Chromosome passenger complexes and bipolar kinesins act together to coordinate spindle elongation, spindle breakdown, and mitotic exit.
Abstract
During mitosis, chromosome passenger complexes (CPCs) exhibit a well-conserved association with the anaphase spindle and have been implicated in spindle stability. However, their precise effect on the spindle is not clear. In this paper, we show, in budding yeast, that a CPC consisting of CBF3, Bir1, and Sli15, but not Ipl1, is required for normal spindle elongation. CPC mutants slow spindle elongation through the action of the bipolar kinesins Cin8 and Kip1. The same CPC mutants that slow spindle elongation also result in the enrichment of Cin8 and Kip1 at the spindle midzone. Together, these findings argue that CPCs function to organize the spindle midzone and potentially switch motors between force generators and molecular brakes. We also find that slowing spindle elongation delays the mitotic exit network (MEN)–dependent release of Cdc14, thus delaying spindle breakdown until a minimal spindle size is reached. We propose that these CPC- and MEN-dependent mechanisms are important for coordinating chromosome segregation with spindle breakdown and mitotic exit.
Introduction
Chromosome passenger complexes (CPCs) are composed of the Aurora B kinase and associated proteins that are believed to regulate the activity and localization of the kinase. The dynamic spatial and temporal localization of CPCs suggests that they perform multiple mitotic functions. Before anaphase, passengers associate with chromosomes and kinetochores where they correct errant kinetochore–microtubule attachments and, thus, ensure the bivalent attachment of chromatids to the mitotic spindle. After anaphase onset, chromosome passengers localize to the anaphase spindle (Cooke et al., 1987; Adams et al., 2000, 2001a,b). Although the precise role on the spindle is not clear, evidence from a variety of systems have implicated CPCs in maintaining the spindle midzone and in ensuring completion of cytokinesis. Despite a large number of functional studies in multiple systems, it is unclear how CPCs carry out their multiple roles during mitosis.
Our understanding of CPC function is largely informed by analysis of the Aurora B kinase (Ipl1 in budding yeast). The physical association of Aurora B with other passengers, including inner centromere protein, Survivin, and Borealin (Sli15, Bir1, and Nbl1, respectively, in budding yeast), has been linked to its catalytic activity (Gassmann et al., 2004; Nakajima et al., 2009). Thus, one idea is that the other passengers act to target Aurora B to its appropriate mitotic substrates. However, not all CPC functions require the Aurora B kinase. For example, in budding yeast, mutations that disrupt the interaction between Sli15 and Bir1 inhibit septin ring disassembly at the end of mitosis, yet ipl1-321 has no effect on septins (Thomas and Kaplan, 2007). Therefore, a second possibility is that discrete CPCs carry out different mitotic functions, some of which may not require the catalytic activity of Aurora B. To further elucidate the mitotic roles and mechanisms of CPC function, we focus on the role of CPCs on the anaphase spindle.
Mutant analysis in budding yeast suggests that CPCs play a role in spindle midzone organization (Mackay et al., 1998; Khmelinskii and Schiebel, 2008). In budding yeast, the spindle midzone is a poorly defined region of overlapping interpolar microtubules where multiple microtubule-associated proteins (MAPs) contribute to stability and outward spindle forces. CPCs associate with the spindle through the simultaneous interaction of Sli15 with microtubules and with Ipl1 and Bir1; Bir1 in turn recruits CBF3 (Kim et al., 1999; Kang et al., 2001; Pereira and Schiebel, 2003; Bouck and Bloom, 2005; Widlund et al., 2006; Thomas and Kaplan, 2007). The sli15-3 mutant prevents the midzone protein Slk19 from properly localizing (Pereira and Schiebel, 2003), which in turn alters the localization of other midzone proteins, such as Cin8 (kinesin-5 family), Bim1 (EB1 family), and Stu2 (Dis1/XMAP215; Khmelinskii et al., 2007). Although the changes in sli15-3 are not sufficient to cause spindle instability in anaphase, spindles become fragile when anaphase is initiated artificially by the engineered cleavage of sister chromatid cohesion (Pereira and Schiebel, 2003). To better understand the role of CPCs during normal anaphase, we used live-cell imaging to quantify spindle behavior in a series of CPC mutants.
Results and discussion
The CBF3–Bir1–Sli15 complex is required for maximal spindle elongation rate
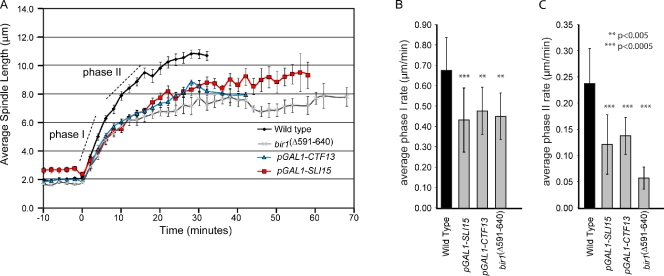
To address the role of CPCs on the spindle, we used pGAL1-SLI15 to transcriptionally deplete the spindle anchoring passenger subunit Sli15. Under repressive conditions (i.e., dextrose-containing medium), Sli15 protein levels were depleted below detection levels in pGAL1-SLI15 (Thomas and Kaplan, 2007). Measurements revealed a shorter final spindle length compared with wild type (9.51 µm compared with 11 µm for wild type) but within the range of reported spindle lengths from different strain backgrounds (6.81 ± 0.65 [Movshovich et al., 2008], 7.2 ± 0.6 [Khmelinskii et al., 2007], 9.1 [Kotwaliwale et al., 2007], and 9.52 ± 0.49 µm [Straight et al., 1998]). Videos of spindle elongation showed no evidence of spindle collapse but rather a decrease in spindle elongation rate. In wild-type cells, we observed the biphasic elongation profile as previously described (Yeh et al., 1995; Straight et al., 1998); a faster phase I elongation (0.68 µm/min from 2.1 to 7.0 µm; Video 1) is followed by a slower phase II rate (0.24 µm/min from 7.0 to 11.0 µm; Fig. 1 A). Depletion of Sli15 significantly slowed phase I and II spindle elongation rates (phase I, 0.43 µm compared with 0.68 µm/min; phase II, 0.12 µm compared with 0.24 µm/min; Fig. 1, Table I, and Video 2).
