA-PK antibody was provided by D.J. Chen (University of Texas Southwestern Medical School, Dallas, TX).
Co-IP and Western blotting
Cells were lysed with NTEN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) containing protease inhibitors on ice for 20 min. The soluble fractions were collected after centrifugation and incubated for 3 h at 4°C with either protein A agarose beads coupled with anti-TopBP1, anti-MDC1 antibodies, or S protein agarose (EMD). The precipitates were then washed and boiled in 2× SDS loading buffer. Samples were resolved on SDS-PAGE and transferred to polyvinylidene fluoride membrane, and immunoblotting was performed with antibodies as indicated.
RNA interference
HeLa cells were transfected twice with a 24-h interval with the indicated siRNAs using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. siRNAs against human TopBP1 or MDC1 have been described previously (Yamane et al., 2002; Kim et al., 2005; Lou et al., 2006). The sequence of control siRNA was 5′-UUCAAUAAAUUCUUGAGGUUU-3′.
Tandem affinity purification
293T cells stably expressing SFB-TopBP1-BRCT4+5 or SFB-MDC1 were used for tandem affinity purification. Those stable cells were lysed with NTEN buffer (see Co-IP and Western Blotting) on ice for 20 min. After cell debris was removal by centrifugation, crude lysates were cleared by centrifugation. The pellets were suspended in nuclease buffer (10 mM Hepes, pH 7.4, 10 mM KCl, 0.5 mM MgCl2, 2 mM CaCl2, and 1 µg/ml of each of pepstatin A and aprotinin) supplemented with 150 U/ml micrococcal nuclease S7 (Roche) and incubated in a 37°C water bath for 5 min until the suspension turned cloudy. Then the chromatin fraction was collected by centrifugation, and the supernatants were incubated with streptavidin Sepharose beads (GE Healthcare) for 3 h at 4°C. The bead-bound proteins were washed three times with NTEN buffer and eluted twice with 2 mg/ml biotin (Sigma-Aldrich) for 1 h at 4°C. The eluates were combined and then incubated with S protein agarose (EMD) for 3 h at 4°C. Beads were washed three times with NTEN buffer. The proteins bound to S protein agarose beads were separated by SDS-PAGE and stained with Coomassie blue. The eluted proteins were identified by mass spectrometric analysis (Taplin Biological Mass Spectrometry Facility, Harvard University).
Immunofluorescence staining
Cells grown on coverslips were mock-treated or treated with 2 mM HU for 2 h. Cells were fixed in 3% paraformaldehyde for 10 min and then permeabilized in 0.5% Triton X-100–containing solution for 5 min on ice. For immunostaining with TopBP1 antibody, cells were fixed in a mixture of acetone and methanol (1:1) at −20°C for 12 min. Cells were incubated with primary antibodies diluted in 5% goat serum at 37°C for 30 min. Cells were washed twice with PBS and then incubated with either fluorescein isothiocyanate (FITC)-conjugated or rhodamine-conjugated secondary antibodies at 37°C for 30 min. Nuclei were counterstained with DAPI. The coverslips were mounted onto glass slides with anti-fade solution and visualized at RT using a fluorescence microscope (Eclipse E800; Nikon) with a 60× NA 1.3 oil objective lens. Images were photographed and analyzed using a Spot 2 Megasample camera (Diagnostic Instruments, Inc.) and Photoshop software (Adobe).
Mitotic spreads
Evidence of premature mitosis in damaged cells relies primarily on the appearance of PCC. Mitotic spreads were prepared. In brief, HeLa cells were transfected with control siRNAs or siRNAs against human TopBP1 or MDC1. Then, 48 h after the first transfection, 2 mM HU and 200 ng/ml nocodazole were added. Cells were harvested for chromosome preparation using a standard protocol 6–8 h after treatment with colcemid treatment (50 ng/ml). Cells were incubated in 0.075 M KCl at 37°C for 20 min and then fixed by multiple changes of Carnoy’s fixative (3:1 methanol/acetic acid). Cells were dropped onto slides and stained with Giemsa. PCC was scored as described previously (Nghiem et al., 2001).
GST pull-down assay
GST fusion proteins were expressed in Escherichia coli and purified as previously described (Hofer et al., 1994). GST fusion proteins were immobilized on glutathione–Sepharose 4B beads and incubated with lysates prepared from cells transiently transfected with plasmids encoding the indicated proteins. The samples were subjected to SDS-PAGE and analyzed by Western blotting.
Online supplemental material
Fig. S1 illustrates that HU-induced TopBP1 and MDC1 focus formation occurs at stalled replication forks but not at fork-derived DNA DSBs. Fig. S2 confirms that TopBP1 acts downstream of H2AX and MDC1 in response to replication stress. Fig. S3 shows the phosphorylation-specific interaction between MDC1 and TopBP1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201010026/DC1.
Acknowledgments
We thank our colleagues in the Chen’s laboratory for insightful discussions and technical assistance. We thank Dr. Jiri Lukas at the Danish Cancer Society for sharing HA-tagged 12A mutant of MDC1. We also thank Dr. David J. Chen at the University of Texas Southwestern Medical School for sharing p-DNAPK antibody.
This work was supported in part by grants from the National Institutes of Health (CA089239, CA092312, and CA100109 to J. Chen). J. Chen is a recipient of an Era of Hope Scholar award from the Department of Defense (W81XWH-05-1-0470) and is a member of the MD Anderson Cancer Center (CA016672).
