The microRNA miR-22 targets CDK6, SIRT1, and Sp1—genes involved in regulation of the senescence program—to suppress cell growth and proliferation.
Abstract
Cellular senescence acts as a barrier to cancer progression, and microRNAs (miRNAs) are thought to be potential senescence regulators. However, whether senescence-associated miRNAs (SA-miRNAs) contribute to tumor suppression remains unknown. Here, we report that miR-22, a novel SA-miRNA, has an impact on tumorigenesis. miR-22 is up-regulated in human senescent fibroblasts and epithelial cells but down-regulated in various cancer cell lines. miR-22 overexpression induces growth suppression and acquisition of a senescent phenotype in human normal and cancer cells. miR-22 knockdown in presenescent fibroblasts decreased cell size, and cells became more compact. miR-22–induced senescence also decreases cell motility and inhibits cell invasion in vitro. Synthetic miR-22 delivery suppresses tumor growth and metastasis in vivo by inducing cellular senescence in a mouse model of breast carcinoma. We confirmed that CDK6, SIRT1, and Sp1, genes involved in the senescence program, are direct targets of miR-22. Our study provides the first evidence that miR-22 restores the cellular senescence program in cancer cells and acts as a tumor suppressor.
Introduction
Tumor progression is a multistep process wherein several defined events are common to cancer cells, such as uncontrolled proliferation and invasion (Hahn and Weinberg, 2002). Cellular senescence is characterized by an irreversible arrest of cell proliferation, so that it can prevent the aberrant and unlimited proliferation of tumor cells (Campisi, 2005). Senescent cells exhibit enlarged morphological changes and less motility than young cells, which may contribute to the suppression of cell migration, invasion, and metastasis (Chen et al., 2000). Oncogene-induced senescence is a cellular response, which can occur in vivo and provides a bona fide barrier to tumorigenesis (Narita and Lowe, 2005). Oncogene-induced senescence was found in premalignant tumors but not in more advanced malignant tumors (Braig et al., 2005; Collado et al., 2005). Therefore, cellular senescence acts as an important barrier to cancer and plays an important role in tumor suppression.
microRNAs (miRNAs) are a class of naturally occurring small noncoding RNAs that negatively regulate the stability and translation of target protein–coding mRNAs at the 3′ untranslated region (UTR). miRNAs typically target a cluster of genes rather than one specific gene (Bartel, 2004), a characteristic which allows them to play critical roles in a variety of biological processes such as cell proliferation, differentiation, apoptosis, and carcinogenesis (He and Hannon, 2004). Recently, a growing number of studies have documented the miRNA expression profiles in human cancers (Calin and Croce, 2006), suggesting that miRNAs emerge as novel biomarkers for various cancers. However, there is currently little information about miRNA profiling studies and biological effects of miRNAs in cellular senescence.
The senescence program is established and maintained by p53 and retinoblastoma protein (pRb) tumor suppressor pathways. The requirements of p53 and pRb for the induction of cellular senescence vary in their prominence depending on the genetic context, species, and cell type (Adams, 2007; Schmitt, 2007; Haferkamp et al., 2009). Recently, various studies have indicated that some miRNAs, such as miR-34a and miR-20a, induce senescence-like growth arrest through regulating cell cycle genes and senescence-associated genes involved in the p53 and/or pRb pathway (Tazawa et al., 2007; Poliseno et al., 2008; Sun et al., 2008). Such miRNAs play a direct role in senescence and are called senescence-associated miRNAs (SA-miRNAs; Lafferty-Whyte et al., 2009).
In the present study, we attempted to screen SA-miRNAs that control cellular senescence in human fibroblasts, and we report here that miR-22 is a novel SA-miRNA that functions in mediating cellular senescence. We studied the role of miR-22 in cellular senescence using human normal cells and cancer cell lines as an in vitro culture system as well as an in vivo mouse breast tumor model. Upon senescence, cells become flattened and enlarged and exhibit biochemical changes such as the increased perinuclear activity of senescence-associated β-galactosidase (SA-β-gal; Dimri et al., 1995; Narita et al., 2003). Another critical event during the cellular senescence process is a decrease in cell growth and cell motility. We found a widespread decrease of miR-22 expression in various human cancer cell lines. Introduction of miR-22 into cancer cells inhibits cell proliferation, accompanied by senescence-like cell morphology and a decrease in cell motility and invasiveness. We predicted the putative direct targets of miR-22 by the computational prediction of targets based on sequence match to the miRNA. We identified three targets, including CDK6, Sp1, and SIRT1, which are directly regulated by miR-22. Furthermore, silencing of these targets resulted in growth arrest and increased SA-β-gal activity, accompanied by pRB dephosphorylation. We confirmed that miR-22 regulated the pRb pathway of cellular senescence through targeting of CDK6 and SIRT1. Ectopic expression of CDK6, SIRT1, or Sp1 could partially rescue the senescence phenotypes in miR-22–transfected cells. Significantly, miR-22 injection suppresses tumor growth and metastasis in vivo by induction of senescence in breast tumor, suggesting that SA-miRNA miR-22 acts as an important barrier to cancer and plays an important role in tumor suppression. Our findings provide new insight for the role of SA-miRNAs between cellular senescence and tumorigenesis.
Results
miR-22 overexpression induces cellular senescence in human fibroblasts
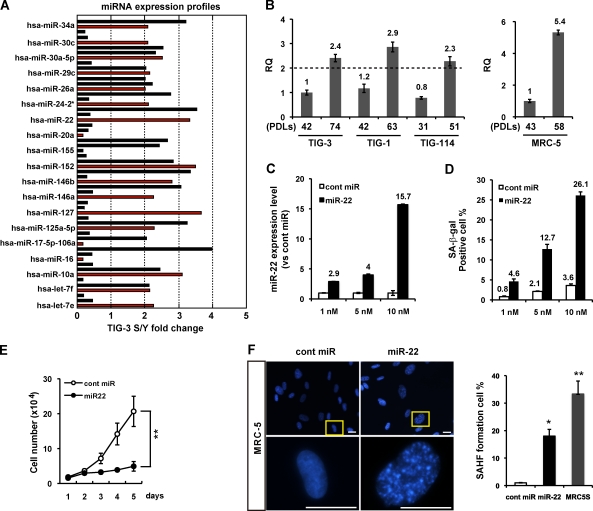
To identify miRNAs that control cellular senescence, we analyzed miRNA expression profiling by miRCURY locked nucleic acid (LNA) miRNA array in young and senescent TIG-3 fibroblasts (Fig. 1 A). We found that a set of altered expression miRNAs has been reported to be involved in cell growth and tumorigenesis (Fig. 1 A and Table S1). Among them, the majority of antigrowth miRNAs were expressed two– to fourfold more in senescent TIG-3 cells than in young cells. To confirm and validate the results, we performed 3D-Gene miRNA microarray to evaluate miRNA expression levels. Two kinds of microarray analysis showed that five miRNAs are uniformly up-regulated by twofold or greater in senescent compared with young cells. These include miR-22, miR-34a, miR-125a-5p, miR-24-2*, and miR-152, several of which, namely miR-34a and miR-125a-5p, are closely associated with senescence-like growth arrest and metastasis in cancer cells (Tazawa et al., 2007; Li et al., 2009; Wang et al., 2009). Here, we focused on miR-22 and evaluated the expression of miR-22 in young and senescent human diploid fibroblast strains. Quantitative (q) RT-PCR analysis confirmed that miR-22 expression was increased in senescent TIG-3 and other fibroblasts and even up-regulated by more than fivefold in senescent MRC-5 cells (Fig. 1 B). These findings suggest that miR-22 up-regulation is universal in senescent human fibroblasts.
Figure 1.
miR-22 is up-regulated in senescent human fibroblasts, mediating cellular senescence. (A) miRNA expression profile of TIG-3 fibroblasts was analyzed by miRNA microarray, presented as fold changes in miRNA expression between TIG-3S (senescent) and young (Y) cells. A set of altered expression miRNAs is indicated by red columns (see Table S1). RQ, relative quantitation. (B) Relative quantitation of miR-22 expression in different PDLs of fibroblasts was analyzed by qRT-PCR analysis. miR-22 expression levels in human fibroblasts were indicated, relative to those in TIG-3 (42 PDL) set at 1 in the left histogram and MRC-5 (43 PDL) set at 1 in the right histogram. U6 was used as an internal normalization control. The dashed line represents the threshold of expression level (twofold vs. TIG-3 42 PDL). (C and D) MRC-5 cells were transfected with cont miR or mature miR-22 (miR-22) for 6 d at indicated concentration. (C) qRT-PCR analysis shows the relative quantitation of miR-22 expression (vs. cont miR) in each transfection group. miR-22 expression levels in miR-22–transfected MRC-5 cells were indicated, relative to that in cont miR–transfected cells set at 1. U6 was used as an internal normalization control. (D) SA-β-gal activity was presented by the percentage of SA-β-gal–positive cells, which was indicated in different dose groups. (E) Cell proliferation assay was performed after transfection of 10 nM miR-22 or cont miR, and cells were counted for the indicated days. Each value was determined in triplicate. **, P < 0.01. (F) Representative photos for SAHF formation in MRC-5 cells at day 6 after transfection. Images were taken with fluorescence microscopy. Enlarged images of the boxed area from the top are shown in the bottom. SAHF formation was quantified by counting 200 cells from >10 random fields, and the results were shown in the right histogram in contrast to MRC5S (senescent; 58 PDL). Data in all the panels represent mean ± SEM (n = 3). *, P < 0.05; **, P < 0.01. Bars, 20 µm.