Figure 1.
Chromosomal passenger proteins slow anaphase spindle elongation. (A) Spindles were filmed as described in Materials and methods, and spindle lengths were measured at each time point. Each graph represents the mean spindle length for multiple cells (Table I), and the error bars in A indicate standard error. (B and C) Calculated spindle elongation rates and the standard deviations from the mean are shown for the indicated strains for phase I (B) and phase II (C). Statistical analyses were performed by comparing the wild type with each mutant (also see Table I).
Table I.
Anaphase spindle elongation rates
| Strains | Phase I rate | Phase II rate | n |
| WT (RT) | 0.68 (0.16) | 0.24 (0.07) | 15 |
| pGAL1-SLI15 | 0.43 (0.16) | 0.12 (0.06) | 20 |
| pGAL1-CTF13 | 0.48 (0.12) | 0.14 (0.04) | 11 |
| bir1Δ591–640 | 0.45 (0.11) | 0.06 (0.02) | 8 |
| ipl1-321 | 0.68 (0.21) | 0.28 (0.11) | 20 |
| kip1Δ pGAL1-SLI15 | 0.50 (0.21) | 0.11 (0.02) | 9 |
| cin8Δ pGAL11-SLI15 | 0.61 (0.14) | 0.25 (0.04) | 22 |
| kip1Δ pGAL1-CTF13 | 0.48 (0.11) | 0.22 (0.09) | 20 |
| cin8Δ pGAL1-CTF13 | 0.58 (0.16) | 0.13 (0.06) | 19 |
| bim1Δ pGAL1-CTF13 | 0.44 (0.12) | 0.21 (0.05) | 9 |
| kip1Δ | 0.49 (0.16) | 0.20 (0.04) | 24 |
| cin8Δ | 0.42 (0.13) | 0.15 (0.07) | 24 |
| bim1Δ | 0.56 (0.12) | 0.25 (0.06) | 21 |
All cells were filmed at 25°C as described in Materials and methods. Mean (standard deviation) elongation rates (µm/min) were calculated using a slope function to fit a linear regression line through each phase. Strains compared with wild type (WT) that were considered statistically significantly at P = 0.05 are shown in bold.
Loss of Sli15 may be equivalent to loss of the Aurora B kinase (Ipl1), as Sli15 has been shown to be necessary to target Ipl1 to the spindle (Kang et al., 2001; Widlund et al., 2006; Thomas and Kaplan, 2007). To test this possibility, we examined the temperature-sensitive allele ipl1-321 to see whether it, too, slowed spindle elongation. Interestingly and consistent with previously published work (Buvelot et al., 2003), ipl1-321 showed no significant decrease in spindle elongation rate compared with wild-type cells (0.68 µm/min for phase I and 0.28 µm/min for phase II). Although we cannot be sure that ipl1-321 is inactive on the spindle, we find that ∼70% of ipl1-321 cells exhibit syntelic kinetochore–microtubule attachments (when a pair of chromatids is attached to microtubules from a single spindle pole), as predicted for loss of Ipl1 function (Biggins et al., 1999; Thomas and Kaplan, 2007). Because we observe a similar level of syntelically attached kinetochores in pGAL1-SLI15 under repressive conditions (unpublished data; Sandall et al., 2006; Thomas and Kaplan, 2007), we conclude that kinetochore misattachments to the spindle that arise in both ipl1-321 and pGAL1-SLI15 are not the cause of slow spindle elongation. We suggest that Sli15 acts independently of Ipl1 to control spindle elongation.
We next analyzed the bir1Δ591–640 allele that specifically disrupts the Bir1–Ndc10 interaction but does not affect the Sli15–Ipl1 complex and has a normal resolution of syntelically attached kinetochores (Thomas and Kaplan, 2007). Similar to pGAL1-SLI15, bir1Δ591–640 spindles elongate slowly (0.45 µm/min for phase I and 0.06 µm/min for phase II; Fig. 1, A–C; Table I; and Video 3). Although Ndc10 is the only CBF3 subunit that directly interacts with Bir1, all of the CBF3 subunits (i.e., Ndc10, Cep3, and Ctf13) associate with anaphase spindles (Thomas and Kaplan, 2009). To address whether the entire CBF3 complex is important for spindle elongation, we analyzed cells depleted for the core CBF3 subunit Ctf13 (pGAL1-CTF13). Under conditions in which no CBF3 is detectable in extracts (Rodrigo-Brenni et al., 2004), we observed a similar decrease in spindle elongation rates (pGAL1-CTF13; Fig. 1, A–C; Table I; and Video 4). We therefore conclude that a CBF3–Bir1–Sli15 complex that is distinct from Ipl1 is required for maximal spindle elongation during anaphase. We favor this interpretation of our data, but these experiments do not rule out that Ipl1 inhibits spindle elongation in the absence of Sli15.
