The cytoplasmic concentration of G-actin is a critical parameter for determining the extent of stimulus-induced G-actin assembly and cell extension.
Abstract
To understand the intracellular role of G-actin concentration in stimulus-induced actin assembly and lamellipodium extension during cell migration, we developed a novel technique for quantifying spatiotemporal changes in G-actin concentration in live cells, consisting of sequential measurements of fluorescent decay after photoactivation (FDAP) of Dronpa-labeled actin. Cytoplasmic G-actin concentrations decreased by ∼40% immediately after cell stimulation and thereafter the cell area extended. The extent of stimulus-induced G-actin loss and cell extension correlated linearly with G-actin concentration in unstimulated cells, even at concentrations much higher than the critical concentration of actin filaments, indicating that cytoplasmic G-actin concentration is a critical parameter for determining the extent of stimulus-induced G-actin assembly and cell extension. Multipoint FDAP analysis revealed that G-actin concentration in lamellipodia was comparable to that in the cell body. We also assessed the cellular concentrations of free G-actin, profilin- and thymosin-β4–bound G-actin, and free barbed and pointed ends of actin filaments by model fitting of jasplakinolide-induced temporal changes in G-actin concentration.
Introduction
The polymerization of actin filaments from monomeric G-actin subunits is essential for driving cell migration and morphogenesis and is controlled spatiotemporally in cells by a variety of actin regulatory proteins. In motile processes, cells extend F-actin–rich lamellipodia, in which polarized and branched arrays of actin filaments are constructed with fast-growing barbed ends near the plasma membrane and slow-growing pointed ends toward the rear. In response to external signals, the number of free barbed ends increases dramatically at the cell periphery, primarily due to Arp2/3 complex–mediated actin nucleation and branching, and actin monomers in the cytoplasm are assembled onto the free barbed ends newly formed at the tip of the lamellipodium, which generates a force for pushing the membrane forward, resulting in the lamellipodium extension (Pollard and Borisy, 2003; Le Clainche and Carlier, 2008). Actin monomers in the cytoplasm are replenished by disassembly of actin filaments from their pointed ends in the rear of the lamellipodium, which is promoted by actin-depolymerizing factor (ADF)/cofilin (Bamburg et al., 1999; Pollard and Borisy, 2003).
In most living cells, the cytoplasmic G-actin concentration (∼100 µM) is extraordinarily higher than the critical concentration (Cc) of actin filaments (∼0.1 µM) (Pollard et al., 2000). This high G-actin concentration is maintained by F-actin disassembly by ADF/cofilin, and via the prevention of spontaneous G-actin nucleation by G-actin–sequestering proteins, profilin and thymosin-β4, and the blockage of F-actin polymerization by F-actin–capping proteins (Pantaloni and Carlier, 1993; Barkalow et al., 1996; Bamburg et al., 1999; Pollard et al., 2000). Major components of cytoplasmic G-actin pool are free ATP–G-actin, ATP–G-actin in complex with thymosin-β4 and profilin, and ADP–G-actin in complex with ADF/cofilin, among which free ATP–G-actin and profilin-bound ATP–G-actin are polymerizable to the barbed ends of F-actin, whereas thymosin-β4–sequestered ATP–G-actin and ADP–G-actin in complex with ADF/cofilin are nonpolymerizable (Safer and Nachmias, 1994; Kang et al., 1999; Carlier and Pantaloni, 2007). Although the large population of cellular G-actin consists of nonpolymerizable ATP–G-actin in complex with thymosin-β4, this complex is in rapid equilibrium with free ATP–G-actin and acts as a reservoir for supplying free ATP–G-actin when it is consumed for stimulus-induced actin assembly (Safer and Nachmias, 1994; Carlier and Pantaloni, 2007). The roles of G-actin and its regulating proteins in actin filament dynamics have been extensively studied in the cell-free system.
Because the rate of actin polymerization is proportional to the concentration of G-actin in cell-free assays (Pollard, 1986), it is assumed that the cytoplasmic G-actin concentration is a critical parameter for the intracellular rate of actin polymerization. However, some previous studies paid little attention to the role of G-actin concentration in stimulus-induced actin polymerization and lamellipodium extension, probably because the cytoplasmic G-actin concentration is usually much higher than the Cc of actin filaments (Cramer, 1999; Chan et al., 2000). To understand the mechanism regulating actin assembly and disassembly during cell migration and morphogenesis, it is essential to measure the spatiotemporal changes in actin filament dynamics and the ratio of G- to F-actin. Fluorescence speckle microscopy directly showed the spatial difference in kinetics of actin assembly and disassembly in live cells (Watanabe and Mitchison, 2002; Ponti et al., 2004; Iwasa and Mullins, 2007). Previous studies also estimated the ratio of G- to F-actin in lamellipodia by cell staining with DNase-I or by comparing FRAP of GFP-actin before and after cell extraction with detergents (Cramer et al., 2002; Koestler et al., 2009). However, these approaches are unable to measure the time-dependent changes in G- and F-actin concentration in individual live cells, and the development of a reliable technique capable of quantifying spatiotemporal changes in G-actin concentration with high time resolution in live cells is worth pursuing.
We previously assessed the cytoplasmic G-actin content by measuring the fluorescence decay after photoactivation (FDAP) of Dronpa (Dp)-labeled actin (Kiuchi et al., 2007). Dp is a GFP-like protein, the fluorescence of which can be reversibly photoactivated and photobleached (Ando et al., 2004); thus, Dp-actin could be used for sequential measurements of FDAP (s-FDAP), allowing quantitative measurements of time-dependent changes in G-actin concentration in live cells. In this study, we describe a novel approach for measuring time-dependent changes in G-actin concentration in live cells and provide evidence that G-actin concentration is an important parameter in determining the extents of stimulus-induced actin assembly and cell expansion. We also show the spatial distribution of G-actin concentration in live cells by multipoint FDAP analysis. Furthermore, we estimate the cellular concentrations of free G-actin, profilin- and thymosin-β4–bound G-actin, and free barbed and pointed ends by model fitting of jasplakinolide (Jasp)-induced changes in G-actin concentration.
Results
Fluorescence decay after single photoactivation of Dp-actin (single FDAP)
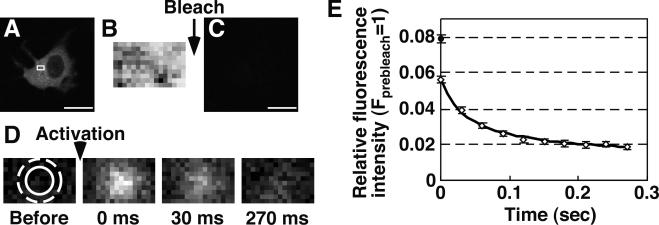
Before s-FDAP analysis to measure temporal changes in G-actin concentration, we examined the optimal conditions of FDAP analysis by measuring fluorescence decay after “single” photoactivation of Dp-actin (single FDAP). Dp-actin, expressed in MCF-7 cells, was photoactivated to the maximum level (Fig. 1 A), and a fluorescence image of a small cytoplasmic region was acquired as a measure of the total amount of Dp-actin in this region (Fig. 1 B). After photobleaching the whole cell to the background level (Fig. 1 C), Dp-actin was locally photoactivated and the fluorescence images were acquired every 30 ms for 270 ms (Fig. 1 D). The relative fluorescence intensities in the photoactivated region were plotted after correcting the values for photomultiplier gain variations and photobleaching (Fig. 1 E and Fig. S1). The normalized fluorescence intensity of Dp-actin at 0 ms was 0.056, whereas the relative fluorescence intensity of immobilized Dp in fixed cells was 0.079 (Fig. 1 E, black circle), indicating that 71% of photoactivated Dp-actin was detected at zero time after photoactivation. The rate of fluorescence decay was much faster than the turnover rate of actin filaments (Theriot and Mitchison, 1991; McGrath et al., 1998; Watanabe and Mitchison, 2002), indicating that the FDAP values measured under these conditions reflect G-actin concentration (but not F-actin disassembly). By fitting the time curve of fluorescence decay (Fig. 1 E), the diffusion coefficient of Dp-actin, the half-life of the mobile fraction, and the mobile fraction were determined to be 13.7 µm2/s, 41 ms, and 79%, respectively. The diffusion coefficient (13.7 µm2/s) was consistent with the reported values for G-actin in the cytoplasm, which ranged from 3 to 30 µm2/s (McGrath et al., 1998; Zicha et al., 2003; McDonald et al., 2006). These results indicate that single FDAP analysis is a useful tool for estimating G-actin concentration in living cells. Based on the half-life of the mobile fraction (41 ms), we set the time point for image acquisition for s-FDAP analysis as 40 ms after photoactivation.
Figure 1.
