Impaired autophagy stabilizes p62 and promotes tumorigenesis through activation of the Nrf2 transcription factor.
Abstract
Suppression of autophagy is always accompanied by marked accumulation of p62, a selective autophagy substrate. Because p62 interacts with the Nrf2-binding site on Keap1, which is a Cullin 3–based ubiquitin ligase adapter protein, autophagy deficiency causes competitive inhibition of the Nrf2–Keap1 interaction, resulting in stabilization of Nrf2 followed by transcriptional activation of Nrf2 target genes. Herein, we show that liver-specific autophagy-deficient mice harbor adenomas linked to both the formation of p62- and Keap1-positive cellular aggregates and induction of Nrf2 targets. Importantly, similar aggregates were identified in more than 25% of human hepatocellular carcinomas (HCC), and induction of Nrf2 target genes was recognized in most of these tumors. Gene targeting of p62 in an HCC cell line markedly abrogates the anchorage-independent growth, whereas forced expression of p62, but not a Keap1 interaction-defective mutant, resulted in recovery of the growth defect. These results indicate the involvement of persistent activation of Nrf2 through the accumulation of p62 in hepatoma development.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a cellular degradation system in which cytoplasmic components, including organelles, are sequestered by double membrane structures called autophagosomes and the sequestered materials are degraded by lysosomal hydrolases for supply of amino acids and for cellular homeostasis (Rubinsztein, 2006; Levine and Kroemer, 2008; Mizushima et al., 2008; Rabinowitz and White, 2010). Although autophagy has generally been considered nonselective, recent studies have shed light on another indispensable role for basal autophagy in cellular homeostasis, which is mediated by selective degradation of a specific substrate(s) (Kirkin et al., 2009; Kraft et al., 2010; Johansen and Lamark, 2011).
p62 is a ubiquitously expressed cellular protein that is conserved in metazoa but not in plants and fungi, and recently it has been known as one of the selective substrates for autophagy (Bjørkøy et al., 2005; Pankiv et al., 2007; Ichimura et al., 2008). This protein is localized at the autophagosome formation site (Itakura and Mizushima, 2011) and directly interacts with LC3, an autophagosome localizing protein (Pankiv et al., 2007; Ichimura et al., 2008). Subsequently, the p62 is incorporated into the autophagosome and then degraded (Pankiv et al., 2007; Ichimura et al., 2008). Therefore, impaired autophagy is accompanied by accumulation of p62 followed by the formation of p62 and ubiquitinated protein aggregates because of the nature of both self-oligomerization and ubiquitin binding of p62 (Komatsu et al., 2007; Nezis et al., 2008).
The Nrf2–Keap1 system is currently recognized as one of the major cellular defense mechanisms against oxidative and electrophilic stresses. Under quiescent conditions, the transcription factor Nrf2 (nuclear factor erythroid 2–related factor 2) is constitutively degraded through the ubiquitin–proteasome pathway as its binding partner Keap1 (kelch-like ECH-associated protein 1) is an adaptor of the ubiquitin ligase complex. Exposure to electrophiles, reactive oxygen species, and nitric oxide instigates modification of the cysteine residues of Keap1, leading to Keap1 inactivation. Hence, Nrf2 becomes stabilized and translocates to the nucleus to induce the transcription of numerous cytoprotective genes through its heterodimerization with small Maf proteins (Motohashi and Yamamoto, 2004; Hayes and McMahon, 2009; Villeneuve et al., 2010).
Recently, we and other groups independently reported that p62 interacts with the Nrf2-binding site on Keap1, and thus, overproduction of p62 or autophagy deficiency competes with the interaction between Nrf2 and Keap1, resulting in stabilization of Nrf2 followed by transcriptional activation of Nrf2 target genes (Copple et al., 2010; Jain et al., 2010; Komatsu et al., 2010; Lau et al., 2010; Riley et al., 2010). Induction of Nrf2 target genes has been recognized in many human cancers (Hayes and McMahon, 2009) that also show p62 accumulation (Zatloukal et al., 2002). However, it is not clear whether p62-induced Nrf2 activation is involved in tumorigenesis and/or tumor progression. In this report, we show that liver-specific Atg7 knockout mice develop hepatocellular adenoma accompanied by excess accumulation of p62 followed by Nrf2 activation, and that persistent activation of Nrf2 through p62 contributes to development in human hepatocellular carcinoma.