Chromosome passenger mutants induce a kinesin-5–dependent spindle brake
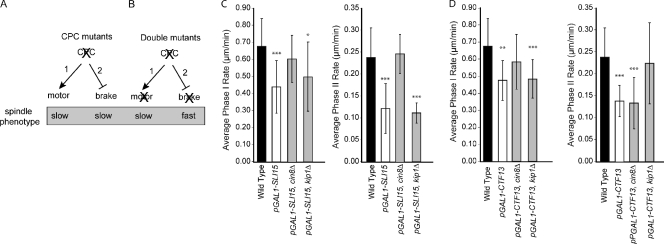
To explain how CPCs affect spindle elongation, we considered two possible mechanisms. First, CPC mutants may reduce outward sliding forces by inhibiting motors (Fig. 2 A, pathway 1). For example, loss of kinesin-5 motors from antiparallel interpolar microtubules in CPC mutants might slow outward sliding and reduce spindle elongation rates. Alternatively, CPC mutants may reduce spindle forces by inducing molecular friction between interpolar microtubules or a frictional brake; a frictional brake can arise when protein cross-links form between antiparallel interpolar microtubules (Fig. 2 A, pathway 2; Civelekoglu-Scholey and Scholey, 2007). For example, it has been shown that kinesin-5 in the Caenorhabditis elegans system can act as a frictional brake to inhibit outward sliding during anaphase B (Saunders et al., 2007). Following the logic diagrammed in Fig. 2 B, we used a double mutant analysis to distinguish between these possibilities. We predict that if CPCs positively contribute to spindle forces, combining a CPC mutant with a mutant in a MAP (e.g., motor) will result in similarly slow or slower spindle elongation (Fig. 2 B, pathway 1). Alternatively, if CPCs act to inhibit a frictional brake, eliminating proteins required for the brake in a CPC mutant will result in faster spindle elongation (Fig. 2 B, pathway 2). We first examined the effect of deleting the genes encoding the kinesin-5 motors, CIN8 and KIP1, in pGAL1-SLI15. Remarkably, loss of Cin8 restored spindle elongation rates in pGAL1-SLI15 to near wild-type values (Fig. 2 C). This is in contrast to the cin8 mutants on their own, which have previously been shown to slow spindle elongation (Table I; Straight et al., 1998; Movshovich et al., 2008). We observed a similar rescue of spindle elongation rates in a pGAL1-CTF13 cin8Δ double mutant (Fig. 2 D), indicating that a CPC that includes CBF3 alters Cin8 action on the spindle. Combining pGAL1-SLI15 with kip1Δ modestly increased the rate of phase I but had little effect on phase II spindle elongation (Fig. 2 B). This is consistent with our observation that the single mutant kip1Δ only slows the phase I elongation rate (Table I). We conclude that, in the presence of CPC, kinesin-5 motors favor outward sliding, and in the absence of CPC function, a population of kinesin-5 motors acts to inhibit spindle elongation. Future work will be required to define the biochemical characteristics of the different populations of kinesin-5 motors.
Figure 2.
Chromosomal passenger proteins inhibit a kinesin-5–dependent brake. (A) The diagram schematizes two possible pathways for CPCs to affect spindle elongation: (1) CPCs positively regulate motors to generate outward sliding forces, or (2) CPCs inhibit a spindle brake. (B) The diagram shows two possible experimental outcomes for double mutant analysis. In pathway 1, a double CPC motor mutant will result in slow (or slower) spindle elongation. In pathway 2, a double CPC mutant will result in faster spindle elongation. (C and D) The indicated double mutants in pGAL1-SLI15 (C) or pGAL1-CTF13 (D) were analyzed as in Fig. 1, and elongation rates are separately compared for each phase. Error bars represent standard deviations from the mean. *, P < 0.5; **, P < 0.005; ***, P < 0.0005.
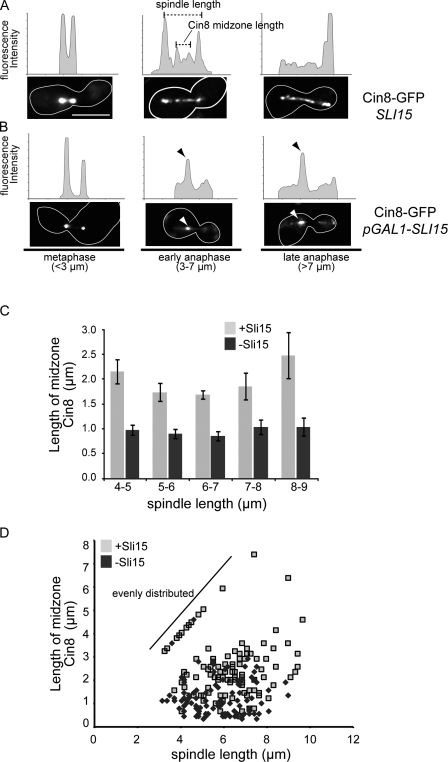
Changes in motor action may arise through their reorganization on the anaphase spindle. To test this possibility, we examined the distribution of Cin8-GFP. In metaphase, the majority of cells exhibited a bilobed distribution as previously described (Fig. 3 A, metaphase; Tytell and Sorger, 2006). In wild-type anaphase cells, Cin8-GFP is found both distributed along the length of the spindle and enriched in a 2–3-µm region in the middle of the spindle, regardless of spindle size (Fig. 3, A and D). Significantly, depletion of Sli15 resulted in the concentration of Cin8-GFP in the middle of spindles of all sizes, with the majority of Cin8-GFP restricted to a region of <1 µm (Fig. 3, B–D). This result is in contrast to the broader distribution reported for Cin8-GFP in sli15-3 (Khmelinskii et al., 2007). However, it should be noted that sli15-3 was isolated in a synthetic lethal screen with ipl1-2, and Sli15-3 supports Ipl1 association with the spindle, whereas Sli15 depletion prevents spindle association of all passengers (Kim et al., 1999; Thomas and Kaplan, 2007). We observed a similar concentration of Kip1-GFP in the middle of spindles in cells depleted for Sli15, although this was most apparent in cells with spindles >7 µm (Fig. S1 D). The enrichment of kinesin-5 motors in the middle of the spindle is not sufficient to completely stall spindle elongation in CPC mutants. We therefore favor the idea that a subpopulation of kinesin-5 motors provides the outward forces in CPC mutants. Regardless, our observations of motor distribution in CPC mutants support a role for CPCs as distribution factors on the anaphase spindle, possibly through direct interaction with other spindle proteins (see Fig. 5 F, i) or by altering the posttranslational modification of midzone proteins (see Fig. 5 F, ii). In the absence of CPCs, motor proteins fail to distribute normally, and outward sliding is inhibited (see Fig. 5 G).