Fluorescence decay after single photoactivation (single FDAP) of Dp-actin. (A) Fluorescence image of Dp-actin in an MCF-7 cell. (B) Fluorescence image of a 4.8 x 3-µm region (A, white box) after photoactivation of the whole cell to the maximum level. (C) Image after photobleaching. (D) Time-lapse images after photoactivation of a 1.8-µm-diameter area (solid circle) for 3 ms. Images were acquired every 30 ms for 270 ms. (E) Time course of Dp-actin fluorescence decay after single photoactivation. The relative intensity in a 3-µm-diameter area (D, dotted circle) was plotted (white diamond), after correcting (Fig. S1) and normalizing to the maximum intensity in the same area, measured as in B. The diffusion coefficient and mobile fraction of Dp-actin were determined by fitting the time curve of fluorescence decay (black line). The black circle indicates the normalized fluorescence intensity of immobilized Dp after single photoactivation in fixed cells. Data are means ± SEM of 22 cells (Dp-actin) and 25 cells (Dp) from triplicate experiments. Bars, 20 µm.
Time-lapse analysis of G-actin concentration by s-FDAP
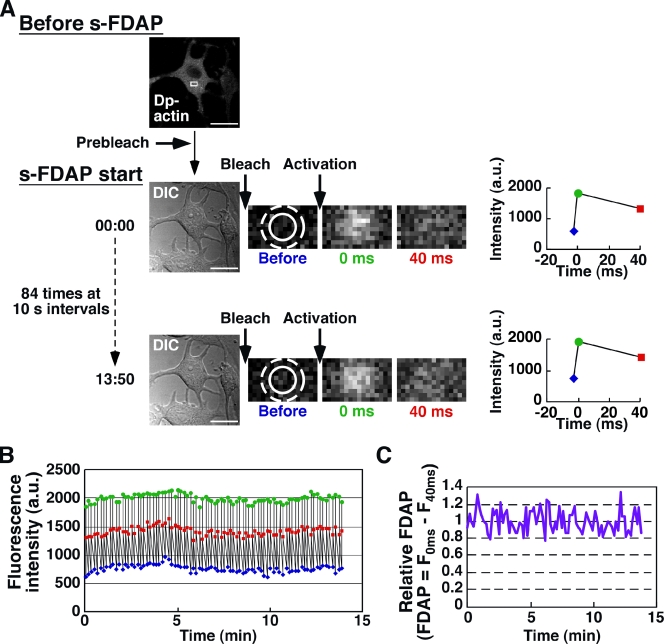
To quantify the time-dependent changes in G-actin concentration, simultaneously with cell morphology, s-FDAP analysis was performed by the following set of procedures: (a) differential interference contrast (DIC) image acquisition; (b) photobleaching of the whole cell; and (c) photoactivation of a small area of the cytoplasm (Fig. 2 A). A set of three fluorescence images of the photoactivated region was acquired before photoactivation and at 0 and 40 ms after photoactivation (Fig. 2 A). Each set of procedure was repeated 84 times at 10-s intervals for 14 min (Fig. 2 B). During s-FDAP analysis, no irreversible photobleaching of Dp-actin was observed (Fig. 2 B). The FDAP value was defined as the difference between the fluorescence intensities at 0 and 40 ms after photoactivation and was normalized to the mean FDAP value during the first 2-min measurements. No significant change was observed in the relative FDAP during the 14-min observation of untreated MCF-7 cells (Fig. 2 C).
Figure 2.
s-FDAP analysis. (A) Procedures of s-FDAP analysis. Dp-actin expressed in MCF-7 cells was prebleached to the background level. s-FDAP analysis consists of DIC image acquisition, photobleaching of the whole cell, photoactivation of the 1.8-µm-diameter region (solid circle), and fluorescence image acquisition of a rectangular region (white box) before and at 0 and 40 ms after photoactivation. The fluorescence intensities in the 3-µm-diameter region (dashed circle) before (blue) and at 0 (green) and 40 ms (red) after photoactivation were measured. Each set of procedures was repeated 84 times at 10-s intervals. Bars, 20 µm. (B) Time course of fluorescence intensities measured as in A. (C) Time course of relative FDAP from the data shown in B. FDAP values are expressed as the difference between fluorescence intensities 0 and 40 ms after photoactivation and are normalized to the mean FDAP value during the first 2-min observation.
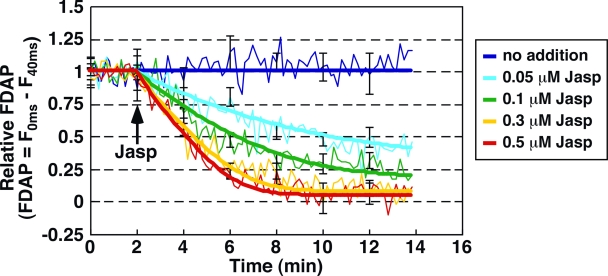
We next used s-FDAP analysis to measure temporal changes in G-actin concentration in cells treated with Jasp, an F-actin–stabilizing drug (Bubb et al., 1994). Jasp decreases G-actin concentration by lowering the rate of F-actin disassembly in cell-free assays (Bubb et al., 1994, 2000). When we analyzed the ratio of G-actin to total actin by F-actin sedimentation assays, Jasp treatment dose-dependently decreased the ratio of G-actin in MCF-7 cells, and Jasp-induced changes in the ratio of Dp–G-actin were comparable to those of endogenous G-actin (Fig. S2), indicating that the changes in Dp–G-actin concentration reflect the changes in endogenous G-actin. Using s-FDAP analysis, we demonstrated that the relative FDAP gradually decreased after Jasp treatment in a time- and dose-dependent manner (Fig. 3, thin lines). Thus, s-FDAP analysis can be used for tracing time-dependent changes in G-actin concentration in live cells.
Figure 3.
s-FDAP analysis of Jasp-induced changes in G-actin concentration. MCF-7 cells were transfected with Dp-actin. The indicated concentrations of Jasp were added 2 min after the start of s-FDAP analysis. Time courses of relative FDAP (thin lines) and the best-fit curves by model fitting (bold lines) are shown. Data are means ± SEM of 13–21 cells from at least three independent experiments.
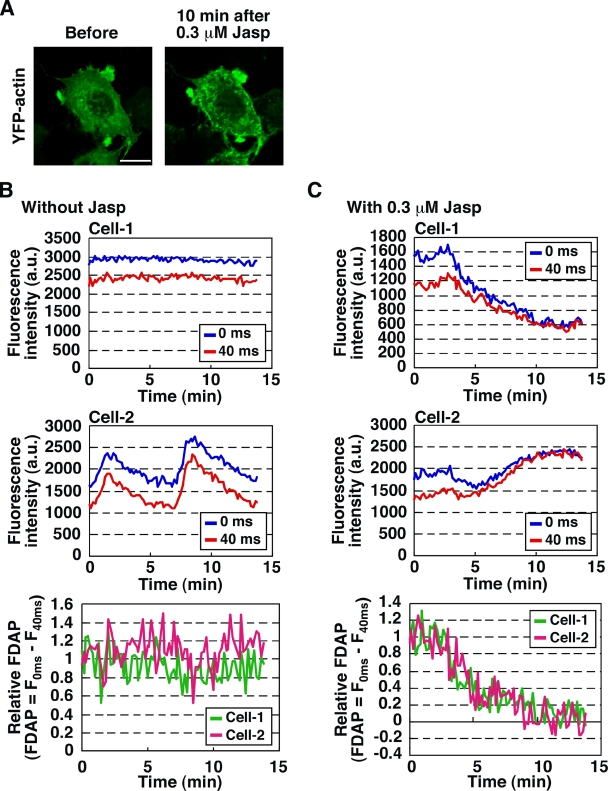
The total fluorescence intensity of Dp-actin in the region of interest (ROI) occasionally fluctuated during s-FDAP analysis, due to the local accumulation or dilution of actin in the cells, particularly in the cells treated with Jasp (Fig. 4 A). However, these variations in Dp-actin fluorescence had no obvious effect on s-FDAP analysis because the FDAP value was given as the difference in fluorescence intensities (Fig. 4, B and C). Thus, s-FDAP analysis can quantitatively measure time-dependent changes in G-actin concentration in live cells, irrespective of local accumulation or dilution of actin.
Figure 4.
Effect of local accumulation or dilution of actin on s-FDAP analysis. (A) Images of YFP-actin before and after Jasp treatment. MCF-7 cells expressing YFP-actin were treated with 0.3 µM Jasp for 10 min. Bar, 20 µm. (B and C) Effects of local accumulation or dilution of actin on s-FDAP analysis. Time courses of Dp-actin fluorescence intensities at 0 (blue) and 40 ms (red) after photoactivation during s-FDAP analysis are shown for two distinct cells (Cell-1 and -2) treated with 0.3 µM Jasp (C) or untreated (B). Time courses of the relative FDAP from Cell-1 (green) and Cell-2 (magenta) are shown in the bottom panels.