To investigate the involvement of miR-22 in cellular senescence in human fibroblasts, we enforced miR-22 expression by introduction of mature miR-22 duplex into young fibroblasts. We first examined miR-22 expression level by qRT-PCR analysis at day 6 after direct transfection with miR-22 duplex into young MRC-5 cells (Fig. 1 C). Compared with the endogenous level of miR-22 in the senescent MRC-5 cells (fivefold higher than young cells), miR-22 was up-regulated by 2.9-fold (1 nM), 4-fold (5 nM), and 15.7-fold (10 nM) in MRC-5 cells, relative to control miRNA (cont miR) in each transfection group. Furthermore, miR-22 also increased SA-β-gal activity (Fig. 1 D), a well known senescence cytosolic biomarker, in a transfection dose-dependent manner, whereas cont miR did not induce SA-β-gal activity. These results imply that up-regulation of miR-22 expression is significant for the induction of senescence phenotypes. Because the cessation of cell proliferation is a hallmark of cellular senescence, we examined whether cell proliferation is altered by overexpression of miR-22. We observed that transfection of 10 nM miR-22 caused a remarkable inhibition of cell proliferation compared with that of cont miR (Fig. 1 E), and this growth inhibition by miR-22 is in a dose-dependent manner (not depicted). Senescence-associated heterochromatin foci (SAHF) formation is thought to be a senescence nuclei biomarker and is often observed in senescent fibroblasts (Dimri et al., 1995; Narita et al., 2003; Adams, 2007). We observed obvious SAHF formation in miR-22–transfected cells, and the percentage of SAHF-positive cells was significantly increased by miR-22 overexpression and was also increased in senescent MRC-5 cells (Fig. 1 F). This result is not cell type specific because other fibroblasts, such as IMR90, transfected with miR-22 appeared to be senescence phenotypes (unpublished data).
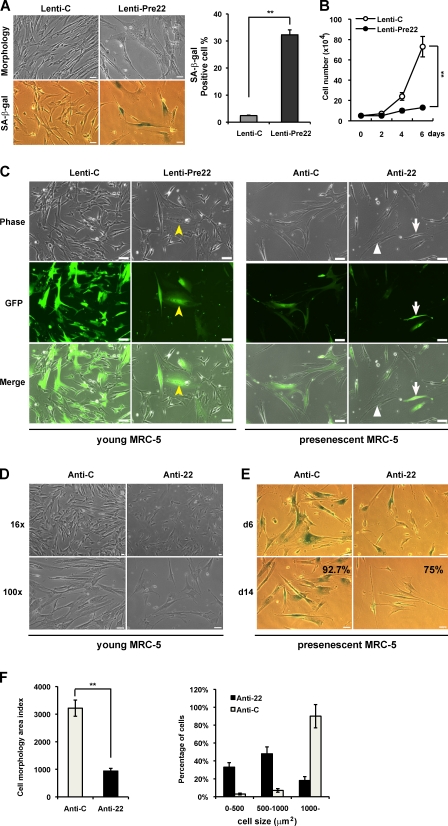
To ensure that the function of mature miRNA is not at a supraphysiological level, we repeated these experiments using a stable miRNA vector that mimics miRNA biological processing. Pre–miR-22 lentiviral construct (Lenti-Pre22), stably expressing miR-22 precursor in its native context, was used to study the effect of miR-22 on cellular senescence in MRC-5 fibroblasts. Lenti-Pre22–infected cells exhibited the enlarged senescence morphology and SA-β-gal–positive staining (Fig. 2 A). Compared with Lentiviral empty vector (Lenti-C), Lenti-Pre22 infection significantly increased SA-β-gal activity and caused growth arrest in MRC-5 cells (Fig. 2, A and B), which is similar to the effect of mature miR-22. Collectively, these results suggest that miR-22 is associated with cellular senescence, accompanied by the induction of major specific senescence-associated markers in human diploid fibroblasts.
Figure 2.
Stable expression and knockdown of miR-22 affect senescence phenotypes in MRC-5 cells. (A) Cell morphology and SA-β-gal activity were analyzed by phase-contrast microscopy at day 6 after infection with empty vector (Lenti-C) or premiR-22 (Lenti-Pre22). The percentage of SA-β-gal–positive cells is presented in the right histogram. (B) Cell proliferation assay was performed after infection of Lenti-Pre22, and cells were counted for the indicated days, compared with control cells. Each value was determined in triplicate. **, P < 0.01. (C) Cell morphology was analyzed with fluorescence microscopy at day 6 after infection. GFP-labeled cells indicate infected cells (middle). Yellow arrowheads and white arrows depict miR-22 overexpression in young MRC-5 cells and miR-22 knockdown in presenescent MRC-5 cells, respectively. Those cells that failed to be infected are marked with white triangles. (D) Cell morphology of anti-22–infected young MRC-5 was analyzed by phase-contrast microscopy and compared with anti-C. (E) Presenescent MRC-5 cells infected with anti-C or anti-22 were subjected to SA-β-gal assay at days 6 and 14 after infection. The percentage of SA-β-gal–positive cells at day 14 after infection is presented in the bottom panels. (F) Cell morphology area and cell size distribution were analyzed using ImageJ by counting GFP-expressed presenescent MRC-5 cells after infection with anti-C or anti-22 for 6 d. Cell morphology area index represents cell size (micrometers squared). Cell size distribution was divided into three groups: 0–500, 500–1,000, and 1,000 μm2 and up. Data in all the panels represent mean ± SEM (n = 3). **, P < 0.01. Bars: (A, D, and E) 50 µm; (C) 20 µm.
Furthermore, we wondered how knockdown of miR-22 impacts cellular senescence phenotypes and whether a reduced expression of miR-22 would impede the progression of senescence and extend the life span in fibroblasts. miRZip anti–miR-22 expression lentivector (anti-22), which can stably express anti–miR-22 and provide permanent miR-22 inhibition, was used to infect young (44 population doubling level [PDL]) and presenescent (54 PDL) MRC-5 cells. We observed that Leni-Pre22–infected young MRC-5 cells demonstrated enlarged senescent morphological changes (Fig. 2 C, GFP-labeled cells marked with yellow arrowheads). In contrast, anti-22–infected young MRC-5 cells seemed smaller and younger than anti-22 empty vector (anti-C)–treated cells (Fig. 2 D). For presenescent MRC-5 cells, stable knockdown of miR-22 caused obvious morphological changes, exhibiting small and thin morphology (Fig. 2 C, GFP-labeled cells marked with arrows), whereas those cells that failed to be infected exhibited enlarged senescent morphology (Fig. 2 C, white triangles) within the same field under the fluorescent microscope. Presenescent MRC-5 cells entered senescence and demonstrated obvious SA-β-gal staining as time passed. Anti-22–infected presenescent MRC-5 cells maintained small and thin morphology for >2 wk with a decreased percentage of SA-β-gal–positive cells (Fig. 2 E), and there appeared to be a significant decrease in cell size and percentage of cells distributed in a large-sized group (Fig. 2 F), compared with senescent anti-C–treated cells. Although stable knockdown of miR-22 exhibited neither the promotion of cell proliferation nor the extension of the life span (not depicted), this might be a result of irreversible growth arrest in senescent cells. These findings suggest the requirement of miR-22 in mediating senescence and indicate that miR-22 inhibition is indeed an obstacle for the progression of senescence in fibroblasts.
miR-22 overexpression induces growth suppression and senescence-like phenotypes in human breast epithelial and cancer cells
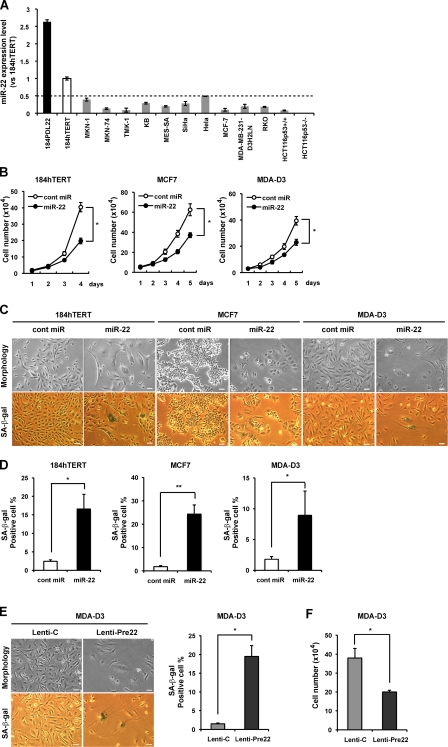
Senescence has been most widely studied in fibroblasts in vitro but is also well defined in other cell types, such as epithelial cells which are the origin of most carcinoma (Narita et al., 2003). Expression of human telomerase reverse transcriptase (hTERT) in certain cell types has been shown to extend cellular life span without malignant transformation. To investigate the effect of miR-22 on cellular senescence in human epithelial cells, we used hTERT-infected HMEC184 (184hTERT) cells that possess the unlimited proliferation capacity of HMEC184 cells and are regarded as immortalized. We found that miR-22 was expressed higher by >2.5-fold in senescent HMEC184 (22 PDL) than in 184hTERT cells that have a similar miR-22 expression level to that of normal young HMEC184 (Fig. 3 A). Compared with immortalized 184hTERT cells, a widespread decrease in miR-22 level was observed in various human cancer cells (Fig. 3 A), indicating that miR-22 may have an intrinsic function in tumor suppression associated with various human malignancies.
Figure 3.
Overexpression of miR-22 induces senescence-like phenotypes in human breast epithelial and breast cancer cells. (A) qRT-PCR analysis shows relative quantitation of miR-22 expression level (vs. 184hTERT) in human epithelial and various cancer cells. Expression levels of miR-22 in various cells were relative to that in 184hTERT cells set at 1. U6 was used as an internal normalization control. The dashed line represents the threshold of expression level (0.5-fold vs. 184hTERT). (B) Cell proliferation assay was performed after transfection of miR-22, and cells were counted for the indicated days, compared with control cells. Each value was determined in triplicate. *, P < 0.05. (C–E) Cell morphology and SA-β-gal activity were analyzed by phase-contrast microscopy at day 6 after transfection (C and D) or infection (E) in indicated cells. SA-β-gal activity was presented by the percentage of SA-β-gal–positive cells. (F) Cell proliferation assay was performed at day 6 after MDA-D3 cells were infected with Lenti-Pre22 and compared with control cells. Data in all the panels represent mean ± SEM (n = 3). *, P < 0.05; **, P < 0.01. Bars, 50 µm.