Figure 3.
Cin8-GFP is concentrated at the spindle midzone after depletion of Sli15. (A and B) Cells expressing Cin8-GFP in the presence (A) or absence (B) of Sli15 were filmed as in Fig. 1, and representative examples of mitotic spindles <3 µm, between 3 and 7 µm, and >7 µm are presented. Line traces show relative fluorescent intensities and are not comparable between cells. The dashed lines indicate how spindle length and Cin8 midzone length were measured for C and D. Arrowheads (black/white) in B indicate the concentration of Cin8-GFP at the spindle midzone and the corresponding fluorescent peak in the line trace. The white outlines indicate the cell border as determined in separate images (not depicted) based on autofluorescence. (C) The lengths of Cin8-GFP midzones were measured as shown in A and plotted for the indicated range of spindle lengths in the presence or absence of Sli15. Error bars represent standard deviations from the mean. (D) A scatter plot compares the length of Cin8-GFP spindle staining to the length of the spindle (CFP-Tub1) in the presence or absence of Sli15. The diagonal line indicates a population of mostly wild-type cells with contiguous Cin8-GFP spindle distribution (i.e., Cin8-GFP midzone length = spindle length). Bar, 5 µm.
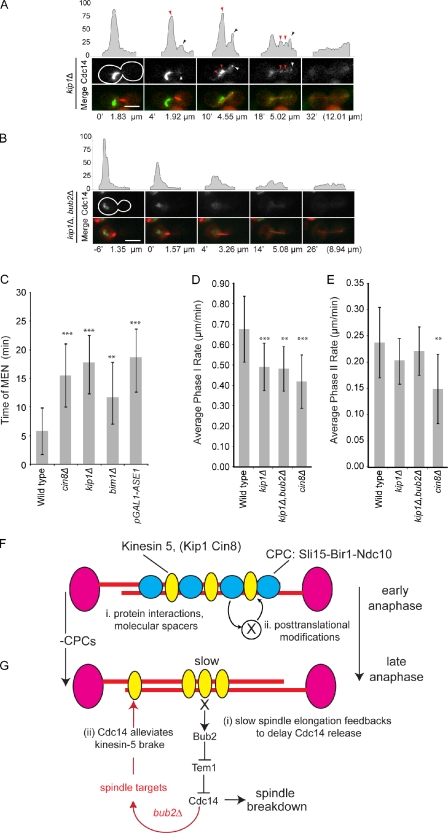
Figure 5.
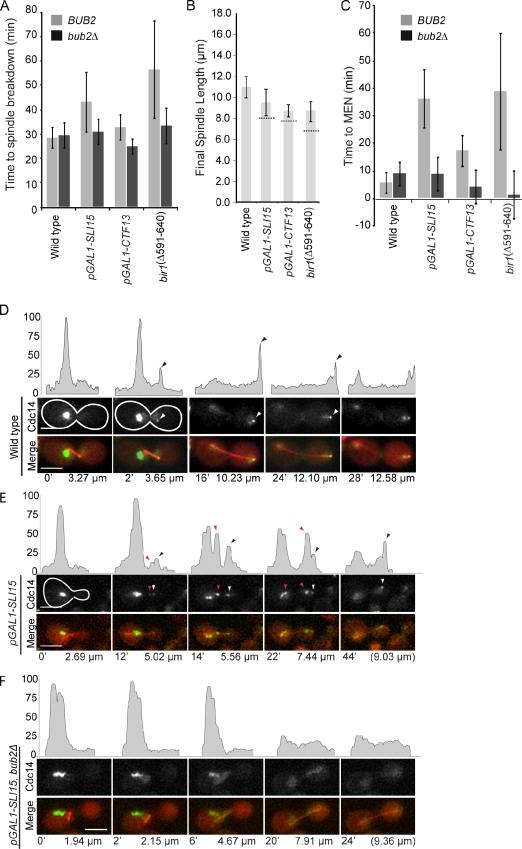
MAP mutants that slow spindle elongation also delay Cdc14 release and prolong anaphase. (A and B) Representative frames and fluorescent intensity line traces from videos of kip1Δ (A) or kip1Δ bub2Δ (B) cells expressing CFP-Tub1 and Cdc14-GFP are presented as in Fig. 3 and Fig. 4. Arrowheads above the intensity profiles represent Cdc14-GFP at the daughter spindle pole (black on intensity profiles and white in the micrographs) or in foci between poles (red) in kip1Δ cells. The time from the start of spindle elongation and the spindle lengths are indicated below the images. The white outlines indicate the cell border as determined in separate images (not depicted) based on autofluorescence. (C) The mean time of MEN as measured by the complete release of Cdc14-GFP is plotted for indicated strains. (D and E) The corresponding values for spindle elongation rates are plotted. The statistical significance was determined by the comparison of mutants with wild type and is annotated here (also see Table II) as follows: **, P < 0.005; ***, P < 0.0005. (F) The model illustrates how CPCs might act to either (i) directly or (ii) indirectly distribute spindle midzone proteins. (G) The model summarizes feedback between spindle elongation and mitotic exit. (i) Release of Cdc14 is delayed when spindle elongation is delayed, a response that requires Bub2. (ii) The eventual release of Cdc14 during the transition from early to late anaphase or in bub2Δ alters spindle organization to alleviate the brake. Error bars represent standard deviations from the mean. Bars, 5 µm.