Estimation of cellular concentrations of free G-actin, profilin- and thymosin-β4–bound G-actin, and free barbed and pointed ends of actin filaments
We analyzed the data shown in Fig. 3 by model fitting to estimate the cellular concentrations of free G-actin, profilin- and thymosin-β4–bound G-actin, and free barbed and pointed ends of actin filaments. Time-dependent and Jasp dose-dependent changes in relative FDAP with high time resolution (at 10-s intervals) were modeled satisfactorily by assuming that Jasp inhibits actin disassembly at free pointed ends, and that the decline in the relative FDAP was caused by the assembly of free and profilin-bound G-actin onto free barbed ends (Fig. 3, bold lines). Assuming that the total concentrations of G-actin, profilin, and thymosin-β4 are 100, 10, and 200 µM (Mogilner and Edelstein-Keshet, 2002), respectively, the cellular concentrations of free G-actin, profilin- and thymosin-β4–bound G-actin, and free barbed and pointed ends of actin filaments under unstimulated conditions were estimated as 1.98 µM, 9.52 µM, 88.5 µM, 4.8 nM, and 5.5 nM, respectively, from the best-fit of the time curve of relative FDAP (see Materials and methods). Thus, the high time resolution and quantitative features of s-FDAP analysis allowed us to estimate the cellular concentrations of key components involved in the regulation of actin dynamics by model analysis.
Stimulus-induced changes in G-actin concentration and cell area
Cells often induce actin assembly in response to external signals. Previous studies have shown that cytoplasmic G-actin decreases after cell stimulation (Southwick and Young, 1990; Edmonds et al., 1996); however, to our knowledge, no report has quantified the stimulus-induced temporal changes in G-actin concentration with simultaneous analysis of cell morphology in individual live cells. Using s-FDAP, we analyzed temporal changes in G-actin concentration and morphology in MCF-7 cells after neuregulin (NRG) stimulation. Morphological changes were analyzed by measuring the total cell area from DIC images acquired during s-FDAP analysis. The relative FDAP of Dp-actin decreased by 41% at 2–4 min after NRG stimulation and then reached a plateau (Fig. 5, A and C), indicating that 41% of G-actin in the cytoplasm polymerized to F-actin in response to NRG. In contrast, no significant change was observed in the relative FDAP of control Dp after NRG stimulation (Fig. 5, B and C). DIC image analyses revealed that the relative cell area increased ∼1.6-fold at 6–8 min after NRG stimulation and then reached a plateau (Fig. 5, A and C). Interestingly, the time course of the increase in cell area was delayed, with a lag time of ∼2–4 min after the decrease in relative FDAP (Fig. 5 A). Time-lapse imaging also showed that YFP-actin accumulated at the cell periphery 2–3 min after cell stimulation, and then the lamellipodium began to extend (Fig. 5 D and Video 1). These results suggest that cytoplasmic G-actin is primarily used for F-actin assembly at the cell periphery in the initial phase after NRG stimulation, and thereafter the lamellipodium begins to extend.
Figure 5.
NRG-induced changes in G-actin concentration and cell area. (A and B) MCF-7 cells expressing Dp-actin (A) or Dp (B) were subjected to s-FDAP analysis. Cells were left untreated or were treated with NRG 2 min after the start of the measurements. Temporal changes in relative FDAP in NRG-treated (green) and untreated (blue) cells and changes in relative cell area in NRG-treated (red) and untreated (yellow) cells are plotted. Data are means ± SEM of 26–28 cells (Dp-actin) or 14–15 cells (Dp) from at least three independent experiments. (C) Quantitative analysis of the relative FDAP between 4 and 6 min (top) and the increase in relative cell area between 10 and 12 min (bottom) after the start of s-FDAP measurement. *, P < 0.05. (D) Time-lapse imaging of YFP-actin in an NRG-stimulated MCF-7 cell. Time is indicated as min:sec. Arrows indicate the positions of F-actin assembly (2:30) and lamellipodium extension (5:30). Bar, 20 µm. See Video 1.
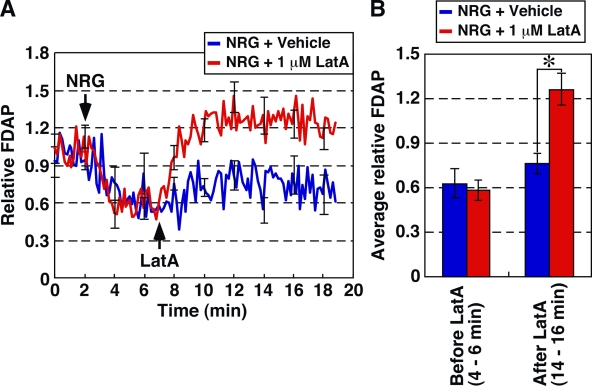
Using s-FDAP analysis, we also analyzed the temporal changes in G-actin concentration in cells treated with latrunculin A (LatA), a drug that induces actin filament disassembly by sequestering G-actin. When MCF-7 cells prestimulated with NRG for 5 min were treated with 1 µM LatA, the relative FDAP of Dp-actin significantly increased to the level over the FDAP value before NRG stimulation at 2–3 min after LatA treatment and then reached a plateau (Fig. 6). Thus, s-FDAP analysis can detect LatA-induced increase in G-actin concentration and revealed that actin filaments polymerized into the lamellipodia by NRG stimulation were disassembled within a few minutes after LatA treatment.
Figure 6.
s-FDAP analysis of LatA-induced increase in G-actin concentration. (A) s-FDAP analysis. MCF-7 cells expressing Dp-actin were treated with NRG 2 min after the start of s-FDAP analysis. At 7 min, cells were treated with 1 µM LatA (red) or DMSO (blue). FDAP values were normalized to the mean value during the first 2-min observation. (B) Quantification of the relative FDAP before and after LatA treatment in NRG-treated cells. Data are means ± SEM of 8 cells (vehicle) and 11 cells (LatA) from at least three independent experiments. *, P < 0.05.
Stimulus-induced G-actin loss and cell extension correlate with G-actin concentration in prestimulated cells
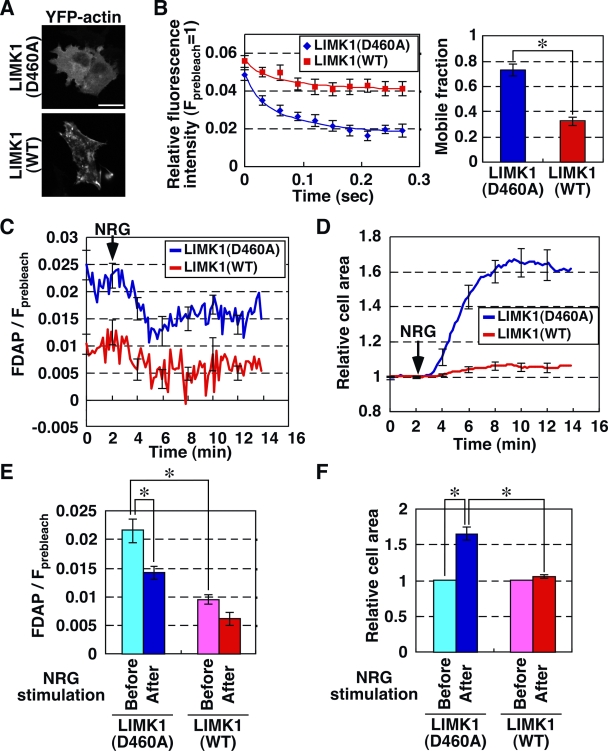
Cofilin inactivation suppresses stimulus-induced actin assembly and lamellipodium extension (Chan et al., 2000; Kiuchi et al., 2007). Therefore, we analyzed the effects of LIM kinase 1 (LIMK1), which phosphorylates and inactivates cofilin (Yang et al., 1998), on NRG-induced changes in G-actin concentration and cell extension. Expression of wild-type (WT) LIMK1, but not its kinase-dead LIMK1(D460A), induced F-actin accumulation (Fig. 7 A). The single FDAP analysis showed that expression of LIMK1(WT) reduced the rate of fluorescence decay of Dp-actin, compared with expression of control LIMK1(D460A) (Fig. 7 B, left). The mobile fractions of Dp-actin in LIMK1(WT)- and control LIMK1(D460A)-expressing cells were estimated as 33 and 73%, respectively, indicating that the mobile fraction of Dp-actin in LIMK1-expressing cells decreased by 55%, before NRG stimulation (Fig. 7 B, right). The s-FDAP analyses showed that the NRG-induced decrease in relative FDAP and the increase in cell area were suppressed in LIMK1(WT)-expressing cells (Fig. 7, C–F), indicating that cofilin is required for NRG-induced actin assembly and cell extension. The G-actin concentration was still much higher than the Cc of actin filaments in LIMK1-expressing cells, and microinjection of G-actin into the cytoplasm of LIMK1-expressing cells resulted in the recovery of NRG-induced actin assembly and lamellipodium extension (Kiuchi et al., 2007). We therefore postulated that the extents of stimulus-induced actin assembly and cell extension correlate with the cytoplasmic G-actin concentration in the prestimulated cell.