To test this notion, we first evaluated the effect of miR-22 on cell growth and SA-β-gal activity in human breast epithelial cells and two breast cancer cell lines. We found that miR-22 significantly inhibited cell growth in 184hTERT, MCF7, and MDA-MB-231-luc-D3H2LN (called MDA-D3 for short; Fig. 3 B). We noted that mature miR-22 caused remarkable and characteristic morphological alterations, including enlarged cellular size and a flattened shape (Fig. 3 C), and significantly increased SA-β-gal activity in the three cells (Fig. 3 D). Furthermore, we confirmed that overexpression of premiR-22 in MDA-D3 cells also significantly induced senescence-specific morphological changes, increased SA-β-gal activity (Fig. 3 E), and inhibited cell growth (Fig. 3 F), similar to the effect of mature miR-22.
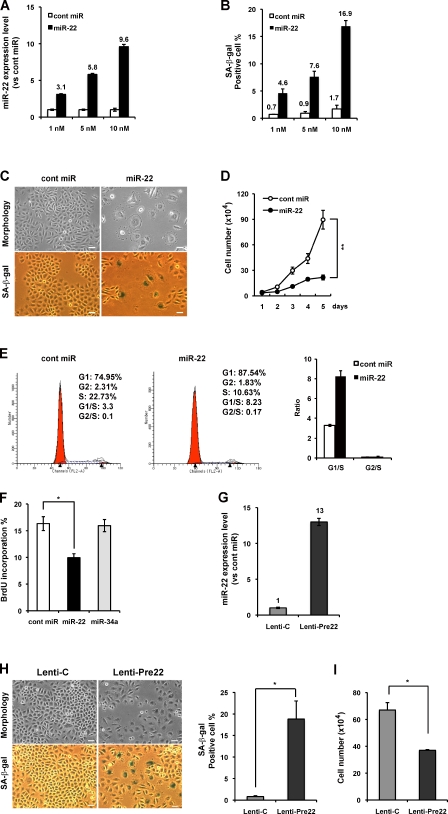
The marked induction of senescence by miR-22 in human breast cancer cells prompted us to investigate whether miR-22 induces cellular senescence in other human cancer cells. Therefore, we further studied the effect of miR-22 in the human cervical carcinoma cell line SiHa. We confirmed the dose-dependent increase in miR-22 expression level (Fig. 4 A) and SA-β-gal activity (Fig. 4 B) after direct transfection of miR-22 in SiHa cells, whereas cont miR affected neither the cell growth nor the SA-β-gal activity. Introduction of 10 nM miR-22 resulted in cells exhibiting a senescence-like flattened shape and SA-β-gal–positive staining (Fig. 4 C), and it induced remarkable growth suppression compared with that of cont miR (Fig. 4 D). Because senescent cells never reenter the cell cycle and appear to decrease in DNA synthesis, we performed FACS analysis and quantity analysis of BrdU incorporation in SiHa cells. miR-22 overexpression induced cell cycle arrest at G1 phase, accompanied by the decrease in percentages of S phase (Fig. 4 E). The inhibitory effect of miR-22 on cell cycle progression has also been recently reported in other cancer cells (Ting et al., 2010). Furthermore, miR-22 resulted in the decrease of BrdU incorporation in comparison with cont miR or miR-34a (Fig. 4 F), indicating impaired DNA replication in miR-22–transfected cells during the S phase of the cell cycle. In addition, we confirmed that miR-22 expression was increased in Lenti-Pre22–infected SiHa cells (Fig. 4 G) close to the expression level of miR-22 (10 nM). Lenti-Pre22 also induced senescence-specific morphological changes, increased SA-β-gal activity, and inhibited cell growth (Fig. 4, H and I), indicating the induction of cellular senescence by miR-22 overexpression in SiHa cells.
Figure 4.
Overexpression of miR-22 induced cellular senescence, cell cycle G1 arrest, and the decrease in BrdU incorporation in SiHa cells. (A and B) SiHa cells were transfected with cont miR or miR-22 at the indicated concentration for 6 d. qRT-PCR results show the relative level of miR-22 expression to cont miR in each transfection group (A). SA-β-gal activity was presented by the percentage of SA-β-gal–positive cells (B). (C) Cell morphology and SA-β-gal activity were analyzed by phase-contrast microscopy at day 6 after transfection of 10 nM miR-22 or cont miR. (D) Cell proliferation assay was performed after transfection of 10 nM miR-22, and cells were counted for the indicated days, compared with control cells. Each value was determined in triplicate. **, P < 0.01. (E) Cell cycle analysis was performed at 48 h after transfection. The percentage of G1, S, and G2 are demonstrated as shown. The histogram displays the relative changes of G1 and G2 phase compared with S phase. (F) BrdU quantitative analysis was performed at 72 h after transfection, presented by the percentage of BrdU incorporation. (G) Stable expression of miR-22 (Lenti-Pre22) was evaluated by qRT-PCR analysis, presented by the relative quantitation of miR-22 expression level at day 6 after infection. Expression level of miR-22 in Lenti-Pre22–transfected cells was relative to that in Lenti-C–transfected cells set at 1. U6 was used as an internal normalization control. (H) Cell morphology and SA-β-gal activity were analyzed by phase-contrast microscopy at day 6 after infection. The percentage of SA-β-gal–positive cells is presented in the right histogram. (I) Cell proliferation assay was performed at day 6 after infection with Lenti-Pre22 and compared with control cells. Data in all the panels represent mean ± SEM (n = 3). *, P < 0.05; **, P < 0.01. Bars, 50 µm.
Moreover, we did not observe any significant increase in TUNEL-positive apoptotic cells in miR-22–transfected SiHa and MDA-D3 cells (Fig. S1, A and B). These findings suggest that the growth suppression induced by miR-22 overexpression was caused by the induction of G1 arrest in cellular senescence rather than apoptosis. In reverse, miR-22 knockdown in cancer cells resulted in various morphological changes and differential timing of apoptosis in different cell lines. Anti-22–infected SiHa cells appeared to be small and underwent apoptosis at day 6 after infection (Fig. S1 C), whereas MDA-D3 cells became rounded and apoptotic cells were observed in anti-22–infected cells from the third day after infection (Fig. S1 D), which might be because the timelines for the progression of different cells through apoptosis vary in different cell lines (Jessel et al., 2002).
Various findings have suggested the critical role of telomere shortening in contributing to cellular senescence in human cells (Deng et al., 2008). Telomeric 3′-overhang (G-tail) is essential for proper telomere function (Tahara et al., 2005). Here, overexpression of miR-22 had no effect on the length of either total telomere or G-tail (Fig. S2), indicating that miR-22–induced senescence is possibly independent of telomere shortening.
SIRT1, Sp1, and CDK6 are direct targets of miR-22
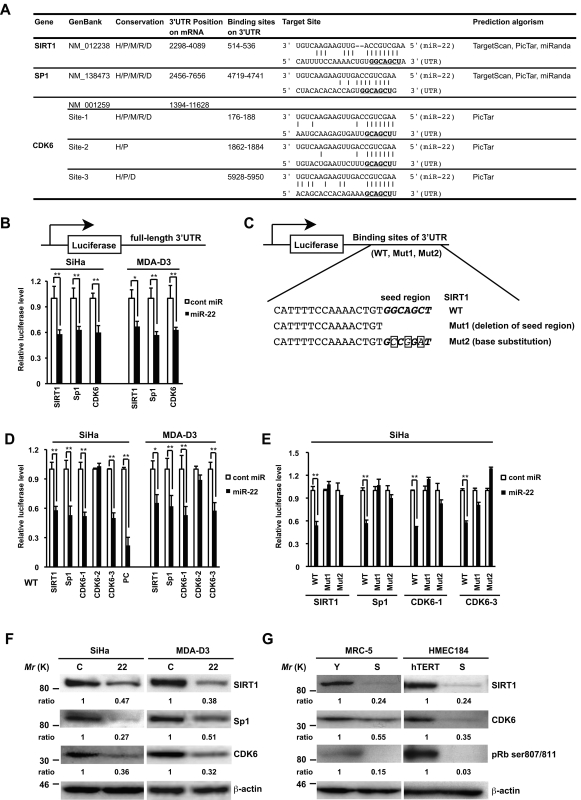
miRNAs are known to suppress hundreds of mRNA targets, resulting in global changes in the cellular phenotype of cells (He and Hannon, 2004). We therefore make an effort to identify potential targets for miR-22 using both in silico and experimental approaches. We used a consensus approach with three widely used types of software (miRanda, TargetScan, and PicTar) to perform the target prediction. After overlapping prediction analysis, we screened those genes that were down-regulated upon senescence and implicated in cell growth and cell cycle regulation. Based on this, SIRT1, Sp1, and CDK6 were selected to be putative miR-22 target genes. The mRNAs of SIRT1 and Sp1 contain putative binding sites for miR-22 in their 3′-UTRs, and each site is broadly conservative among mammals. CDK6 mRNA contains three miR-22 binding sites in the 3′-UTR, whereas it is different in conservation for each site (Fig. 5 A).
Figure 5.
SIRT1, Sp1, and CDK6 are direct targets of miR-22. (A) Summary of miR-22 target sites in the 3′-UTR of SIRT1, Sp1, and CDK6. H, human; P, chimp; M, mouse; R, rat; D, dog. The underlined bold nucleotides indicate target sites. (B–E) Target validation of SIRT1, Sp1, and CDK6 was confirmed in the luciferase reporter assay. (C) Scheme of the luciferase reporter constructs containing conserved miR-22 target sites (WT), deletion of seed region (Mut1), or base substitution of seed region (Mut2) was indicated using SIRT1 as an example. The seed region is italic bold, and point mutations are boxed in the seed region. Each construct, including full-length 3′-UTR (B), WT (D), or Muts (E), was cotransfected with miR-22 or cont miR into SiHa or MDA-D3 cells. Relative luciferase level = (Sluc/Srenilla)/(Cluc/Crenilla) in relative light units (RLUs). Sluc, RLUs of firefly luciferase activity in miR-22–transfected sample. Srenilla, RLUs of renilla luciferase activity in miR-22–transfected sample. Cluc, RLUs of firefly luciferase activity in cont miR–transfected groups. Crenilla, RLUs of renilla luciferase activity in cont miR–transfected groups. PC, positive control. (F) Representative Western blot analysis of SIRT1, Sp1, and CDK6 in SiHa and MDA-D3 cells transfected with cont miR (C) and mature miR-22 (22) at 72 h after transfection. (G) The expression levels of SIRT1, CDK6, and pRB phosphorylation in MRC-5 and HMEC184hTERT cells were analyzed by immunoblotting. β-actin was used as a loading control and the relative density of bands was densitometrically quantified. Y, young; S, senescent; hTERT, 184hTERT. Data in all the panels represent mean ± SEM (n = 3). *, P < 0.05; **, P < 0.01.