The spindle elongation rate is coupled to mitotic exit through Cdc14 release
The CPC mutants that slow spindle elongation also exhibit a dramatic increase in the duration of anaphase, as measured from the initiation of spindle elongation to spindle breakdown. In wild-type cells grown at 25°C, the mean duration of anaphase is 28.5 min (Fig. 4 A and Table II) but lasts 49.9 min in CPC mutants (Fig. 4 A; mean times for the CPC alleles are listed in Table II). It is interesting to note that the extended times for anaphase are sufficient to explain the relatively normal length of spindles found in mutants with slow spindle elongation rates (Fig. 4 B, dashed lines, indicating the length of spindles if anaphase durations were equivalent to wild type). One way to extend anaphase is to delay the mitotic exit network (MEN)–regulated release of Cdc14 from the nucleolus. Consistent with this possibility, CPC mutants exhibit a delay in the release of Cdc14 compared with wild-type cells (5.8 min for wild type and 36 min for pGAL1-SLI15; Fig. 4, C–E; and Table II). The multiple Cdc14-GFP foci observed in pGAL1-SLI15 have 71% of the Cdc14-GFP fluorescence signal found in metaphase, a very similar level to that observed in the MEN mutant cdc15-2 (67%) and significantly higher than wild-type cells at a similar stage of anaphase (30%; Fig. S2 B). Importantly, CPC mutants retain Cdc14 in the nucleolus for a period of time that is consistent with the transient delay in mitotic exit. We therefore argue that the transient delay in Cdc14 release is sufficient to explain the delay in mitotic exit observed in CPC mutants, although we cannot rule out that inhibition of upstream MEN regulators also contributes to delayed mitotic exit.
Figure 4.
CPC mutants delay Cdc14 release and prolong anaphase. The indicated CPC mutants, with or without BUB2, expressing CFP-Tub1 and Cdc14-GFP were filmed in anaphase. (A) The time to spindle breakdown was measured from the first frame of spindle elongation to spindle collapse. (B) The final spindle length reached before spindle breakdown is shown for each indicated strain. The dashed lines across the bar for the mutants indicate the projected spindle length, taking into account the slower rate of elongation and wild-type anaphase duration. (C) Time to mitotic exit was determined by monitoring the distribution of Cdc14-GFP. Full MEN activation was defined by the loss of fluorescence signal in the nucleolus. Error bars represent standard deviations from the mean. (D–F) Representative frames are shown from videos of wild type (D), pGAL1-SLI15 (E), or the double pGAL1-SLI15 bub2Δ (F) cells expressing CFP-Tub1 (red) and Cdc14-GFP (grayscale at the top and green in the merge). The time from the start of spindle elongation and the spindle lengths are indicated below the images. Line traces above the images show relative fluorescent intensities for Cdc14-GFP. Black arrowheads above the fluorescent intensity traces indicate Cdc14-GFP at the spindle pole (white arrowheads in the micrograph). The red arrowheads between spindle poles indicate Cdc14-GFP foci in pGAL1-SLI15 (E) but not the double pGAL1-SLI15 bub2Δ (F). The white outlines indicate the cell border as determined in separate images (not depicted) based on autofluorescence. Bars, 5 µm.
Table II.
Anaphase spindle and mitotic exit timing
| Strains | Time of MEN | Time of final breakdown | Final spindle length | n |
| WT | 5.8 (4.2) | 28.5 (4.5) | 11.00 (0.95) | 15 |
| pGAL1-SLI15 | 36.3 (11.0) | 43.3 (12.7) | 9.51 (1.32) | 20 |
| pGAL1-CTF13 | 17.4 (6.1) | 32.9 (5.6) | 8.73 (0.51) | 11 |
| bir1Δ591–640 | 38.9 (21.6) | 56.6 (20.4) | 8.64 (0.97) | 8 |
| bub2Δ | 9.1 (4.7) | 29.5 (5.5) | 9.87 (1.60) | 8 |
| bub2Δ pGAL1-SLI15 | 8.9 (6.4) | 31.0 (5.5) | 10.01 (1.67) | 8 |
| bub2Δ pGAL1-CTF13 | 4.2 (6.4) | 25.0 (3.4) | 8.75 (1.16) | 9 |
| bub2Δ bir1Δ591–640 | 1.3 (9.0) | 33.5 (7.7) | 9.48 (2.82) | 5 |
| cin8Δ | 15.5 (5.6) | 37.5 (7.0) | 9.52 (1.79) | 24 |
| kip1Δ | 17.8 (4.8) | 35.1 (4.1) | 10.66 (1.35) | 24 |
| bub2Δ kip1Δ | 5.4 (4.5) | 32.6 (3.7) | 9.89 (1.08) | 10 |
| bim1Δ | 11.7 (6.2) | 28.1 (6.68) | 9.10 (1.16) | 21 |
All cells were filmed at 25°C as described in Materials and methods. All numbers reported are means (standard deviation). Time of MEN shows times after anaphase initiation (in minutes) of MEN (see Materials and methods). Final spindle lengths (µm) were measured from the final frame before breakdown. All strains that were compared to wild type (WT) and considered statistically significantly at P = 0.05 are shown in bold.