Figure 7.
Effects of LIMK1 on NRG-induced G-actin loss and cell extension. (A) Images of YFP-actin in MCF-7 cells expressing CFP-LIMK1(WT) or -LIMK1(D460A). Bar, 20 µm. (B) Effect of LIMK1 on the mobile fraction of Dp-actin. MCF-7 cells cotransfected with Dp-actin and mDsRed-LIMK1(WT) (red) or -LIMK1(D460A) (blue) were subjected to single FDAP analysis. Time curves of fluorescence decay were fitted using the diffusion coefficient of Dp-actin (13.7 µm2/s; thin lines). Data are means ± SEM of 15 cells (LIMK1(WT)) and 16 cells (LIMK1(D460A)) from at least three independent experiments. Right panel shows the mobile fractions. *, P < 0.05. (C) Effect of LIMK1 on NRG-induced G-actin loss. MCF-7 cells coexpressing Dp-actin and mDsRed-LIMK1(D460A) or -LIMK1(WT) were treated with NRG and analyzed by s-FDAP. FDAP values were normalized to the total intensity of Dp-actin in the same area as in Fig. 1 E. (D) Effect of LIMK1 on NRG-induced cell area extension. Cell area was measured using DIC imaging. (E) Quantification of the relative FDAP before (0–2 min) and after (4–6 min) NRG stimulation, calculated from data in C. *, P < 0.05. (F) Quantification of the increase in the relative cell area before (0–2 min) and after (10–12 min) NRG stimulation, calculated from data in D. Data are means ± SEM of 16 cells (LIMK1(D460)) and 15 cells (LIMK1(WT)) from at least three independent experiments. *, P < 0.05.
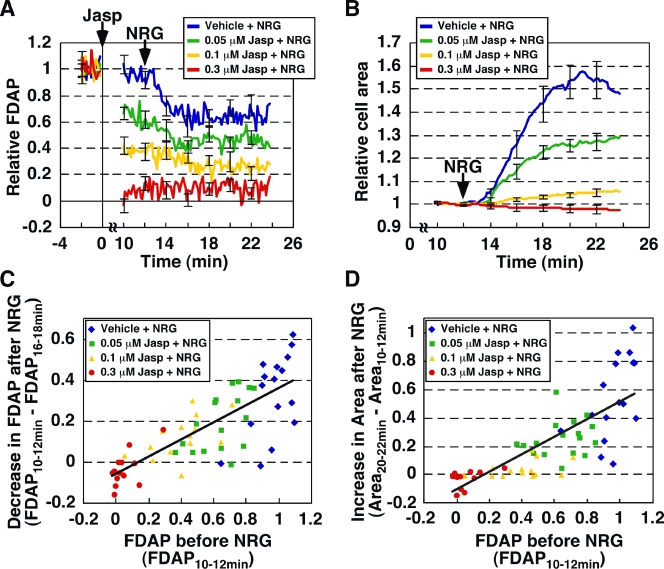
To address this issue quantitatively, we analyzed, using s-FDAP, the effects of Jasp-mediated reduction in G-actin concentration in prestimulated cells on NRG-induced actin assembly and cell size extension (Fig. 8, A and B). MCF-7 cells were pretreated with various concentrations of Jasp and were then treated with NRG. The s-FDAP analysis revealed that Jasp pretreatment decreased the relative FDAP (i.e., the G-actin concentration) before NRG stimulation in a dose-dependent manner, and showed that the relative FDAP decreased further after NRG treatment (Fig. 8 A). The decrease in FDAP after NRG treatment showed a good correlation with the relative FDAP before NRG treatment (Fig. 8 C, correlation coefficient = 0.78), which indicated that the decrease in G-actin (i.e., the amount of G-actin assembled to F-actin) after NRG stimulation strongly depends on the G-actin concentration in the prestimulated cell. Jasp pretreatment also suppressed NRG-induced cell area expansion dose-dependently (Fig. 8 B), and the increase in cell area after NRG stimulation exhibited a strong correlation with the relative FDAP in the prestimulated cell (Fig. 8 D, correlation coefficient = 0.79). Such significant correlations over a wide range of G-actin concentrations indicate that NRG-induced actin assembly and cell extension strongly depend on the G-actin concentration before cell stimulation, even though the G-actin concentration is in a range much higher than the Cc of actin filaments.
Figure 8.
NRG-induced G-actin loss and cell extension correlate with G-actin concentration in prestimulated cells. (A) Effect of Jasp pretreatment on NRG-induced loss of G-actin. MCF-7 cells expressing Dp-actin were treated with Jasp and then with NRG. Changes in G-actin concentration were measured by s-FDAP. FDAP values were normalized to the mean value at −2 to 0 min. (B) Effect of Jasp on NRG-induced cell area extension. Cell area was measured by DIC imaging and normalized to the mean value at 10–12 min. Data are means ± SEM of 16 cells (vehicle), 17 cells (0.05 µM Jasp), 15 cells (0.1 µM Jasp), and 13 cells (0.3 µM Jasp) from at least three independent experiments. (C) Correlation between G-actin concentration in prestimulated cells (measured by relative FDAP at 10–12 min) and NRG-induced loss of G-actin (calculated as the difference between relative FDAP at 10–12 min and 16–18 min). Data are from A. (D) Correlation between G-actin concentration in prestimulated cells (measured as above) and NRG-induced cell area extension (calculated as the difference between relative cell areas at 20–22 min and 10–12 min). Data are from A and B.
Role of G-actin concentration in stimulus-induced free barbed end formation
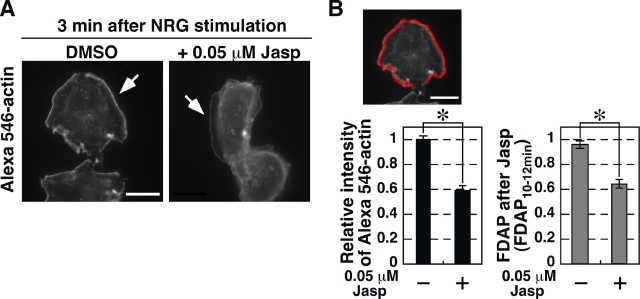
The number of free barbed ends increases dramatically at the cell periphery immediately after cell stimulation (Chan et al., 2000; Kiuchi et al., 2007), indicating that stimulus-induced actin assembly and lamellipodium extension depend on the generation of free barbed ends. To examine the role of G-actin concentration in stimulus-induced free barbed end formation, we analyzed the effect of Jasp pretreatment on the NRG-induced increase in free barbed ends. The number of free barbed ends was assessed by measuring the fluorescence intensity of Alexa 546–labeled G-actin incorporated into the periphery of saponin-permeabilized cells (Kiuchi et al., 2007). When MCF-7 cells were pretreated with 0.05 µM Jasp and then stimulated with NRG, the amount of free barbed ends formed in the cell periphery was decreased by 40% in Jasp-pretreated cells, compared with untreated cells (Fig. 9, A and B). The reduction in free barbed ends was comparable to the reduction in the relative FDAP after treatment with the same concentration of Jasp (Fig. 8 A and Fig. 9 B), suggesting that the G-actin concentration in the prestimulated cell is critical for the generation of stimulus-induced free barbed ends.
Figure 9.
Effect of Jasp treatment on NRG-induced free barbed end formation. (A) Images of Alexa 546–actin. MCF-7 cells were treated with 0.05 µM Jasp or DMSO for 10 min and then with NRG for 3 min. Free barbed end formation was visualized by Alexa 546–actin incorporation into the cell periphery (arrows). Bar, 20 µm. (B) Quantification of free barbed ends. The mean fluorescence intensity of Alexa 546–actin in a region 1 µm from the cell edge (red region) of Jasp-treated cells was compared with that of control cells (left). Right panel shows the relative FDAP of 0.05 µM Jasp-treated or untreated cells, obtained from Fig. 8 A. Data are means ± SEM of 26–38 cells (Alexa 546–actin) and 16–17 cells (FDAP). *, P < 0.05.