To address those genes directly regulated by miR-22, we performed luciferase reporter assay. We constructed pmirGLO full-length 3′-UTR of both SIRT1 and Sp1. Because the full-length 3′-UTR of CDK6 is too long (∼10 kbp), human CDK6 3′-UTR containing three binding sites (6 kbp) was amplified and cloned into pmirGLO vector. Furthermore, to study which site in the 3′-UTR of each gene is important, we engineered luciferase reporters that have exact binding sites of 3′-UTRs of these genes including the wild type (WT) and two mutant UTRs (Mut1 and Mut2), as shown in Fig. 5 C. In SiHa and MDA-D3 cells, miR-22 significantly reduced the luciferase activities of the full-length 3′-UTR (Fig. 5 B) or WT SIRT1, SP1, and CDK6 (site 1 and site 3) reporters (Fig. 5 D), compared with the negative cont miR. In contrast, neither Mut1 (deletion of seed region) nor Mut2 of mutant reporters was repressed by miR-22, which indicates that these target sites directly mediate the repression (Fig. 5 E). In addition, this down-regulation was not seen in WT construct of CDK6 site 2, possibly because of poorly conserved sites among mammals. These results provide experimental evidence that miR-22 can directly repress translation initiation of SIRT1, SP1, and CDK6.
Western blot analysis showed that overexpression of miR-22 markedly down-regulated SIRT1, SP1, and CDK6 in SiHa and MDA-D3 cells (Fig. 5 F). Moreover, we confirmed that SIRT1 and CDK6 were down-regulated in senescent MRC-5 fibroblasts and HMEC184 epithelial cells, accompanied by a decrease in pRB phosphorylation at ser807/811 (Fig. 5 G). This indicates that SIRT1 and CDK6 are senescence-associated genes that are involved in cellular senescence possibly through the pRb pathway. Sp1 was not detected because of its low expression level in these cells.
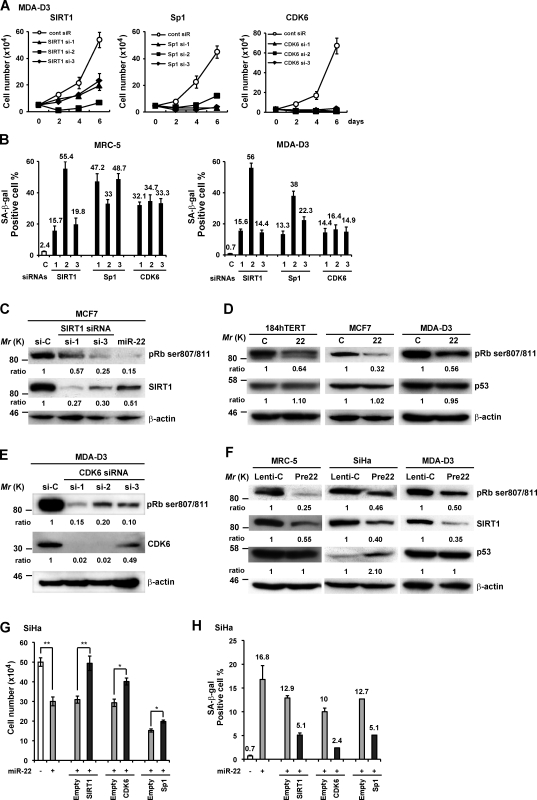
Furthermore, silencing of these target genes by siRNAs resulted in growth arrest (Fig. 6 A) and increased SA-β-gal activity in MRC-5 fibroblasts and MDA-D3 cells (Fig. 6 B), as well as morphological changes (not depicted), similar with miR-22–induced senescence phenotypes. We confirmed that the siRNAs against SIRT1 and CDK6 knocked down the expression of SIRT1 and CDK6 and caused dephosphorylation of pRB at ser807/811 (Fig. 6, C and E), which was also induced by either mature miR-22 or premiR-22 overexpression in various different cells (Fig. 6, D and F). In addition, p53 protein level did not change in MRC-5 and MDA-D3 cells, but p53 expression is up-regulated in miR-22–transfected SiHa cells (Fig. 6 F). These findings indicate that miR-22 may affect the pRb pathway of cellular senescence by targeting SIRT1 and CDK6.
Figure 6.
SIRT1, Sp1, and CDK6 are potentially involved in miR-22–mediated cellular senescence. (A) Representative growth curves corresponding to MDA-D3 cells transfected with cont siR or siRNAs against SIRT1, Sp1, and CDK6. Each value was determined in triplicate. (B) The histograms display the percentage of SA-β-gal–positive cells in MRC-5 and MDA-D3 cells at day 6 after siRNA transfection. (C–F) Western blot analysis was performed in those cells as indicated in each panel at 72 h after siRNA/miRNA transfection (C–E) or day 6 after infection (F). β-actin was used as a loading control and the relative density of bands was densitometrically quantified. (G and H) SiHa cells were first transfected with miR-22 duplex and, 24 h later, sequentially transfected with expressing plasmids or empty vectors as indicated. Cell proliferation (G) and SA-β-gal (H) activity was evaluated at day 6 after miRNA transfection. Data in all the panels represent mean ± SEM (n = 3). *, P < 0.05; **, P < 0.01.
Lastly, we attempted to determine whether overexpression of any of the three genes could rescue the senescence phenotype in cells overexpressing miR-22. We forced SiHa or MDA-D3 cells to express SIRT1, Sp1, or CDK6 using plasmid constructs lacking 3′-UTRs of these genes. Indeed, miR-22–induced cell growth repression and SA-β-gal activity were partially rescued by the introduction of SIRT1, CDK6, or Sp1 in either SiHa cells (Fig. 6, G and H) or MDA-D3 cells (not depicted), although it seemed weak for the effect of Sp1 overexpression on cell growth, which might be because of the indirect role of Sp1 in the pRb pathway of senescence. Collectively, these findings suggest that SIRT1, Sp1, and CDK6 play an important role in miR-22–induced senescence.
miR-22 alters tumor cell morphology and suppresses cell invasiveness in vitro
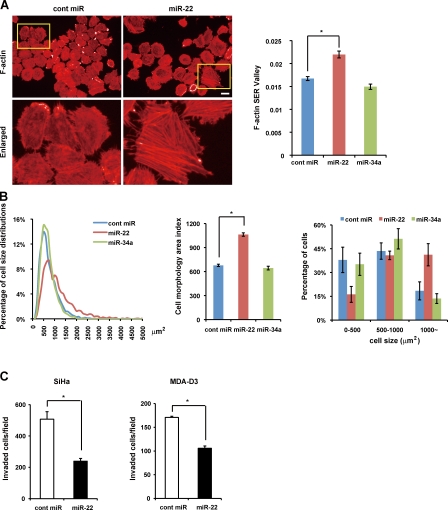
Changes in cell morphology are important parameters of cancer invasion and metastasis. We performed high content analysis using Operetta to measure the morphology area of all the cells transfected with miR-22, miR-34a, or cont miR on each well of a 96-well plate (Fig. 7 A and Fig. S3 B) and analyzed cell size distribution (Fig. 7 B). We found there were significant differences in cell size distribution between miR-22–treated cells and other cells. miR-22 remarkably increased cell morphology area up to 1.6-fold overall and raised percentages of cells distributed in large cell size groups in comparison with cont miR or miR-34a, revealing that miR-22 triggers senescence morphological changes in tumor cells. In addition, miR-22–treated cells contain enhanced actin stress fibers (Fig. 7 A) similar to senescent fibroblasts, indicating that those cells are less motile (Chen et al., 2000; Belguise et al., 2005). We observed that senescent fibroblasts and Lenti-Pre22–infected cancer cells exhibiting large flattened senescence-like morphology appeared to decrease in cell movement (Fig. S4 and Videos 1–4, large GFP-positive cells), indicating that miR-22–induced senescence morphology in cancer cells could be attributed to the suppression of cell motility. Matrigel invasion assay showed that miR-22 significantly reduced the number of invaded cells in SiHa and MDA-D3 cells (Fig. 7 C), indicating that the invasive potential of cancer cells was severely affected by miR-22. Therefore, the inhibition of cancer cell invasion by miR-22 may be a result of the induction of cellular senescence.
Figure 7.