It is intriguing to speculate that CPC mutants inhibit upstream MEN activators and, in doing so, influence multiple aspects of anaphase progression. We first examined the possibility that CPC mutants inhibit MEN at the level of the Tem1 GTPase by deleting its GAP, BUB2 (Sullivan and Morgan, 2007). In pGAL1-SLI15 bub2Δ, we observed the complete release of Cdc14-GFP soon after anaphase initiation (36 min for pGAL1-SLI15 compared with 2.9 min for pGAL1-SLI15 bub2Δ; Fig. 4, C and E; and Table II). In addition, bub2Δ restores normal anaphase duration (time to spindle breakdown of 31 min in pGAL1-SLI15 bub2Δ compared with 29 min for wild type; Fig. 4 A) and rescues the slow spindle elongation in pGAL1-SLI15 and other CPC mutants (Fig. S2, C and D). The rescue of spindle elongation is consistent with the reports that Cdc14 can act on multiple spindle targets (Pereira and Schiebel, 2003; Higuchi and Uhlmann, 2005; Khmelinskii et al., 2007, 2009). We propose that early Cdc14 release alters the spindle in the absence of CPCs to disengage the kinesin-5–dependent brake (Fig. 5 G, ii, model). Together, our data argue that slow spindle elongation engages a feedback pathway that delays MEN at the level of Bub2 regulation of Tem1 and, thus, the full release of Cdc14 through a Bub2-dependent pathway (Fig. 5 G, i), possibly to ensure stable elongating spindles and prevent premature mitotic exit.
A prediction of this feedback pathway is that any mutant that slows spindle elongation should also delay Cdc14 release. To test this prediction, we measured the release of Cdc14-GFP in kip1Δ or cin8Δ strains. As observed previously (Saunders et al., 1997; Straight et al., 1998; Movshovich et al., 2008), spindle elongation is slowed in both the kip1Δ and cin8Δ (Fig. 5, D and E; Table I; and Video 5). The slower spindle elongation rate caused by the loss the kinesin-5 motors was also accompanied by a delay in the full release of Cdc14-GFP (Fig. 5, A and C, red arrows, for kip1Δ; Fig. 5 C and Table II for cin8Δ). The remaining nucleolar Cdc14-GFP is similar to the population observed in CPC mutants and is Bub2 dependent (full release of Cdc14 is 5.4 and 17.8 min for kip1Δ bub2Δ and kip1Δ, respectively; Table II and Video 6) The cin8Δ bub2Δ is inviable and could not be analyzed, perhaps indicating the importance of MEN inhibition in slowly elongating spindles. Other mutants that slow spindle elongation (bim1Δ and the depletion of Ase1 using pGAL1-ASE1) also delay Cdc14-GFP release (Fig. 5 C and Video 7), arguing that the delay in turning off the Bub2 checkpoint arises from a general property of the spindle rather than the loss of a specific protein. Importantly, we do not detect any defect in spindle positioning in the CPC or spindle mutants analyzed here (unpublished data), making it unlikely that Bub2 inhibits Cdc14 release caused by mispositioned spindles. We therefore propose that the Bub2 checkpoint delays Cdc14 release until anaphase B spindles are fully elongated in addition to its better recognized role in delaying mitotic exit in response to mispositioned spindles.
The ability of CPCs to modulate spindle elongation through changes in motor activity raises the possibility that such a pathway is important to ensure proper chromosome segregation. One possibility is that CPCs respond to the changing state of the spindle by coordinating midzone changes with the length of the spindle, thus ensuring that cells do not exit mitosis before spindles have reached a minimal size. Another possibility is that CPCs alter the spindle midzone in response to unresolved chromosomes. This possibility has precedence in the “no-cut” pathway where chromatin activates Ipl1 to delay abscission (Norden et al., 2006; Mendoza et al., 2009). In this scenario, Ipl1 only becomes relevant to the spindle brake when there is chromatin at the midzone, a prediction that will be important to test. We propose a model whereby CPCs link unsegregated chromosomes to a spindle brake and that slower spindle elongation delays spindle breakdown until chromosomes have been completely segregated (Fig. 5, F and G, models).
Materials and methods
Yeast growth and strain construction
Yeast strains were grown in complete media (YEP [yeast extract/bactopeptone] media supplemented with 1% adenine and tryptophan) plus 2% dextrose (YPD) or a mixture of 2% raffinose and galactose (YEP + R/G; Guthrie and Fink, 1991). The GFP and CFP-Tub1 strains and constructs were provided by A. Straight (Stanford University, Stanford, CA) and K. Bloom (University of North Carolina, Chapel Hill, NC). New strains were created using gene disruption, and promoter insertions were performed using a one-step PCR-mediated technique (Longtine et al., 1998; Thomas and Kaplan, 2007) and are listed in Table S1.