Spatial distribution of G-actin concentration measured by multipoint FDAP analysis
The s-FDAP analysis is useful for measuring temporal changes in G-actin concentration in a single ROI of a living cell. We next examined whether the FDAP technique is applicable to measure spatial distribution of G-actin concentration by sequentially performing single FDAP analysis at multiple points of a living cell. In motile cells, actin filaments are highly enriched in the lamellipodium, where actin assembly takes place preferentially at the tip of the lamellipodium and disassembly in the middle to the rear (Watanabe and Mitchison, 2002; Ponti et al., 2004). However, whether the spatial difference in actin assembly and disassembly sites would create the spatial gradient of G-actin concentration in cells and whether G-actin concentration would differ within the lamellipodium and between the lamellipodium and the cell body remain to be elucidated.
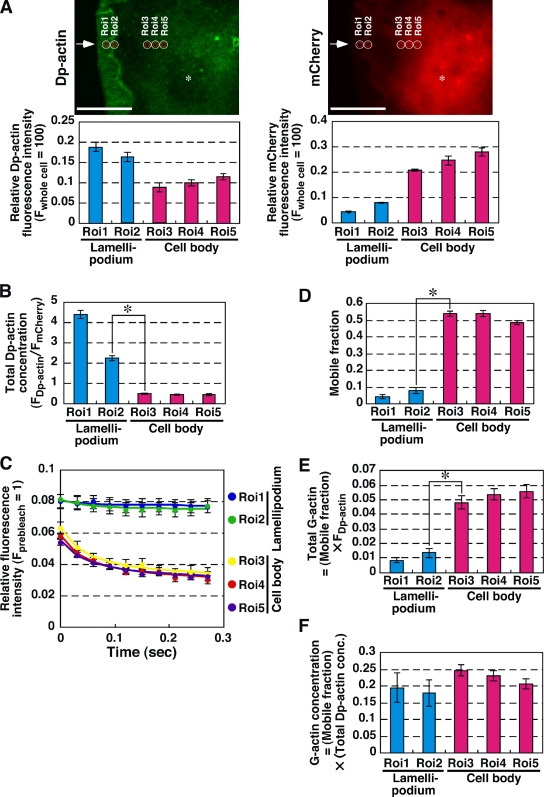
To address these issues, we analyzed the distribution of G-actin concentration in five different ROIs (ROI-1 and -2 in the middle and the rear of the lamellipodium and ROI-3 to -5 in the cell body) by multipoint single FDAP analysis (Fig. 10 A). We used N1E-115 cells expressing active RacV12 because they extended the stable lamellipodia constitutively and did not locomote. N1E-115 cells were cotransfected with RacV12, Dp-actin, and mCherry, and single FDAP analysis was performed sequentially at five ROIs. The relative fluorescence intensity of Dp-actin and mCherry in each ROI was measured to estimate the total amount of Dp-actin (including F- and G-actin) and the volume in the photoactivation area, respectively (Fig. 10 A, bottom panels). The total Dp-actin concentration in each ROI was estimated by dividing the fluorescence intensity of Dp-actin by that of mCherry (Fig. 10 B). This indicates that total actin concentration is significantly high in the lamellipodium. Multipoint FDAP analysis revealed that the rate of fluorescence decay of Dp-actin was very slow in the lamellipodium (ROI-1 and -2) compared with that in the cell body (ROI-3 to -5; Fig. 10 C). The values of mobile fraction in ROI-1 and -2 were 4.5 and 8.1%, respectively, whereas those in ROI-3 to -5 were 54, 54, and 49%, respectively (Fig. 10 D). We also measured the fluorescence decay of Dp-actin in the tip of the lamellipodium, but the decay was very little and insufficient to determine the mobile fraction in this region (not depicted). The total G-actin content (the amount of total Dp-actin multiplied by the mobile fraction) in each ROI was estimated as 0.0084 and 0.013 in ROI-1 and -2, and 0.048, 0.054, and 0.056 in ROI-3 to -5, respectively (Fig. 10 E), which indicates that a large population of G-actin is localized in the cell body. Finally, the G-actin concentration (the total Dp-actin concentration multiplied by the mobile fraction) was estimated as 0.2 and 0.18 in ROI-1 and -2 in the lamellipodium, and 0.25, 0.23, and 0.21 in ROI-3 to -5 in the cell body, respectively (Fig. 10 F). Thus, no significant difference was observed between G-actin concentrations in the middle and the rear of the lamellipodium and those in the cell body. The high concentration of G-actin in the lamellipodium probably contributes to the effective actin polymerization at the tip of the lamellipodium.
Figure 10.
Multipoint FDAP analysis showing spatial distribution of G-actin concentration in lamellipodium and cell body. (A) Fluorescence images of Dp-actin (left) and mCherry (right) in Rac(V12)-expressing N1E-115 cells. Positions of ROIs (ROI-1 and -2 in the middle and rear of the lamellipodium and ROI-3 to -5 in the cell body) are indicated as white circles. Red circles indicate the areas of photoactivation. Relative fluorescence intensities of Dp-actin and mCherry in each ROI are shown on the bottom. Arrows and asterisks indicate the tip of the lamellipodium and the nucleus, respectively. Bar, 20 µm. (B) Total Dp-actin concentration, calculated from the fluorescence intensities of Dp-actin and mCherry. *, P < 0.05. (C) Multipoint single FDAP analysis. Time courses of Dp-actin fluorescence decay after single photoactivation were measured in each ROI. Fluorescence intensity was normalized to total Dp-actin fluorescence intensity in each ROI and fitted using the diffusion coefficient of Dp-actin (13.7 µm2/s). Data are means ± SEM of 28 cells from at least three independent experiments. (D) Mobile fraction of Dp-actin in each ROI, calculated from the time curve of the mean fluorescence intensity of Dp-actin in C. Error bars are standard errors of mobile fraction. *, P < 0.05. (E) The population of G-actin in each ROI, calculated by the total Dp-actin fluorescence intensity and the mobile fraction. Error bars were calculated by the law of propagation of errors, using the error bars in A and D. *, P < 0.05. (F) The concentration of G-actin in each ROI, calculated by the total Dp-actin concentration and the mobile fraction. Error bars were calculated by the law of propagation of errors, using the error bars in B and D.
Discussion
The spatiotemporal regulation of actin assembly and disassembly is a basic mechanism for driving cell migration and morphogenesis. This study provides new microscopic techniques, s-FDAP and multipoint FDAP, capable of measuring temporal and spatial changes in G-actin concentration in live cells. Using these techniques, we demonstrated that: (a) G-actin concentration in the cytoplasm was reduced by 41% after NRG stimulation and thereafter the cell area extended; (b) The extents of stimulus-induced G-actin loss and cell extension were linearly correlated with the G-actin concentration before stimulation over a wide range of G-actin concentrations; and (c) G-actin concentration in the lamellipodium was comparable to that in the cell body. These findings suggest that the cytoplasmic G-actin concentration is a key parameter for determining the extents of stimulus-induced actin polymerization and cell extension and that the concentration of G-actin in the lamellipodium is sufficiently high to support actin polymerization at the tip of the lamellipodium.
Spatiotemporal changes in G-actin concentration measured by s-FDAP and multipoint FDAP analysis
In single FDAP and multipoint FDAP analysis, we measured the FDAP of Dp-actin with high frame rate (4.8 ms/frame) and short observation time (0.27 s) to minimize the effect of F-actin disassembly on FDAP analysis. We previously showed that the length of lamellipodia was ∼7 µm and the rate of actin retrograde flow was 6.9 µm/min in N1E-115 cells (Kiuchi et al., 2007), indicating that the turnover time of F-actin and the rate of F-actin disassembly in lamellipodia are 61 s and 1.6%/s, respectively. Therefore, even if all actin molecules are F-actin in the photoactivated area, the ratio of G-actin disassembled from F-actin during single FDAP analysis for 0.27 s will be only 0.43% of total actin. This value is much lower than the mobile fraction in the cell body (54%) or in the rear of lamellipodium (8.1%; Fig. 10 D). This strongly suggests that the fluorescence decay of Dp-actin observed in this study is mostly caused by diffusion of G-actin in the photoactivated area, and F-actin disassembly contributes, if any, only faintly to FDAP analysis.
Previous studies showed the biochemical, cell-staining, and microscopic methods for estimating the intracellular G-actin concentration (Southwick and Young, 1990; Edmonds et al., 1996; McGrath et al., 1998; Cramer et al., 2002; Hotulainen et al., 2005; Koestler et al., 2009). Compared with these methods, s-FDAP analysis has advantages in that it can measure temporal changes in G-actin concentration in live cells quantitatively and repeatedly (84 iterations every 10 s), and it can also measure changes in individual cell morphology simultaneously. Because FDAP values in s-FDAP are given as the difference in fluorescence intensities, variations in total Dp-actin fluorescence in ROI does not overtly affect s-FDAP analysis. Additionally, because photoactivation is performed only briefly in a small region, the vast majority of Dp-actin in the cell is in the nonfluorescent state (with little absorbance at 488 nm) (Ando et al., 2004) during photobleaching; thus, the laser-induced photodamage and irreversible photobleaching of Dp-actin are minimized in s-FDAP analysis.