Overexpression of miR-22 induces cell enlargement and inhibits cell invasion in vitro. (A) Cell morphology area and actin stress fiber formation (stained with phalloidin) in miR-22–transfected SiHa cells were examined by confocal microscopy and compared with control cells. Enlarged images of the boxed area from the top are shown in the bottom. Bar, 20 µm (top, left and right). The histogram shows that F-actin formation was quantified using the texture analysis, and F-actin SER Valley represented occurrence of stress fiber structures within cells. (B) Cell size distribution (left), overall morphology area (middle), and percentage of cells in three groups of cell size (right) were calculated and analyzed by automated image analysis in SiHa cells. (C) Effect of miR-22 on SiHa and MDA-D3 cell invasion was measured by Matrigel invasion assay for 48 h, presented by quantitative determination of the number of invaded cells. Data in all the panels represent mean ± SEM (n = 3). *, P < 0.05.
miR-22 inhibits tumor growth and metastatic potential of aggressive breast cancer in vivo
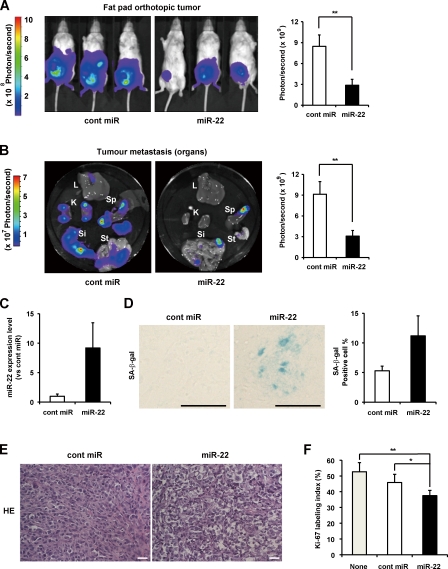
We next asked whether miR-22 overexpression would also induce senescence in vivo and suppress tumor growth and metastasis in vivo. Using breast cancer tumor models, the cont miR–treated mice showed the apparent presence of primary tumor, whereas those injected with miR-22 complex exhibited no increase in the luminescence of primary tumor during the same observation period (Fig. 8 A, left). Judging from photon count between the cont miR– and miR-22–treated groups at the two points of the experiment, miR-22 treatment resulted in a mean decrease in tumor growth of 41.3% at day 39 (P = 0.04; not depicted) and 66% at day 46 (P = 0.005; Fig. 8 A, right), suggesting that miR-22 exerted significant tumor growth suppression in vivo. We also compared tumor metastasis with important organs in the two groups and were surprised to find that miR-22 delivery resulted in the inhibition of distant metastasis in the liver, kidney, spleen, stomach, and small intestine (Fig. 8 B). There were significant differences in the whole body of mice between the cont miR– and miR-22–treated groups on day 46 (P = 0.004), indicating the inhibition of metastasis by injection with miR-22 in vivo.
Figure 8.
Synthetic miR-22 delivery induces cellular senescence in vivo and inhibits breast tumor growth and metastasis in vivo. (A and B) Representative fat pad orthotopic tumor (A) and selected organ (B) images of mice on day 46 after inoculation. Quantitation of bioluminescence emitted from the primary tumor (A) or whole body (B) of mice was presented as the mean ± SEM (n = 6). L, liver; K, kidney; Sp, spleen; Si, small intestine; St, stomach. (C) Relative quantitation of miR-22 level in primary tumor tissues was analyzed by qRT-PCR. Total RNA was isolated from five 10-µm-thick optimal cutting temperature compound–embedded frozen tissue sections. Expression level of miR-22 in miR-22–treated tumors was relative to that in cont miR–treated tumors set at 1. U6 was used as an internal normalization control. Data represent mean ± SEM (n = 5). (D and E) Representative histochemical detection of SA-β-gal activity in vivo (D) and hematoxylin and eosin (HE) staining (E) in primary tumor on day 46 after inoculation. Images were taken with light microscopy. Seven independent fields were chosen, and the percentage of SA-β-gal–positive cells is presented in the histogram. Data represent mean ± SEM (n = 4). Bars, 50 µm. (F) Effect of miR-22 on cell proliferation in primary tumor tissues was evaluated by Ki-67 immunostaining. The histogram displays the percentage of Ki-67 labeling index in these groups. Data represent mean ± SEM (n = 3). *, P < 0.05; **, P < 0.01.
We also confirmed that synthetic miR-22 was delivered into primary tumor by quantitation of the miR-22 level in tumor (Fig. 8 C). Furthermore, the number of SA-β-gal–positive cells in miR-22–treated tumor dramatically increased, correlated with the amount of miR-22 in tumor (Fig. 8 D). In addition, we did not observe SA-β-gal–positive cells in nontumor cells such as vascular endothelial cells and other stroma cells of mice. Moreover, we found that the morphologies of the tumors were different; the cells in cont miR–treated groups were densely packed and slightly elongated, whereas those in miR-22–treated groups were larger and more irregularly shaped (Fig. 8 E). Pathologically, vacuolation and nuclear pyknosis associated with degradation of tumor cells were found in miR-22–treated tumor tissues. Therefore, it is possible that induction of cellular senescence in tumor cells may result in these pathological changes.
The progression toward metastasis formation requires proliferation of tumor cells at primary sites and distant sites. Notably, the expression of Ki-67 in miR-22–treated groups was significantly lower than that in cont miR–treated groups and nontreated groups (Fig. 8 F), indicating the inhibition of cell proliferation in tumor tissues treated with miR-22. Together, these findings suggest that miR-22 significantly induced cellular senescence in breast cancer in vivo, consequently inhibiting tumor growth and metastasis in vivo.
Discussion
The present study for the first time reported the functional effect of miR-22 as a novel regulator of cellular senescence in normal human and cancer cells and addressed the inhibitory role of miR-22 in tumor growth and metastasis, suggesting that miR-22–induced senescence acts as a barrier to cancer progression in vitro and in vivo. Our study extends the current understanding of SA-miRNAs in tumorigenesis by unraveling the role of miR-22 in cellular senescence and tumor suppression. miRNAs present a mechanism in which genes involved in a variety of different signaling pathways can be regulated simultaneously (He and Hannon, 2004; Lafferty-Whyte et al., 2009). Not only does miR-22–induced senescence inhibit unlimited tumor cell proliferation, senescence morphological changes also suppress tumor cell motility and invasion, partially because of enhanced actin stress fibers in cancer cells, indicating that the induction of senescence contributes to the suppression of tumor cell growth, invasion, and metastasis in vitro and in vivo.
Our study shows that miR-22 is differentially expressed in normal human and cancer cells. Consistent with our results, miR-22 dysregulation in cancer and human diseases has been documented in several miRNA profiling experiments. miR-22 is down-regulated in intrahepatic cholangiocarcinoma (Kawahigashi et al., 2009) and silenced in acute lymphoblastic leukemia (Li et al., 2010) and hepatocellular carcinoma (Calin et al., 2004) but up-regulated in osteoarthritis (Iliopoulos et al., 2008). These varying levels of miR-22 expression may suggest different requirements for alterations in their target gene pathways. miR-22 regulates a repertoire of cancer-related genes; however, there is no indication of the importance of miR-22 in tumorigenesis through targeting senescence-associated genes. In the present study, gain-of-function and loss-of-function phenotypes of miR-22 demonstrated the role of miR-22 as a positive regulator of cellular senescence and identified SIRT1, Sp1, and CDK6 as critical targets of miR-22 in the senescence signaling pathway.
Previous studies have documented that SIRT1, Sp1, and CDK6 might act on the p53 and/or pRb pathway. (Ota et al., 2006; Ruas et al., 2007; Tapias et al., 2008; Brooks et al., 2009) SIRT1 plays an important role in the longevity and cellular senescence of most organisms through directly modulating the p16–pRb signaling pathway (Huang et al., 2008) as well as p53 and other proteins (Solomon et al., 2006; Brooks and Gu, 2009). The Sp1 transcription factor regulates the expression of multiple cell cycle genes, including the p53 and Sp1 gene itself (Koutsodontis et al., 2001; Tapias et al., 2008). CDK6 protein has been understood to phosphorylate pRB and delay senescence (Ruas et al., 2007; Ohtani et al., 2009). Our study demonstrated that miR-22 induced the dephosphorylation of pRB by targeting SIRT1 and CDK6. Collectively, we suppose that miR-22 regulates cellular senescence through connecting SIRT1, Sp1, and CDK6 to affect the pRb pathway and might coordinate p53 and other signaling pathways of cellular senescence in a cell type– and genetic context–dependent manner.
The ability of miRNAs to affect many mRNAs is similar to the ability of transcription factors to regulate many promoters simultaneously. Recent studies demonstrate that miR-22 constitutes a feedback loop with c-myc and MYCBP and forms a regulatory loop in the phosphatase and tensin homolog–AKT pathway (Iliopoulos et al., 2008; Pandey and Picard, 2009; Bar and Dikstein, 2010; Xiong et al., 2010). The human miR-22 gene is located in a minimal loss of heterozygosity region on chromosome 17 close to p53 (Calin et al., 2004). We predict that miR-22 might induce complex changes and the extensive cooperation between miR-22 and p53 known to be involved in the senescence program, which remains to be elucidated.
miR-22 has been shown to be ubiquitously expressed in human cells and various tissues (Neely et al., 2006). Expression of miR-22 increases after the induction of stem cell differentiation and in erythropoiesis, which indicates that it might have an important role in cell development and differentiation (Choong et al., 2007; Gangaraju and Lin, 2009). miR-22 is highly expressed in mammary progenitor cells, implicating its possible role in progenitor self-renewal (Ibarra et al., 2007). To date, there is no indication of miR-22 function in normal human cells. In our study, miR-22 is particularly up-regulated upon cellular senescence in human fibroblasts and epithelial cells, and overexpression of miR-22 induced senescence phenotypes in these cells, indicating that miR-22 might emerge as the principal regulator that controls cell functions in physiological and pathophysiological settings. However, it is obscure in the mechanism that controls the increased expression of miR-22 during senescence. We attempted stable knockdown of miR-22 in early-passage MRC-5 (Fig. 2 D) and IMR90 cells (unpublished data), which resulted in both of the cells seeming younger than untreated cells. Furthermore, we investigated how stable knockdown of miR-22 affected late-passage MRC-5 cells. Our results demonstrated that those anti-22–infected presenescent MRC-5 cells appeared to be small, thin, and young with the decrease in cell morphology area and fewer SA-β-gal–positive cells (Fig. 2, C–F), indicating that inhibition of miR-22 is an obstacle for cells to undergo senescence. Given our finding that miR-22 induces cellular senescence in human fibroblasts in vitro, it will be interesting to determine whether miR-22 is involved in human aging in vivo.