For time-lapse microscopy experiments, cultures were grown at 30°C in YEP + 2% dextrose to log phase before arresting with 10 µg/ml α-factor (American Peptide, Inc.). For experiments using pGAL1 transcriptional depletion, cultures were grown in YEP + 2% galactose/2% raffinose before arresting in α-factor. To shut off transcription, cells were washed in YPD + α-factor, incubated 60 min, and then released from the α-factor block in YPD for 60 min before mounting for imaging. Temperature-sensitive alleles were grown and arrested as with cultures at a permissive temperature and then shifted to 37°C for 60 min and released from arrest into YPD at 37°C. Cells were prepared for imaging by washing with synthetic defined (SD) media (3% yeast nitrogen base + amino acids [Sigma-Aldrich]) at the appropriate temperature. The cells were resuspended in SD complete medium at a 1:1 ratio (cell pellet/medium). The cell solution was applied to a depression slide (Fisherbrand Hanging Drop Slide; Thermo Fisher Scientific) filled with a low-melt agarose pad (4% agarose in SD complete + 2% dextrose). Slides were sealed with VALAP (1:1:1 vaseline/lanolin/paraffin). For analysis of temperature-sensitive alleles, slides were mounted on a 37°C heated stage, and the objective lens was heated to 37°C. All other strains analyzed using time-lapse microscopy were filmed at 25°C.
Microscopy and image analysis
Time-lapse images were acquired using an inverted microscope (IX70; Olympus) and a 60×/1.40 NA Plan Apochromat oil immersion lens configured on a microscope (DeltaVision Deconvolution; API) with a camera (CoolSnap HQ; Roper Scientific). SoftWoRx software (Resolve 3D; Applied Precision) was used to capture 4-µm projections every 2 min for 3 h at an ambient temperature unless otherwise noted. Images were processed using SoftWoRx software (v3.5.1) projection deconvolution, and spindles were manually measured using the straight-line measurement in ImageJ image processing and analysis software (National Institutes of Health). The spindle elongation rate was calculated using the slope function to fit a linear regression line through each range of spindle lengths. Each condition was filmed on at least two separate sessions, and 15 fields of view for each slide were captured to ensure reproducibility. All means are reported for the indicated number and corresponding standard deviation reported in Table I and Table II; statistically significant differences were determined by two-tailed homoscedastic Student’s t test at the level of P = 0.05.
The timing of cell cycle events was determined as follows. The activation of the Cdc14 early anaphase release pathway was assigned to the first time frame in which Cdc14-GFP was detected at the daughter spindle pole body. The activation of the MEN was assigned the time frame in which Cdc14-GFP is no longer enriched (less than twofold of background fluorescence intensity) in the nucleolus or in foci associated with late nucleolar. The time to spindle breakdown was measured from the first frame of spindle elongation to the disappearance of CFP-Tub1 fluorescence between the spindle poles, typically also accompanied by the inward repositioning of the spindle poles. Estimations of spindle lengths in CPC mutants in the absence of increased anaphase duration (Fig. 4 B) were made by multiplying mutant spindle rates by the mean wild-type duration of anaphase (28.5 and 34.0 min at 30 and 37°C, respectively).
Analysis of 3D fluorescent images
Cells were prepared and mounted as in the time-lapse imaging experiment. 50 0.2-µm slices were acquired using a microscope (IX70) and a 60×/1.40 NA Plan Apochromat oil immersion lens configured on a microscope (DeltaVision Deconvolution). Images were deconvolved with SoftWoRx software using a parameter-calculated point-spread function. Spindle length and fluorescent intensity measurements were made using ImageJ image processing and analysis software. Individual cell volumes were converted to 2D maximum projections for cells containing mitotic spindles >2 µm in length. The extent of midzone distribution for motor-GFP fusions was defined as the longest region of continuous fluorescence between the spindle poles (Fig. 3 A, dashed lines). Data for intensity line-scan plots (kinesin-5 motor and Cdc14 intensity plots) were compiled from the mean intensity of 1 × 15–pixel regions.
Online supplemental material
Fig. S1 shows that Kip1-GFP is concentrated at the spindle midzone of anaphase spindles. Fig. S2 shows MEN, Cdc14 release, and spindle elongation. Video 1 shows anaphase in wild-type cells expressing Cdc14-GFP and CFP-Tub1. Video 2 shows anaphase in pGAL1-SLI15 cells expressing Cdc14-GFP and CFP-Tub1. Video 3 shows anaphase in bir1Δ591–640 cells expressing Cdc14-GFP and CFP-Tub1. Video 4 shows anaphase in pGAL1-CTF13 cells expressing Cdc14-GFP and CFP-Tub1. Video 5 shows anaphase in kip1Δ cells expressing Cdc14-GFP and CFP-Tub1. Video 6 shows anaphase in kip1Δ bub2Δ cells expressing Cdc14-GFP and CFP-Tub1. Video 7 shows anaphase in pGAL1-ASE1 cells expressing Cdc14-GFP and CFP-Tub1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201011002/DC1.
Acknowledgments
We would like to acknowledge Brandon Zipp, Frank McNally, Jonathan Scholey, and Daniel Starr for helpful comments on this manuscript. We are grateful to Kerry Bloom and Aaron Straight for providing Tub1-GFP strains and constructs.
This work was funded by grants from the American Cancer Society (RSG-02-035-01 and RSG-02-035-05-CCG) to K.B. Kaplan.
Note added in proof. Recent work from Roostalu et al. (2011) has demonstrated that the kinesin-5 motor Cin8 exhibits bidirectionality on microtubules in vitro. The switch between directions of movement depends on its configuration on microtubules and may be relevant to the reorganization of Cin8 we observe in vivo when associated with slow spindle elongation (Roostalu et al. 2011. Science. doi:10.1126/science.1199945).