The high time resolution of s-FDAP analysis is sufficient to use the data of Jasp-induced changes in G-actin concentration for model analysis. In the best-fit model, cellular concentrations of free barbed and pointed ends were estimated as 4.8 and 5.5 nM, respectively, the values being much lower than the cellular concentration of actin filaments (Cano et al., 1991). Because the cellular concentrations of capping protein and Arp2/3 complex are in the micromolar range and their affinities to barbed and pointed ends are in the nanomolar range (Schafer et al., 1996; Mullins et al., 1998), it is predicted that most of the barbed and pointed ends are capped in resting cells. The free barbed and pointed end concentrations estimated from our model analysis were consistent with this prediction. Thus, the data on temporal changes in G-actin concentration by s-FDAP analysis are useful for model analysis for estimating the cellular concentrations of key components involved in the regulation of actin filament dynamics, including free G-actin, profilin- and thymosin-β4–bound G-actin, and free barbed and pointed ends, and will provide basic parameters for understanding actin filament dynamics during cell migration and morphogenesis.
Role of G-actin concentration in stimulus-induced actin assembly and cell extension
Using s-FDAP analysis, we showed the temporal changes in G-actin concentration after NRG stimulation and the linear correlation of the extents of NRG-induced G-actin loss and cell extension with the G-actin concentration before NRG stimulation. The cytoplasmic G-actin concentration decreased by 41% after NRG stimulation and then the cell extended. Time-lapse imaging of YFP-actin also visualized NRG-induced F-actin accumulation in the cell margin before the extension of lamellipodium. These results suggest that, in the initial phase after NRG stimulation, cytoplasmic G-actin is used primarily for F-actin assembly at the cell margin, which does not bring about cell extension directly, and thereafter lamellipodium extension begins probably by the mechanism depending on the extent of the initial assembly of F-actin.
Previous studies showed that the free barbed ends dramatically increased in the cell edge immediately after cell stimulation (Chan et al., 2000; Kiuchi et al., 2007). Arp2/3 complex–mediated nucleation and branching of actin filaments appear to play a major role in stimulus-induced free barbed end formation (Sun et al., 2007). Arp2/3 complex is activated by the WASP/WAVE family of proteins and preexisting actin filaments (Pantaloni et al., 2000; Amann and Pollard, 2001; Takenawa and Miki, 2001). In the dendritic nucleation model, Arp2/3 complex binds to preexisting actin filaments and promotes the growth of a new branch, which in turn becomes a new binding site for Arp2/3 complex, resulting in the formation of dendritic actin filaments (Pollard and Borisy, 2003). Because each branch grows until it is capped, the number of free barbed ends generated after cell stimulation depends on the growth rate of actin filaments. In addition to the Arp2/3 complex–mediated barbed end formation, the high concentration of free G-actin can also promote spontaneous actin nucleation for polymerization (Carlier and Pantaloni, 2007). We showed that stimulus-induced loss of G-actin (due to assembly to F-actin) and free barbed end generation strongly depend on the G-actin concentration in prestimulated cells. Additionally, the time course of G-actin loss is similar to that of free barbed end formation (Kiuchi et al., 2007). These results suggest that cytoplasmic G-actin concentration is a critical parameter in determining the rate of actin assembly and the level of free barbed end generation, and that cytoplasmic G-actin is used principally for Arp2/3 complex–assisted barbed end formation and spontaneous actin nucleation for actin polymerization in the initial phase after cell stimulation. Although the major component of cellular G-actin is the nonpolymerizable ATP–G-actin in complex with thymosin-β4, this complex is in rapid equilibrium with free ATP–G-actin and can release it when it is consumed (Carlier and Pantaloni, 2007).
Multipoint FDAP analysis showed that G-actin concentration in the middle and rear of lamellipodia is comparable to that in the cell body. Given that G-actin is continuously released from the middle and rear of lamellipodia, it is presumable that the amount of G-actin disassembled from lamellipodia is much lower than the amount of G-actin preexisting in this region. To evaluate the effect of local production of G-actin (by F-actin disassembly) on the spatial distribution of G-actin concentration, we estimated the gradient of G-actin concentration between ROI-2 and the surrounding region and compared it with G-actin concentration in ROI-2. Assuming that ROI-2 is a cylinder with a radius of r and a height of h, the rate of increase in the amount of G-actin in ROI-2 (Vin) is estimated by the following equation:
where Ctotal, Fimmobile, vturnover, and πr2h are the total concentration of Dp-actin, the immobile fraction, the F-actin turnover rate, and the volume of the cylinder, respectively. On the other hand, G-actin produced in ROI-2 immediately diffuses to the surrounding space from the side surface of the cylinder in ROI-2. According to Fick’s first law of diffusion, the rate of decrease in the amount of G-actin (Vout) can be represented by the following equation:
where D, ∇c, and 2πrh are the diffusion coefficient of G-actin, the gradient of G-actin concentration between ROI-2 and the surrounding space, and the area of the side surface of the cylinder, respectively. Because RacV12-expressing N1E-115 cells extend stable lamellipodia constitutively, G-actin concentration in ROI-2 should be constant, with the rate of increase in the amount of G-actin (Vin) being equal to the rate of decrease (Vout) in ROI-2. Thus, the gradient of G-actin concentration between ROI-2 and the surrounding space is expressed by the following equation:
∇c was determined as 0.0018 using the values: Ctotal = 2.22, Fimmobile = 0.92, vturnover = 0.016 /s, D = 13.7 µm2/s, and r = 1.5 µm. Thus, the gradient corresponds to only 1% of G-actin concentration in ROI-2 (0.18). This estimation strongly suggests that G-actin disassembled in the rear of lamellipodia contributes little to the gradient formation of G-actin concentration.
To estimate the diffusion of G-actin between the lamellipodium and the cell body, we plotted time evolution of G-actin diffusion on a 2D plane. G-actin diffused from a position (r = 0) spread quickly (Fig. S3 A). The leading edge of G-actin diffusion reaches a distance of 9 µm until after 0.3 s (Fig. S3 B), which corresponds to the distance between ROI-2 and ROI-3 in Fig. 10 A. The root mean square diffusion length of G-actin in one second on a 2D plane was calculated as 7.4 µm, using the diffusion coefficient of Dp-actin (13.7 µm2/s). Consistent with these estimations, time-lapse imaging showed that G-actin disassembled from the rear of lamellipodia rapidly diffused into the cytoplasm, and G-actin in the cytoplasm was effectively incorporated into F-actin at the tip of lamellipodia (Kiuchi et al., 2007). Taking into account these results, rapid diffusion of a large amount of cytoplasmic G-actin, which is much greater than the amount of G-actin generated by F-actin disassembly, equalizes G-actin concentrations in the lamellipodium and in the cell body.
Recently, G-actin concentration in lamellipodia was estimated as 150 µM by combination of FRAP and electron microscopy experiments (Koestler et al., 2009). They concluded that G-actin concentration in lamellipodia is extremely higher than the Cc of actin filaments. We have shown that G-actin concentration in lamellipodia was comparable to that in the cytoplasm. We also previously showed that actin polymerization rate at the tip of lamellipodia was linearly correlated with the amount of G-actin in the cytoplasm (Kiuchi et al., 2007). These results suggest that G-actin concentration in lamellipodia is sufficiently high to induce F-actin assembly at the leading edge, and the rate of actin polymerization depends on G-actin concentration in lamellipodia (and also in the cytoplasm). Thymosin-β4–bound ATP-G-actin is nonpolymerizable, but this complex can release free ATP-G-actin when it is decreased; thus, the higher the concentration of total G-actin, the higher the concentration of free ATP–G-actin (Carlier and Pantaloni, 2007). These results indicate that total G-actin concentration is a critical parameter for determining the extent of actin polymerization.
We have developed techniques involving s-FDAP and multipoint FDAP that are capable of quantifying spatiotemporal changes in G-actin concentration and cell shape in individual live cells. We have provided clear evidence that the levels of stimulus-induced actin assembly and cell extension correlate strongly with the G-actin concentration in prestimulated cells, despite the fact that the cellular G-actin concentration is much greater than the Cc of actin filaments. This observation indicates that the status of the cytoplasmic G-actin concentration is one of the key elements for determining the level of actin polymerization and is critically involved in the mechanisms underlying a variety of cell phenomena driven by actin assembly. s-FDAP and multipoint FDAP analyses can be applied to measure spatiotemporal changes in the concentrations of mobile fractions of various cytoskeletal and membrane-bound proteins in live cells and will shed light on their involvements in diverse cell activities, including cell migration, division, morphogenesis, and polarity formation.