It has been reported that the regulation of proliferation signaling, cytoskeletal remodeling, and apoptosis survival pathways would logically be required to create the senescent cell phenotype, which can be regulated by SA-miRNAs with differential timings of these complex signaling cascades (Lafferty-Whyte et al., 2009). These might give an explanation to the distinct effects of miR-22 knockdown on cell proliferation, morphology, and viability in various cells. We observed that actin stress fibers were enhanced in miR-22–transfected enlarged cells (Fig. 7 A), whereas anti-22–infected cells appeared to be small and to have fewer stress fibers (unpublished data). Moreover, we noted that cytoskeleton arrangement and focal adhesion turnover occurred with differential timing in various anti-22–infected cells and thus might affect cell growth, adhesion, and survival, which needs to be confirmed in a further study. In addition, miR-22 expression level is quite low in various cancer cells, and permanent inhibition of miR-22 induced apoptosis, indicating that miR-22 kept to a certain level seems to be essential for cellular survival. It has been reported that miR-22 targets the phosphatase and tensin homolog and anti-22 induces apoptosis in different cancer cells (Bar and Dikstein, 2010; Liu et al., 2010), which is consistent with our results.
In this study, miR-22 inhibits the proliferation of human breast cancer metastasis cell lines (MDA-D3) both in vitro and in vivo through induction of genetic reprogramming of the senescent pathway. Synthetic miR-22 injection significantly suppresses tumor growth and metastasis in a mouse model of breast cancer metastasis, indicating the therapeutic potential of miR-22 in breast cancer metastasis. Currently, the emergence of new technologies that use synthetic miRNA mimics or anti–miRNA oligonucleotides holds great promise in clinical miRNA therapy (Garofalo and Croce, 2011). The first clinical trial for LNA-antimiR-122–based hepatitis C therapeutics was initiated after the successful therapeutic application of miR-122 antagonism in mice (Elmén et al., 2008), whereas the clinical application of synthetic miRNA mimics has not been reported. Synthetic miR-22 mimic treatment in cancer will become a significant scientific and therapeutic challenge.
SA-miRNAs, such as miR-22, induce cellular senescence without causing apoptosis in cancer cells and subsequently result in tumor suppression (Fig. S5), providing a novel promising approach for the next generation of cancer therapy using nucleic acid biomedicine. We propose that permanent micromanaging of cancer cells by inducing senescence phenotypes using SA-miRNAs may be novel and efficient for protecting recurrent cancer after conventional cancer treatments without causing side effects.
Materials and methods
Cell culture
MRC-5 cells were grown in DME/F12 (1:1, vol/vol) supplemented with SAP (0.2 mM serine, 0.1 mM aspartic acid, and 1.0 mM pyruvate). MDA-MB-231-luc-D3H2LN is a luciferase-expressing cell line that was derived from MDA-MB-231 cells, maintained in RPMI1640 medium. Other human fibroblasts and cancer cells were cultured in DME. All media were supplemented with 10% FBS (vol/vol). HMEC184 cells were cultured in MEGM BulletKit (Takara Bio Inc.). TIG-3 (74–76 PDL), TIG-1 (63 PDL), TIG-114 (51 PDL), MRC-5 (58–60 PDL), and HMEC184 (22 PDL) were cultured for 2–4 wk as senescent normal cells. MRC-5 (41–51 PDL) and 184hTERT (84–100 PDL) were used as young fibroblasts and immortalized epithelial cells, respectively.
miRNA microarray
Total RNAs were harvested from young (42 PDL) and senescent (74 PDL) TIG-3 fibroblasts using the traditional acid guanidinium-phenol-chloroform extraction method and quantified with a spectrophotometer (NanoDrop; Thermo Fisher Scientific). Microarray analysis of miRNA expression was performed by both miRCURY LNA miRNA Array (Exiqon) and 3D-Gene microarray (Toray).
Real-time qPCR
Total RNA was extracted from the cells and tissues using the miRNeasy Mini kit (QIAGEN). The expression of miRNA was quantified by miRNA assays (TaqMan; Applied Biosystems). Real-time qRT-PCR was performed using a real-time PCR system (ABI StepOne and StepOnePlus; Applied Biosystems) and LightCycle 480 (Roche). Expression of miRNA was defined from the threshold cycle, and relative expression levels were calculated using the 2−ΔΔCt method after normalization with reference to the expression of U6 small nuclear RNA.
Transient miRNA/siRNA transfection and plasmid transfection
Hsa-miR-22 duplex and negative control were obtained from QIAGEN. siRNAs targeting SIRT1, Sp1, CDK6, and negative control siRNA were purchased from Invitrogen. Cells were transfected with 10 nM of either miRNA or siRNA (except as mentioned) using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s protocol. Transfection efficiency under the conditions we adopted in this study was estimated to be >90% according to our observations made using a fluorescence-labeled double-stranded siRNA.
For rescue experiments, the expression plasmids used were pcDNA3.1-SIRT1 (a gift from B. Marshall, Gladstone Institute, San Francisco, CA), pCMVneo-CDK6 (provided by S. Van den Heuvel, Utrecht University, Utrecht, Netherlands), and CMV-Sp1 (Addgene). These constructs contain the encoding region of mRNA but lack the 3′-UTR of these genes. Cells were first transfected with miRNA in a 60-mm dish for 24 h and sequentially transfected with 0.25 µg of plasmid DNA using Lipofectamine LTX Plus reagent (Invitrogen) according to the manufacturer’s protocol. The overexpression of SIRT1, Sp1, and CDK6 was confirmed by Western blotting. After a 24-h incubation, cells were seeded to 35-mm-diameter dishes and incubated for 4 d until cell proliferation assay and SA-β-gal assay.
Lentivirus infection
Lentiviruses were generated by cotransfecting 0.9 µg of lentiviral vector (premiR-22, miRZip, anti–miR-22, or empty vectors; System Biosciences) and 2.7 µg of packaging plasmid mix (1:1:1 for 0.9 µg pPACK-H1-GAG, pPACK-H1-Rev, and pVSV-G) in 293T cells using Lipofectamine LTX Plus reagent. Supernatants were collected 48 h after transfection, filtered through a 0.45-µm membrane, and directly used to infect cells. Cells were observed and images were acquired using a 10× objective with fluorescent microscopy (Axiovert 200M; Carl Zeiss) in combination with a camera (AxioCam; Carl Zeiss) and AxioVision software (Carl Zeiss) at room temperature.
Cell proliferation and SA-β-gal assay
For cell proliferation assay, 48 h after transfection or infection, 3–5 × 104 cells were seeded in a series of 35-mm-diameter dishes and counted for the indicated days. For cytochemical and histochemical detection of SA-β-gal activity, SA-β-gal staining was performed as described previously (Debacq-Chainiaux et al., 2009). SA-β-gal–positive cells were quantified by counting positive and negative cells at 100× magnification in at least five random independent fields. Pictures were taken with a 10× phase-contrast objective on a light microscope (IMT-2; Olympus) with a camera using Image saver (AE-6905; ATTO) at room temperature.
Automated image acquisition
SiHa cells were seeded in a 96-well Viewplate (PerkinElmer) and transfected with cont miR, miR-22, or miR-34a for 72 h. To measure cell size and F-actin, cells were fixed by 4% PFA and stained with the actin marker Rhodamine phalloidin (1:40; Invitrogen; provided by S. Kobayashi and H. Kishi, Yamaguchi University, Ube, Japan). For BrdU quantitative analysis, cells were pulse labeled with 10 µM BrdU (Sigma-Aldrich) for 1 h at 37°C, incubated with 5% CO2, and fixed by 70% ice-cold ethanol for 30 min at room temperature. Cells were treated with 2N HCl for 20 min, neutralized with 0.2 M Tris-HCl, pH 7.5, and permeabilized with 0.1% Triton X-100 for 5 min, followed by the mouse anti-BrdU (1:200; Dako) and incubation for 1 h. Cells were then stained with anti–mouse AF488 (1:500; Invitrogen). DAPI (1 µg/ml; Dojindo) images were used for nuclear recognition and cell counting. Images were acquired in a fully automated and unbiased manner using a 10× objective with a spinning disk confocal microscope (Operetta; PerkinElmer) at room temperature. Eight images per well were collected to obtain a sufficient number of cells for reliable statistical analysis. Image correction and analysis were performed using custom-designed image analysis software (Harmony; PerkinElmer). The histogram in Fig. 7 A shows that F-actin was quantified using the texture analysis by Harmony software (PerkinElmer), and F-actin SER Valley represented the occurrence of stress fiber structures within cells.
FACS analysis
48 h after transfection, cells were fixed in 70% ice-cold ethanol and stained with PBS containing 50 µg/ml propidium iodide and 100 µg/ml RNase A for DNA content analysis by flow cytometry analysis on a FACSCalibur system (BD). The percentage of cells in the various cell cycle phases was calculated using ModFitLT v2.0 software (Verity Software House).
Apoptosis assays
Cells were plated in an 8-well CultureSlide (BD), and apoptotic cells were detected with traditional or modified TUNEL assay using the DeadEnd Fluorometric TUNEL System (Promega) according to the manufacturer’s protocol. In modified TUNEL assay, Cy5-dUTP (GE Healthcare) was substituted for fluorescein-dUTP in a standard TUNEL reaction to detect apoptotic cells of GFP-expressed cells. Images were acquired using a 40× objective with fluorescent microscopy (Axiovert 200M) in combination with a camera (AxioCam) and AxioVision software at room temperature.
Hybridization protection assay (HPA) and Southern blot analysis
Cells were plated in a 100-mm culture dish and transfected, and DNA was extracted with a traditional phenol-chloroform 72 h after transfection. The lengths of total telomere and G-tail were determined using Southern blotting and HPA methods as described previously (Tahara et al., 2005).