Footnotes
Abbreviations used in this paper:
- CPC
- chromosome passenger complex
- MAP
- microtubule-associated protein
- MEN
- mitotic exit network
References
- Adams R.R., Wheatley S.P., Gouldsworthy A.M., Kandels-Lewis S.E., Carmena M., Smythe C., Gerloff D.L., Earnshaw W.C. 2000. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10:1075–1078 10.1016/S0960-9822(00)00673-4 [DOI] [PubMed] [Google Scholar]
- Adams R.R., Carmena M., Earnshaw W.C. 2001a. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49–54 10.1016/S0962-8924(00)01880-8 [DOI] [PubMed] [Google Scholar]
- Adams R.R., Maiato H., Earnshaw W.C., Carmena M. 2001b. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880 10.1083/jcb.153.4.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544 10.1101/gad.13.5.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D.C., Bloom K.S. 2005. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA. 102:5408–5413 10.1073/pnas.0405925102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S.Y., Vermaak D., Biggins S. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329–339 10.1083/jcb.200209018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Scholey J.M. 2007. Mitotic motors: kinesin-5 takes a brake. Curr. Biol. 17:R544–R547 10.1016/j.cub.2007.05.030 [DOI] [PubMed] [Google Scholar]
- Cooke C.A., Heck M.M., Earnshaw W.C. 1987. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J. Cell Biol. 105:2053–2067 10.1083/jcb.105.5.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Carvalho A., Henzing A.J., Ruchaud S., Hudson D.F., Honda R., Nigg E.A., Gerloff D.L., Earnshaw W.C. 2004. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166:179–191 10.1083/jcb.200404001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G., 1991. Guide to yeast genetics and molecular biology. Methods in Enzymology, Vol. 194 San Diego: Academic Press; [PubMed] [Google Scholar]
- Higuchi T., Uhlmann F. 2005. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 433:171–176 10.1038/nature03240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Cheeseman I.M., Kallstrom G., Velmurugan S., Barnes G., Chan C.S. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763–774 10.1083/jcb.200105029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Schiebel E. 2008. Assembling the spindle midzone in the right place at the right time. Cell Cycle. 7:283–286 10.4161/cc.7.3.5349 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J., Schiebel E. 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177:981–993 10.1083/jcb.200702145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Roostalu J., Roque H., Antony C., Schiebel E. 2009. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell. 17:244–256 10.1016/j.devcel.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kang J.S., Chan C.S. 1999. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145:1381–1394 10.1083/jcb.145.7.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale C.V., Frei S.B., Stern B.M., Biggins S. 2007. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev. Cell. 13:433–445 10.1016/j.devcel.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Mackay A.M., Ainsztein A.M., Eckley D.M., Earnshaw W.C. 1998. A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J. Cell Biol. 140:991–1002 10.1083/jcb.140.5.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M., Norden C., Durrer K., Rauter H., Uhlmann F., Barral Y. 2009. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat. Cell Biol. 11:477–483 10.1038/ncb1855 [DOI] [PubMed] [Google Scholar]
- Movshovich N., Fridman V., Gerson-Gurwitz A., Shumacher I., Gertsberg I., Fich A., Hoyt M.A., Katz B., Gheber L. 2008. Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. J. Cell Sci. 121:2529–2539 10.1242/jcs.022996 [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Tyers R.G., Wong C.C., Yates J.R., III, Drubin D.G., Barnes G. 2009. Nbl1p: a Borealin/Dasra/CSC-1-like protein essential for Aurora/Ipl1 complex function and integrity in Saccharomyces cerevisiae. Mol. Biol. Cell. 20:1772–1784 10.1091/mbc.E08-10-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden C., Mendoza M., Dobbelaere J., Kotwaliwale C.V., Biggins S., Barral Y. 2006. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 125:85–98 10.1016/j.cell.2006.01.045 [DOI] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 302:2120–2124 10.1126/science.1091936 [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni M.C., Thomas S., Bouck D.C., Kaplan K.B. 2004. Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Mol. Biol. Cell. 15:3366–3378 10.1091/mbc.E03-12-0887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandall S., Severin F., McLeod I.X., Yates J.R., III, Oegema K., Hyman A., Desai A. 2006. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 127:1179–1191 10.1016/j.cell.2006.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A.M., Powers J., Strome S., Saxton W.M. 2007. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr. Biol. 17:R453–R454 10.1016/j.cub.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M.A. 1997. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 8:1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Sedat J.W., Murray A.W. 1998. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143:687–694 10.1083/jcb.143.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Morgan D.O. 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8:894–903 10.1038/nrm2276 [DOI] [PubMed] [Google Scholar]
- Thomas S., Kaplan K.B. 2007. A Bir1p Sli15p kinetochore passenger complex regulates septin organization during anaphase. Mol. Biol. Cell. 18:3820–3834 10.1091/mbc.E07-03-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Kaplan K.B. 2009. Kinetochore regulation of anaphase and cytokinesis. The Kinetochore: From Molecular Discoveries to Cancer Therapy. De Wulf P., Earnshaw W.C., Springer, New York/London: 371–394 [Google Scholar]
- Tytell J.D., Sorger P.K. 2006. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 172:861–874 10.1083/jcb.200509101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund P.O., Lyssand J.S., Anderson S., Niessen S., Yates J.R., III, Davis T.N. 2006. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol. Biol. Cell. 17:1065–1074 10.1091/mbc.E05-07-0640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Skibbens R.V., Cheng J.W., Salmon E.D., Bloom K. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687–700 10.1083/jcb.130.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]