Materials and methods
Reagents and plasmids
Jasp and LatA were purchased from Invitrogen. NRG was purchased from R&D Systems. Plasmids for Dp- and YFP-tagged actin and CFP- and mDsRed-tagged LIMK1 and its kinase-negative mutant (D460A) were constructed as described previously (Kiuchi et al., 2007).
Cell culture and transfection
MCF-7 cells were cultured in DME supplemented with 10% FCS. Cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen) and cultured for 48 h before FDAP analysis. Serum-starved cells were treated with 50 ng/ml NRG.
Fluorescence decay after single photoactivation (single FDAP analysis)
Photoactivation of Dp-actin and fluorescence imaging were performed with a laser-scanning confocal microscope (LSM 510; Carl Zeiss, Inc.) equipped with a PlanApochromat 63x objective lens (NA 1.4) using LSM510 software v3.2 (Carl Zeiss, Inc.) at 37°C in phenol red–free DME. Laser power was minimized to reduce photobleaching during image acquisition. Dp-actin was expressed in MCF-7 cells and a whole cell was photoactivated to the maximum level by intense irradiation with a 458-nm laser for ∼10 s, and then a fluorescence image in a 16 x 10-pixel (4.8 x 3 µm) rectangular region was acquired as a measure of the total amount of Dp-actin in this region. After photobleaching the whole cell to the background level by intense irradiation with a 488-nm laser, Dp-actin was photoactivated locally in a 1.8-µm-diameter circular region by intense irradiation with a 458-nm laser for 3 ms. Fluorescence images (12 bit) of a 4.8 x 3-µm rectangular region were acquired every 30 ms for 270 ms by weak irradiation with the 488-nm laser at a frame rate of 4.8 ms/frame using the bidirectional scanning mode of LSM 510. An additional 20 fluorescence images were collected before photoactivation for subtracting the background level. The pinhole was set to obtain an optical slice depth of 2.1 µm, which corresponds to about half the cell body thickness. The relative fluorescence intensities in the 3-µm-diameter circular region determined by the profile of the fluorescence intensity in the photoactivation area were plotted after correcting the values for photomultiplier gain variations and photobleaching (Fig. S1), and normalizing to the maximum fluorescence intensity in the same area.
s-FDAP analysis
MCF-7 cells were transfected with Dp-actin and a whole cell was photobleached to the background level by intense irradiation with a 488-nm laser. Then a DIC image of the whole cell was acquired. After photobleaching the whole cell, Dp-actin was photoactivated locally in a 6-pixel-diameter (1.8 µm) circular region by intense irradiation with a 458-nm laser for 3 ms. Three fluorescence images (12 bit) of a 16 x 10-pixel (4.8 x 3 µm) rectangular region were acquired before photoactivation and at 0 and 40 ms after photoactivation by weak irradiation with the 488-nm laser, as described above for single FDAP analysis. After image acquisition 40 ms after photoactivation, the next round of procedures was performed. Each set of procedures was repeated 84 times at 10 s intervals, using the “MultiTimeSeries version 32” macro in the LSM 510 software. The mean fluorescence intensity at 40 ms after photoactivation was corrected for photobleaching. The amount of FDAP was defined as the difference between the fluorescence intensities at 0 and 40 ms after photoactivation for each time point. ImageJ software was used to analyze the fluorescence images and DIC images.
Multipoint single FDAP analysis
For multipoint single FDAP analysis, N1E-115 cells were transfected with Rac(V12), Dp-actin, and mCherry. Dp-actin and mCherry fluorescence images in the whole cell were acquired as a measure of the total amount of Dp-actin and mCherry. Multipoint single FDAP analysis consists of photobleaching of the whole cell, photoactivation of a 1.8-µm-diameter region (Fig. 10 A, red circle), and fluorescence image acquisition of a rectangular region (4.8 x 3 µm), as described above for single FDAP analysis, except for the pinhole size. The pinhole was set to obtain an optical slice depth of 7.3 µm to measure FDAP in both the thin lamellipodium and thick cell body region. The relative fluorescence intensity in a 3-µm-diameter area (Fig. 10 A, white circle) was plotted for each ROI after correcting and normalizing to the maximum intensity in the same region. Single FDAP analysis was repeated at 5 ROIs using the MultiTimeSeries version 32 macro. The order of measurements was ROI-2, -5, -1, -4, and -3. ROI-2 was 9 µm away from ROI-3.
Determination of diffusion coefficient and mobile fraction of Dp-actin
To determine the diffusion coefficient and mobile fraction for Dp-actin, single FDAP data were analyzed using a protocol reported previously (Matsuda et al., 2008). The profile of the fluorescence intensity in the photoactivation area was fitted using equation 1 in Matsuda et al. (2008), by which the photoactivation constant (K) and the half-width of the laser at 1/e2 intensity (w) were determined as 0.24 ± 0.01 and 1.49 ± 0.02 µm, respectively (mean ± SEM, n = 25). The time course of the normalized fluorescence intensity after photoactivation was fitted using equation 3 in Matsuda et al. (2008). Fitting was performed by Prism 5 (GraphPad Software).
Model analysis of Jasp-induced change in G-actin concentration
Because the Cc of an ATP-bound actin monomer is lower at the free barbed ends (B) than at the free pointed ends (P), and the concentration of ATP in the cytoplasm is high, it is presumed that, in vivo, free G-actin (G) disassembles from the pointed ends and assembles onto the barbed ends (G-actin disassembly from barbed ends and G-actin assembly on the pointed ends can be ignored [Eq. 1 and 2]; Pollard and Borisy, 2003; Novak et al., 2008). In the cytoplasm, profilin (Pr) and thymosin-β4 (T) form complexes with free G-actin to produce profilin–G-actin (PrG) and thymosin-β4–G-actin complexes (TG) (Eq. 3 and 4). Profilin–G-actin complexes can also assemble onto the barbed ends at the same rate as free G-actin (Eq. 5; Kang et al., 1999). Jasp (J) is bound to F-actin in a molar ratio of 1:1, and inhibits actin disassembly (Bubb et al., 1994, 2000). In this study, the decline in Jasp concentration due to its binding to F-actin is ignored because a large amount of Jasp exists in the extracellular solution ([J] = constant). Inhibition of actin disassembly by Jasp is represented by the binding of Jasp to the pointed end of the actin filaments (JP) in rapid equilibrium (Eq. 6), where Kvi is the equilibrium dissociation constant for the Jasp-pointed end complex.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Before the addition of Jasp, the rate of actin assembly is equal to the rate of actin disassembly. This steady state is represented by Eq. 7–12:
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
where [Gtotal], [Prtotal], and [Ttotal] are the total concentrations of G-actin, profilin, and thymosin-β4, respectively, in the cytoplasm before the addition of Jasp. These equations lead to the following equation:
| (13) |
The following parameter values were used: k−i = 100 s−1, k+ii = 10 µM−1 s−1, k+iii = 50 µM−1 s−1, k−iii = 5 s−1, k+iv = 1 µM−1 s−1, k−iv = 2.5 s−1, k+v = 10 µM−1 s−1, [Gtotal] = 100 µM, [Prtotal] = 10 µM, and [Ttotal] = 200 µM (Mogilner and Edelstein-Keshet, 2002; Novak et al., 2008). In [G] ≥ 0 and [B] = 0∼100 µM, Eq. 13 has a solution to [G], which is determined by [B]. [P], [Pr], [PrG], [T], and [TG] at the steady state before the addition of Jasp are also determined by [B]. These values were used as the initial concentrations and Eq. 1–6 were used for fitting the total G-actin concentration ([G]+[PrG]+[TG]) into the time course of the relative FDAP after treatment with various concentrations of Jasp [J] from the data in Fig. 3 using KINSIM/FITSIM. The best-fit values were [B] = 4.8 nM and Kvi = 120 nM. The value of Kvi is consistent with that previously reported (Kvi = 15∼300 nM) estimated by in vitro assay (Bubb et al., 1994). The initial concentration of each component was also determined by the best-fit value for [B], as follows: [G] = 1.98 µM, [P] = 5.52 nM, [Pr] = 0.48 µM, [PrG] = 9.52 µM, [T] = 111.5 µM, and [TG] = 88.5 µM. When the fitting was done without the assumption that Jasp binds to the pointed end in rapid equilibrium, the best-fit values were almost identical to those with this assumption, which indicates that the binding of Jasp to the pointed ends reaches equilibrium rapidly, and the rate of Jasp-induced decrease in G-actin concentration is not dependent on the rate of Jasp binding to the pointed end, but probably on the rate of G-actin assembly to the barbed end.
Quantification of free barbed ends
To visualize free barbed ends, 0.45 µM Alexa 546–labeled actin monomers and 0.025% saponin were applied to the outside of the cells, according to the procedures described previously (Chan et al., 2000; Kiuchi et al., 2007). Incorporation of Alexa 546–actin into the cell periphery was quantified by measuring the average fluorescence intensity in a region 1 µm from the cell edge, using a customized macro in ImageJ.