Western blotting
72 h after transfection or at day 6 after infection, cells were homogenized in lysis buffer (50 mM Tris-HCl, pH 8.0, 120 mM NaCl, 1% NP-40, 100 mM NaF, 0.2 mM Na3VO4, and Complete mini protein inhibitor cocktail [Roche]). 30 µg of proteins in the total cell lysate was separated by SDS-PAGE and transferred to polyvinylidene fluoride membrane. Antibodies to p53 (clone BP53-12; Millipore), phospho-Rb (ser807/811; Cell Signaling Technology), and β-actin (Sigma-Aldrich) were purchased, and anti-CDK6 (C-21), SIRT1 (H-300), and Sp1 (PEP2) antibodies were purchased from Santa Cruz Biotechnology, Inc. The secondary antibodies were HRP-conjugated anti–rabbit (NA 934V) and –mouse (NA 931V) antibodies (GE Healthcare). Immunoreactive bands were visualized using an ECL Plus kit (GE Healthcare), followed by exposure to x-ray film (RX-U; Fujifilm). The density of bands was densitometrically quantified using ImageJ (National Institutes of Health).
Luciferase reporter assay
The full-length 3′-UTRs of human SIRT1 and Sp1 were amplified by PCR from genomic DNA and cloned at the SacI and XhoI sites into pmirGLO vector (Promega). The 3′-UTR fragments of human CDK6 containing three putative miR-22 binding sites were also amplified from genomic DNA and cloned at the XhoI and SalI sites into pmirGLO vector. The sense and antisense oligonucleotides for the putative miR-22 binding site at the 3′-UTR of potential targets were annealed and cloned at the SacI and XbaI sites into pmirGLO vector. An internal NotI site was added to the oligonucleotides for clone confirmation. A positive control construct contains complete complementary mature miR-22 sequence. PCR primers and oligonucleotide sequences for constructs are provided in Table S2. All the constructs were further confirmed by sequencing.
For luciferase activity analysis, each construct was cotransfected with miRNA duplex in a 96-well plate using DharmFECT Duo transfection reagent (Thermo Fisher Scientific) for 72 h, and luciferase assays were performed with the Dual-Luciferase reporter system (Promega) according to the manufacturer’s instructions. Luminescent signal was quantified by luminometer (Glomax; Promega), and each value from firefly luciferase construct was normalized by Renilla luciferase assay.
Cell motility observation by fluorescence microscopy
Cells were seeded in a 4-well 35-mm dish (Greiner Bio-One) at a density of 1,000 cells/well and grown for 48 h in culture medium. Before recordings were initiated, the multidishes were left for at least 30 min on the microscope stage for temperature equilibration. Time-laspe video recordings of live cells were performed for determination of cell motility. In brief, live cells from several nonoverlapping areas were recorded in 30-min or 1-h intervals over a period of 8–24 h using a CFl Plan Apo 10× objective with a fluorescence microscope (BIOREVO BZ-9000; KEYENCE) equipped with a motorized movable microscope stage. Recordings were stored as 8-bit 680 × 512 pixel images. The microscope stage contained a thermostatically controlled heating element and was surrounded by a Plexiglas incubator, thereby ensuring that live specimens could be maintained at 37°C during recordings.
In vitro invasion assay
48 h after transfection, cells were resuspended in culture medium without serum and seeded at densities of 1.5 × 105 cells/well in 24-well Transwell inserts (8-µm pores; BD) coated with 50 µg Matrigel (BD). The lower chamber was supplemented with a medium with 10% FBS. After 48 h of incubation, the cells on the upper surface were scraped off, whereas the invasive cells attached to the lower surface of the membrane inserts were fixed and stained with hematoxylin. The invading cells were observed and counted from nine images (including at least 2,000 cells) in three fields of three membranes using a 10× phase-contrast objective under light microscopy (U-PMTVC; Olympus) at room temperature.
Tumor imaging in vivo
5-wk-old female C.B17/Icr-scid (Scid/scid) mice (CLEA Japan, Inc.) were inoculated with MDA-MB-231-luc-D3H2LN cells into the fat pad on day 0 as a 1:1 mixture of ECM gel complex and cells at 2 × 106 cells/50 µl/site. The hsa-miR-22 duplex and cont miR with RNA-jetPEI (Polyplus Transfection) complex at the ratio of 1:1 in a volume of 100 µl (20 µg/site) were injected intratumorally every other day from day 13 to 31 after inoculation. The development of subsequent tumor growth and metastasis was monitored once a week by in vivo imaging. In brief, mice were injected with 150 mg/kg D-luciferin (Promega) intraperitoneally and imaged immediately to count the photons from the whole bodies using the IVIS imaging system (Xenogen) according to the manufacturer’s instructions. 10 min later, photons from firefly luciferase were counted. Data were analyzed using LivingImage software (version 2.5; Xenogen).
Immunohistochemistry
Immunohistochemical staining was performed with anti–Ki-67 monoclonal antibody (1:50; Dako) after antigen retrieval by microwave treatment in citrate buffer, pH 6.0, and detection by a streptavidin–biotin peroxidase system using the LSAB kit (Dako). The sections were incubated with primary antibody at 4°C overnight. A labeling index percentage of Ki-67 was determined by examining at least 500 tumor cells at 200× magnification in three representative and intensely stained areas using a 20× objective under light microscopy (U-PMTVC) at room temperature. The expression of Ki-67 was graded as high (>50% of positive cells) and low (<50% of positive cells).
Statistical analysis
Significance of differences between the treated samples and controls was determined by two-tailed t tests using Excel (Microsoft). P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows the opposite effect of miR-22 overexpression and knockdown on apoptosis in human cancer cells. Fig. S2 shows that miR-22 has no effect on the length of total telomere or G-tail in human cancer cells. Fig. S3 shows an examination of BrdU quantity analysis, cell morphology area, and F-actin formation by automated image analysis. Fig. S4 shows the images of cell motility observation in senescent fibroblasts and Lenti-Pre22–infected MDA-D3 cells. Fig. S5 shows a scheme for the role of miR-22–induced senescence in cancer cells. Table S1 shows altered expression miRNAs identified by miRNA microarray function in cell growth and tumorigenesis. Table S2 shows PCR primers and oligonucleotide sequences for each construct of miR-22 putative targets in luciferase reporter assay. Videos 1–4 show cell motility observation of young and senescent MRC-5 cells, Lenti-C, and Lenti-Pre22–infected MDA-D3 cells, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201010100/DC1.
Acknowledgments
We thank Maki Yoshida for her assistance with the invasion assay experiments, Ryo Shioda (PerkinElmer, Japan) for his technical help with automated image acquisition, Sei Kobayashi and Hiroko Kishi (Yamaguchi University) for the provided reagents, and Anno Kumiko (Hiroshima University) and Takayuki Mizutani (IZM, Inc., Japan) for their technical support.
This work was supported by the Grant-in-Aid for Scientific Support Programs for Cancer Research; Grant-in-Aid for Scientific Research on Innovative Areas; and Ministry of Education, Culture, Sports, Science, and Technology of Japan (H. Tahara). This work was partially supported by the Science and Technology Incubation Program in Advanced Regions, Japan Science and Technology Agency, and Takeda Science Foundation (H. Tarara) and the Program for the Promotion of Fundamental Studies in Health Science of the National Institute of Biomedical Innovation (T. Ochiya).
Footnotes
Abbreviations used in this paper:
- cont miR
- control miRNA
- HPA
- hybridization protection assay
- hTERT
- human telomerase reverse transcriptase
- LNA
- locked nucleic acid
- miRNA
- microRNA
- PDL
- population doubling level
- pRb
- retinoblastoma protein
- SA-β-gal
- senescence-associated β-galactosidase
- SAHF
- senescence-associated heterochromatin foci
- SA-miRNA
- senescence-associated miRNA
- UTR
- untranslated region
- WT
- wild type
References
- Adams P.D. 2007. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene. 397:84–93 10.1016/j.gene.2007.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre X., Jiménez-Velasco A., San José-Enériz E., Garate L., Bandrés E., Cordeu L., Aparicio O., Saez B., Navarro G., Vilas-Zornoza A., et al. 2008. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol. Cancer Res. 6:1830–1840 10.1158/1541-7786.MCR-08-0167 [DOI] [PubMed] [Google Scholar]
- Akao Y., Nakagawa Y., Naoe T. 2006. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 29:903–906 10.1248/bpb.29.903 [DOI] [PubMed] [Google Scholar]
- Bar N., Dikstein R. 2010. miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PLoS ONE. 5:e10859 10.1371/journal.pone.0010859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Belguise K., Kersual N., Galtier F., Chalbos D. 2005. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene. 24:1434–1444 10.1038/sj.onc.1208312 [DOI] [PubMed] [Google Scholar]
- Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dörken B., Jenuwein T., Schmitt C.A. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 436:660–665 10.1038/nature03841 [DOI] [PubMed] [Google Scholar]
- Brooks C.L., Gu W. 2009. How does SIRT1 affect metabolism, senescence and cancer? Nat. Rev. Cancer. 9:123–128 10.1038/nrc2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 6:857–866 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C.M. 2004. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 101:2999–3004 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 120:513–522 10.1016/j.cell.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Chen Q.M., Tu V.C., Catania J., Burton M., Toussaint O., Dilley T. 2000. Involvement of Rb family proteins, focal adhesion proteins and protein synthesis in senescent morphogenesis induced by hydrogen peroxide. J. Cell Sci. 113:4087–4097 [DOI] [PubMed] [Google Scholar]
- Chen Y., Song Y., Wang Z., Yue Z., Xu H., Xing C., Liu Z. 2010. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J. Gastrointest. Surg. 14:1170–1179 10.1007/s11605-010-1202-2 [DOI] [PubMed] [Google Scholar]
- Choong M.L., Yang H.H., McNiece I. 2007. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp. Hematol. 35:551–564 10.1016/j.exphem.2006.12.002 [DOI] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguría A., Zaballos A., Flores J.M., Barbacid M., et al. 2005. Tumour biology: senescence in premalignant tumours. Nature. 436:642 10.1038/436642a [DOI] [PubMed] [Google Scholar]