Footnotes
Abbreviations used in this paper:
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM and Rad3-related
- co-IP
- coimmunoprecipitation
- DSB
- double-strand break
- HU
- hydroxyurea
- IP
- immunoprecipitation
- IR
- ionizing radiation
- MEF
- mouse embryonic fibroblast
- PCC
- premature chromosome condensation
- RPA
- replication protein A
- SFB
- streptavidin-binding peptide
- ssDNA
- single-stranded DNA
- WT
- wild type
References
- Burrows A.E., Elledge S.J. 2008. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev. 22:1416–1421 10.1101/gad.1685108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescutti R., Negrini S., Kohzaki M., Halazonetis T.D. 2010. TopBP1 functions with 53BP1 in the G1 DNA damage checkpoint. EMBO J. 29:3723–3732 10.1038/emboj.2010.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Jackson S.P. 2008. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 9:795–801 10.1038/embor.2008.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K.A., Cortez D. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616–627 10.1038/nrm2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S., Wagner J.M., Kobayashi M., Yamamoto K., Karnitz L.M. 2007. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 21:1472–1477 10.1101/gad.1547007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Kim J.E., Leung C.C., Glover J.N., Chen J. 2010. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell. 37:438–446 10.1016/j.molcel.2010.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer B., Backhaus S., Timmis K.N. 1994. The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene. 144:9–16 10.1016/0378-1119(94)90196-1 [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S., Stucki M. 2010. MDC1: The art of keeping things in focus. Chromosoma. 119:337–349 10.1007/s00412-010-0266-9 [DOI] [PubMed] [Google Scholar]
- Kim J.E., McAvoy S.A., Smith D.I., Chen J. 2005. Human TopBP1 ensures genome integrity during normal S phase. Mol. Cell. Biol. 25:10907–10915 10.1128/MCB.25.24.10907-10915.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W.G. 2006. How cells activate ATR. Cell Cycle. 5:1265–1268 10.4161/cc.5.12.2834 [DOI] [PubMed] [Google Scholar]
- Kumagai A., Lee J., Yoo H.Y., Dunphy W.G. 2006. TopBP1 activates the ATR-ATRIP complex. Cell. 124:943–955 10.1016/j.cell.2005.12.041 [DOI] [PubMed] [Google Scholar]
- Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G. 2010. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 140:349–359 10.1016/j.cell.2009.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W.G. 2007. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 282:28036–28044 10.1074/jbc.M704635200 [DOI] [PubMed] [Google Scholar]
- Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M.A., Celeste A., Manis J.P., van Deursen J., Nussenzweig A., Paull T.T., et al. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 21:187–200 10.1016/j.molcel.2005.11.025 [DOI] [PubMed] [Google Scholar]
- Mäkiniemi M., Hillukkala T., Tuusa J., Reini K., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Mäkelä T.P., Syväoja J.E. 2001. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276:30399–30406 10.1074/jbc.M102245200 [DOI] [PubMed] [Google Scholar]
- Melander F., Bekker-Jensen S., Falck J., Bartek J., Mailand N., Lukas J. 2008. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 181:213–226 10.1083/jcb.200708210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messick T.E., Greenberg R.A. 2009. The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 187:319–326 10.1083/jcb.200908074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem P., Park P.K., Kim Y., Vaziri C., Schreiber S.L. 2001. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc. Natl. Acad. Sci. USA. 98:9092–9097 10.1073/pnas.161281798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y., Fantes P., Sutani T., McInerny C., Creanor J., Yanagida M. 1994. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 13:5319–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam C.L., Cruz N.M., Danielian P.S., Amsterdam A., Lau M.L., Hopkins N., Lees J.A. 2010. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 24:183–194 10.1101/gad.1860310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C., Miller E.S., Townsend K., Pavic L., Morrice N.A., Janscak P., Stewart G.S., Stucki M. 2008. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell Biol. 181:227–240 10.1083/jcb.200709008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 445:328–332 10.1038/nature05465 [DOI] [PubMed] [Google Scholar]
- Van Hatten R.A., Tutter A.V., Holway A.H., Khederian A.M., Walter J.C., Michael W.M. 2002. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 159:541–547 10.1083/jcb.200207090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Elledge S.J. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 96:3824–3829 10.1073/pnas.96.7.3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Chen J. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759–47762 10.1074/jbc.M009785200 [DOI] [PubMed] [Google Scholar]
- Wood J.L., Chen J. 2008. DNA-damage checkpoints: location, location, location. Trends Cell Biol. 18:451–455 10.1016/j.tcb.2008.07.006 [DOI] [PubMed] [Google Scholar]
- Wu L., Luo K., Lou Z., Chen J. 2008. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 105:11200–11205 10.1073/pnas.0802885105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R.R., Axton J.M., Yamamoto Y., Saunders R.D., Glover D.M., Henderson D.S. 2000. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics. 156:711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Wu X., Chen J. 2002. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 22:555–566 10.1128/MCB.22.2.555-566.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Jetten A.M. 2008. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett. 271:179–190 10.1016/j.canlet.2008.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Michael W.M. 2009. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle. 8:2877–2884 10.4161/cc.8.18.9485 [DOI] [PubMed] [Google Scholar]
- Yoo H.Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G. 2009. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol. Biol. Cell. 20:2351–2360 10.1091/mbc.E08-12-1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Diffley J.F. 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 445:281–285 10.1038/nature05432 [DOI] [PubMed] [Google Scholar]