Statistical analysis
All statistical analysis was performed by Student’s t test. Unless otherwise noted, results are expressed as the means ± SEM and data were considered statistically significant at P < 0.05.
Online supplemental material
Fig. S1 shows the correction of fluorescence intensity for photomultiplier gain variations and photobleaching. Fig. S2 shows the effect of Jasp on the ratio of G-actin to total actin. Fig. S3 shows the estimation of G-actin diffusion in a 2D plane. Video 1 shows the time-lapse imaging of YFP-actin in an NRG-treated MCF-7 cell. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201101035/DC1.
Acknowledgments
We thank Dr. A. Miyawaki (RIKEN, Japan) for providing Dronpa plasmids and Dr. H. Miyata (Tohoku University, Japan) and Dr. N. Shayeghi (King’s College London, UK) for comments on the spatial distribution of G-actin.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K. Mizuno.
Footnotes
Abbreviations used in this paper:
- ADF
- actin-depolymerizing factor
- Cc
- critical concentration
- DIC
- differential interference contrast
- Dp
- Dronpa
- FDAP
- fluorescence decay after photoactivation
- Jasp
- jasplakinolide
- LatA
- latrunculin A
- LIMK1
- LIM kinase 1
- NRG
- neuregulin
- ROI
- region of interest
- s-FDAP
- sequential FDAP
- WT
- wild-type
References
- Amann K.J., Pollard T.D. 2001. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat. Cell Biol. 3:306–310 10.1038/35060104 [DOI] [PubMed] [Google Scholar]
- Ando R., Mizuno H., Miyawaki A. 2004. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 306:1370–1373 10.1126/science.1102506 [DOI] [PubMed] [Google Scholar]
- Bamburg J.R., McGough A., Ono S. 1999. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 9:364–370 10.1016/S0962-8924(99)01619-0 [DOI] [PubMed] [Google Scholar]
- Barkalow K., Witke W., Kwiatkowski D.J., Hartwig J.H. 1996. Coordinated regulation of platelet actin filament barbed ends by gelsolin and capping protein. J. Cell Biol. 134:389–399 10.1083/jcb.134.2.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb M.R., Senderowicz A.M.J., Sausville E.A., Duncan K.L.K., Korn E.D. 1994. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 269:14869–14871 [PubMed] [Google Scholar]
- Bubb M.R., Spector I., Beyer B.B., Fosen K.M. 2000. Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 275:5163–5170 10.1074/jbc.275.7.5163 [DOI] [PubMed] [Google Scholar]
- Cano M.L., Lauffenburger D.A., Zigmond S.H. 1991. Kinetic analysis of F-actin depolymerization in polymorphonuclear leukocyte lysates indicates that chemoattractant stimulation increases actin filament number without altering the filament length distribution. J. Cell Biol. 115:677–687 10.1083/jcb.115.3.677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.-F., Pantaloni D. 2007. Control of actin assembly dynamics in cell motility. J. Biol. Chem. 282:23005–23009 10.1074/jbc.R700020200 [DOI] [PubMed] [Google Scholar]
- Chan A.Y., Bailly M., Zebda N., Segall J.E., Condeelis J.S. 2000. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148:531–542 10.1083/jcb.148.3.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer L.P. 1999. Role of actin-filament disassembly in lamellipodium protrusion in motile cells revealed using the drug jasplakinolide. Curr. Biol. 9:1095–1105 10.1016/S0960-9822(99)80478-3 [DOI] [PubMed] [Google Scholar]
- Cramer L.P., Briggs L.J., Dawe H.R. 2002. Use of fluorescently labelled deoxyribonuclease I to spatially measure G-actin levels in migrating and non-migrating cells. Cell Motil. Cytoskeleton. 51:27–38 10.1002/cm.10013 [DOI] [PubMed] [Google Scholar]
- Edmonds B.T., Wyckoff J., Yeung Y.G., Wang Y., Stanley E.R., Jones J., Segall J., Condeelis J. 1996. Elongation factor-1 alpha is an overexpressed actin binding protein in metastatic rat mammary adenocarcinoma. J. Cell Sci. 109:2705–2714 [DOI] [PubMed] [Google Scholar]
- Hotulainen P., Paunola E., Vartiainen M.K., Lappalainen P. 2005. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 16:649–664 10.1091/mbc.E04-07-0555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa J.H., Mullins R.D. 2007. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 17:395–406 10.1016/j.cub.2007.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F., Purich D.L., Southwick F.S. 1999. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem. 274:36963–36972 10.1074/jbc.274.52.36963 [DOI] [PubMed] [Google Scholar]
- Kiuchi T., Ohashi K., Kurita S., Mizuno K. 2007. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J. Cell Biol. 177:465–476 10.1083/jcb.200610005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler S.A., Rottner K., Lai F., Block J., Vinzenz M., Small J.V. 2009. F- and G-actin concentrations in lamellipodia of moving cells. PLoS ONE. 4:e4810 10.1371/journal.pone.0004810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C., Carlier M.-F. 2008. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 88:489–513 10.1152/physrev.00021.2007 [DOI] [PubMed] [Google Scholar]
- Matsuda T., Miyawaki A., Nagai T. 2008. Direct measurement of protein dynamics inside cells using a rationally designed photoconvertible protein. Nat. Methods. 5:339–345 [DOI] [PubMed] [Google Scholar]
- McDonald D., Carrero G., Andrin C., de Vries G., Hendzel M.J. 2006. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J. Cell Biol. 172:541–552 10.1083/jcb.200507101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J.L., Tardy Y., Dewey C.F., Jr, Meister J.J., Hartwig J.H. 1998. Simultaneous measurements of actin filament turnover, filament fraction, and monomer diffusion in endothelial cells. Biophys. J. 75:2070–2078 10.1016/S0006-3495(98)77649-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A., Edelstein-Keshet L. 2002. Regulation of actin dynamics in rapidly moving cells: a quantitative analysis. Biophys. J. 83:1237–1258 10.1016/S0006-3495(02)73897-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R.D., Heuser J.A., Pollard T.D. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 95:6181–6186 10.1073/pnas.95.11.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I.L., Slepchenko B.M., Mogilner A. 2008. Quantitative analysis of G-actin transport in motile cells. Biophys. J. 95:1627–1638 10.1529/biophysj.108.130096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D., Carlier M.F. 1993. How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell. 75:1007–1014 10.1016/0092-8674(93)90544-Z [DOI] [PubMed] [Google Scholar]
- Pantaloni D., Boujemaa R., Didry D., Gounon P., Carlier M.F. 2000. The Arp2/3 complex branches filament barbed ends: functional antagonism with capping proteins. Nat. Cell Biol. 2:385–391 10.1038/35017011 [DOI] [PubMed] [Google Scholar]
- Pollard T.D. 1986. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103:2747–2754 10.1083/jcb.103.6.2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Blanchoin L., Mullins R.D. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576 10.1146/annurev.biophys.29.1.545 [DOI] [PubMed] [Google Scholar]
- Ponti A., Machacek M., Gupton S.L., Waterman-Storer C.M., Danuser G. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786 10.1126/science.1100533 [DOI] [PubMed] [Google Scholar]
- Safer D., Nachmias V.T. 1994. Beta thymosins as actin binding peptides. Bioessays. 16:473–479 10.1002/bies.950160706 [DOI] [PubMed] [Google Scholar]
- Schafer D.A., Jennings P.B., Cooper J.A. 1996. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135:169–179 10.1083/jcb.135.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwick F.S., Young C.L. 1990. The actin released from profilin—actin complexes is insufficient to account for the increase in F-actin in chemoattractant-stimulated polymorphonuclear leukocytes. J. Cell Biol. 110:1965–1973 10.1083/jcb.110.6.1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.X., Magalhães M.A.O., Glogauer M. 2007. Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J. Cell Biol. 179:239–245 10.1083/jcb.200705122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T., Miki H. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114:1801–1809 [DOI] [PubMed] [Google Scholar]
- Theriot J.A., Mitchison T.J. 1991. Actin microfilament dynamics in locomoting cells. Nature. 352:126–131 10.1038/352126a0 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Mitchison T.J. 2002. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 295:1083–1086 10.1126/science.1067470 [DOI] [PubMed] [Google Scholar]
- Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., Nishida E., Mizuno K. 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 393:809–812 10.1038/31735 [DOI] [PubMed] [Google Scholar]
- Zicha D., Dobbie I.M., Holt M.R., Monypenny J., Soong D.Y.H., Gray C., Dunn G.A. 2003. Rapid actin transport during cell protrusion. Science. 300:142–145 10.1126/science.1082026 [DOI] [PubMed] [Google Scholar]