- Debacq-Chainiaux F., Erusalimsky J.D., Campisi J., Toussaint O. 2009. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4:1798–1806 10.1038/nprot.2009.191 [DOI] [PubMed] [Google Scholar]
- Deng Y., Chan S.S., Chang S. 2008. Telomere dysfunction and tumour suppression: the senescence connection. Nat. Rev. Cancer. 8:450–458 10.1038/nrc2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz R., Silva J., García J.M., Lorenzo Y., García V., Peña C., Rodríguez R., Muñoz C., García F., Bonilla F., Domínguez G. 2008. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 47:794–802 10.1002/gcc.20580 [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 92:9363–9367 10.1073/pnas.92.20.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J., Lindow M., Silahtaroglu A., Bak M., Christensen M., Lind-Thomsen A., Hedtjärn M., Hansen J.B., Hansen H.F., Straarup E.M., et al. 2008. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 36:1153–1162 10.1093/nar/gkm1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C., et al. 2007. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA. 104:15805–15810 10.1073/pnas.0707628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju V.K., Lin H. 2009. MicroRNAs: key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 10:116–125 10.1038/nrm2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M., Croce C.M. 2011. microRNAs: Master regulators as potential therapeutics in cancer. Annu. Rev. Pharmacol. Toxicol. 51:25–43 10.1146/annurev-pharmtox-010510-100517 [DOI] [PubMed] [Google Scholar]
- Haferkamp S., Tran S.L., Becker T.M., Scurr L.L., Kefford R.F., Rizos H. 2009. The relative contributions of the p53 and pRb pathways in oncogene-induced melanocyte senescence. Aging (Albany NY). 1:542–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn W.C., Weinberg R.A. 2002. Rules for making human tumor cells. N. Engl. J. Med. 347:1593–1603 10.1056/NEJMra021902 [DOI] [PubMed] [Google Scholar]
- He L., Hannon G.J. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5:522–531 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- Huang J., Gan Q., Han L., Li J., Zhang H., Sun Y., Zhang Z., Tong T. 2008. SIRT1 overexpression antagonizes cellular senescence with activated ERK/S6k1 signaling in human diploid fibroblasts. PLoS ONE. 3:e1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel R., Hussey D.J., Haier J. 2010. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur. J. Cancer. 46:298–311 10.1016/j.ejca.2009.10.027 [DOI] [PubMed] [Google Scholar]
- Ibarra I., Erlich Y., Muthuswamy S.K., Sachidanandam R., Hannon G.J. 2007. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 21:3238–3243 10.1101/gad.1616307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Malizos K.N., Oikonomou P., Tsezou A. 2008. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS ONE. 3:e3740 10.1371/journal.pone.0003740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessel R., Haertel S., Socaciu C., Tykhonova S., Diehl H.A. 2002. Kinetics of apoptotic markers in exogeneously induced apoptosis of EL4 cells. J. Cell. Mol. Med. 6:82–92 10.1111/j.1582-4934.2002.tb00313.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katakowski M., Zheng X., Jiang F., Rogers T., Szalad A., Chopp M. 2010. MiR-146b-5p suppresses EGFR expression and reduces in vitro migration and invasion of glioma. Cancer Invest. 28:1024–1030 10.3109/07357907.2010.512596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahigashi Y., Mishima T., Mizuguchi Y., Arima Y., Yokomuro S., Kanda T., Ishibashi O., Yoshida H., Tajiri T., Takizawa T. 2009. MicroRNA profiling of human intrahepatic cholangiocarcinoma cell lines reveals biliary epithelial cell-specific microRNAs. J. Nippon Med. Sch. 76:188–197 10.1272/jnms.76.188 [DOI] [PubMed] [Google Scholar]
- Kong W., He L., Coppola M., Guo J., Esposito N.N., Coppola D., Cheng J.Q. 2010. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J. Biol. Chem. 285:17869–17879 10.1074/jbc.M110.101055 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W., Chang T.C., Vivekanandan P., Torbenson M., Clark K.R., et al. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 137:1005–1017 10.1016/j.cell.2009.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsodontis G., Tentes I., Papakosta P., Moustakas A., Kardassis D. 2001. Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J. Biol. Chem. 276:29116–29125 10.1074/jbc.M104130200 [DOI] [PubMed] [Google Scholar]
- Lafferty-Whyte K., Cairney C.J., Jamieson N.B., Oien K.A., Keith W.N. 2009. Pathway analysis of senescence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim. Biophys. Acta. 1792:341–352 [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Dutta A. 2007. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 21:1025–1030 10.1101/gad.1540407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Fu H., Tie Y., Hu Z., Kong W., Wu Y., Zheng X. 2009. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 275:44–53 10.1016/j.canlet.2008.09.035 [DOI] [PubMed] [Google Scholar]
- Li X., Liu J., Zhou R., Huang S., Huang S., Chen X.M. 2010. Gene silencing of MIR22 in acute lymphoblastic leukaemia involves histone modifications independent of promoter DNA methylation. Br. J. Haematol. 148:69–79 10.1111/j.1365-2141.2009.07920.x [DOI] [PubMed] [Google Scholar]
- Liu L., Jiang Y., Zhang H., Greenlee A.R., Yu R., Yang Q. 2010. miR-22 functions as a micro-oncogene in transformed human bronchial epithelial cells induced by anti-benzo[a]pyrene-7,8-diol-9,10-epoxide. Toxicol. In Vitro. 24:1168–1175 10.1016/j.tiv.2010.02.016 [DOI] [PubMed] [Google Scholar]
- Liu Q., Fu H., Sun F., Zhang H., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. 2008. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 36:5391–5404 10.1093/nar/gkn522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M., Lowe S.W. 2005. Senescence comes of age. Nat. Med. 11:920–922 10.1038/nm0905-920 [DOI] [PubMed] [Google Scholar]
- Narita M., Nũnez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 113:703–716 10.1016/S0092-8674(03)00401-X [DOI] [PubMed] [Google Scholar]
- Neely L.A., Patel S., Garver J., Gallo M., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J. 2006. A single-molecule method for the quantitation of microRNA gene expression. Nat. Methods. 3:41–46 10.1038/nmeth825 [DOI] [PubMed] [Google Scholar]
- Ohtani N., Mann D.J., Hara E. 2009. Cellular senescence: its role in tumor suppression and aging. Cancer Sci. 100:792–797 10.1111/j.1349-7006.2009.01123.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H., Tokunaga E., Chang K., Hikasa M., Iijima K., Eto M., Kozaki K., Akishita M., Ouchi Y., Kaneki M. 2006. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 25:176–185 [DOI] [PubMed] [Google Scholar]
- Pandey D.P., Picard D. 2009. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol. Cell. Biol. 29:3783–3790 10.1128/MCB.01875-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Pitto L., Simili M., Mariani L., Riccardi L., Ciucci A., Rizzo M., Evangelista M., Mercatanti A., Pandolfi P.P., Rainaldi G. 2008. The proto-oncogene LRF is under post-transcriptional control of MiR-20a: implications for senescence. PLoS ONE. 3:e2542 10.1371/journal.pone.0002542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricarte-Filho J.C., Fuziwara C.S., Yamashita A.S., Rezende E., da-Silva M.J., Kimura E.T. 2009. Effects of let-7 microRNA on cell growth and differentiation of papillary thyroid cancer. Transl Oncol. 2:236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas M., Gregory F., Jones R., Poolman R., Starborg M., Rowe J., Brookes S., Peters G. 2007. CDK4 and CDK6 delay senescence by kinase-dependent and p16INK4a-independent mechanisms. Mol. Cell. Biol. 27:4273–4282 10.1128/MCB.02286-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C.A. 2007. Cellular senescence and cancer treatment. Biochim. Biophys. Acta. 1775:5–20 [DOI] [PubMed] [Google Scholar]
- Solomon J.M., Pasupuleti R., Xu L., McDonagh T., Curtis R., DiStefano P.S., Huber L.J. 2006. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 26:28–38 10.1128/MCB.26.1.28-38.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Fu H., Liu Q., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. 2008. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 582:1564–1568 10.1016/j.febslet.2008.03.057 [DOI] [PubMed] [Google Scholar]
- Tahara H., Kusunoki M., Yamanaka Y., Matsumura S., Ide T. 2005. G-tail telomere HPA: simple measurement of human single-stranded telomeric overhangs. Nat. Methods. 2:829–831 10.1038/nmeth797 [DOI] [PubMed] [Google Scholar]
- Takakura S., Mitsutake N., Nakashima M., Namba H., Saenko V.A., Rogounovitch T.I., Nakazawa Y., Hayashi T., Ohtsuru A., Yamashita S. 2008. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 99:1147–1154 10.1111/j.1349-7006.2008.00800.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita F., Patrawala L., Osaki M., Takahashi R.U., Yamamoto Y., Kosaka N., Kawamata M., Kelnar K., Bader A.G., Brown D., Ochiya T. 2010. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol. Ther. 18:181–187 10.1038/mt.2009.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapias A., Ciudad C.J., Roninson I.B., Noé V. 2008. Regulation of Sp1 by cell cycle related proteins. Cell Cycle. 7:2856–2867 10.4161/cc.7.18.6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. 2007. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. USA. 104:15472–15477 10.1073/pnas.0707351104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting Y., Medina D.J., Strair R.K., Schaar D.G. 2010. Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochem. Biophys. Res. Commun. 394:606–611 10.1016/j.bbrc.2010.03.030 [DOI] [PubMed] [Google Scholar]
- Tryndyak V.P., Ross S.A., Beland F.A., Pogribny I.P. 2009. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol. Carcinog. 48:479–487 10.1002/mc.20484 [DOI] [PubMed] [Google Scholar]
- Visone R., Pallante P., Vecchione A., Cirombella R., Ferracin M., Ferraro A., Volinia S., Coluzzi S., Leone V., Borbone E., et al. 2007. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 26:7590–7595 10.1038/sj.onc.1210564 [DOI] [PubMed] [Google Scholar]
- Wang G., Mao W., Zheng S., Ye J. 2009. Epidermal growth factor receptor-regulated miR-125a-5p—a metastatic inhibitor of lung cancer. FEBS J. 276:5571–5578 10.1111/j.1742-4658.2009.07238.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Du Q., Liang Z. 2010. Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene. 29:4980–4988 10.1038/onc.2010.241 [DOI] [PubMed] [Google Scholar]