Results and discussion
Loss of Atg7 in mouse liver causes hepatocellular adenoma
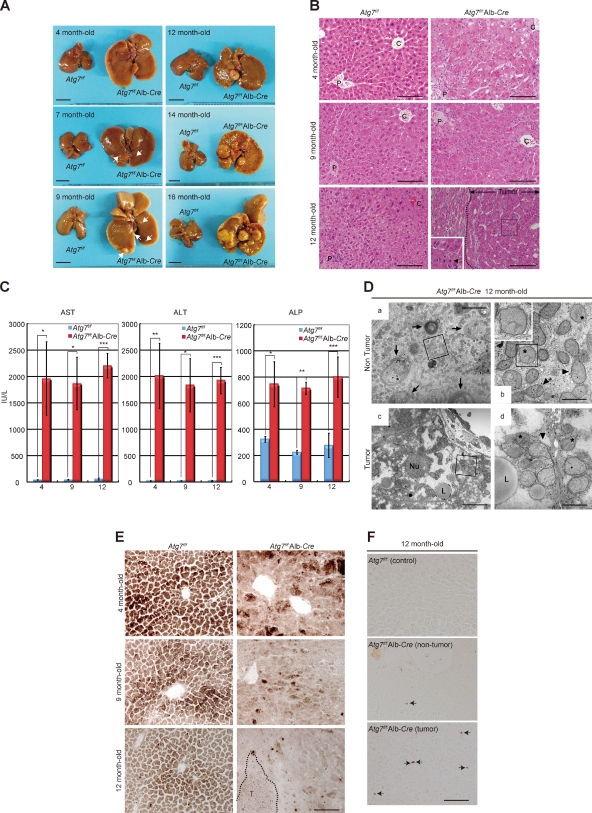
Recently, we found that aged mice with liver-specific Atg7 deficiency develop multiple tumors in the livers similar to systemically mosaic Atg5-deficient mice (Takamura et al., 2011), but the exact mechanism of tumorigenesis is still unclear. Thus, we first examined the chronologic changes in the liver-specific Atg7-deficient mice (Atg7f/f;Alb-Cre; Komatsu et al., 2007), including control littermates (Atg7f/f). Atg7 is an E1-like enzyme for both the Atg12 and Atg8 conjugation systems, and is essential for autophagy (Tanida et al., 1999). The mutant mice exhibited severe hepatomegaly with hepatocyte hypertrophy, as reported previously (Komatsu et al., 2005; Fig. 1, A and B). Loss of Atg7 in mouse livers was also accompanied by inflammation, as judged by both hematoxylin and eosin (H&E) staining (Fig. 1 B) and measuring leakage liver enzymes (Fig. 1 C). Microtumors (diameter less than ∼1 mm) developed randomly throughout the liver of 7-mo-old mutant mice but not in age-matched control mice (Fig. 1 A). The number and size of the tumors increased with age, and several tumors in livers of 12-mo-old mutant mice were >5 mm in diameter (Fig. 1 A and Table S1). H&E staining indicated that the tumors in Atg7-deficient livers were in most cases well demarcated with little fibrous capsule, composed of irregular hepatic plates with tumor cells showing large nuclear cytoplasmic ratio and occasional nuclear atypia (Fig. 1 B). Thus, they were pathologically diagnosed as hepatocellular adenoma.
Figure 1.
Loss of Atg7 eventually triggers hepatocellular adenoma. (A) Chronological changes in gross anatomy of livers of Atg7f/f;Alb-Cre and Atg7f/f mice. Arrows indicate microtumors. Bars, 1 cm. (B) H&E-stained liver sections of Atg7f/f and Atg7f/f;Alb-Cre mice aged from 4 to 12 mo. Dashed line: the tumor region in the liver of a representative 12-mo-old Atg7f/f;Alb-Cre mouse. The inset shows higher magnification view of the boxed region containing a mitotic cell (arrowhead). C, central vein; P, portal vein. Bars, 100 µm. (C) Liver function tests of Atg7f/f (blue bars) and Atg7f/f;Alb-Cre (red bars) mice aged from 4 to 12 mo. Serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALP) were measured. Data are means ± SD of Atg7f/f (n = 3) and Atg7f/f;Alb-Cre (n = 4) at 4 mo of age, of Atg7f/f (n = 3) and Atg7f/f;Alb-Cre (n = 3) at 9 mo of age, and of Atg7f/f (n = 5) and Atg7f/f;Alb-Cre (n = 5) at 12 mo of age. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Electron microscopy of nontumor hepatocytes (a and b) and tumor cells (c and d) in Atg7f/f;Alb-Cre mouse liver. The boxed regions in a and c are enlarged and shown as b and d, respectively. The inset in b shows the higher magnification view of a peroxisome (boxed region in b). Note numerous peroxisomes (asterisks) and accumulation of deformed mitochondria (arrowheads) in the peripheral area. Arrows: concentric membranous structures that were consistently present in Atg7-deficient hepatocytes. Nu, nucleus; L, lipid droplet. Bars: (a and c) 5 µm; (b and d) 1 µm. (E) Histochemical detection of COX activity in liver sections of Atg7f/f and Atg7f/f;Alb-Cre mice aged from 4 to 12 mo. Dashed line: liver tumor region in a representative 12-mo-old Atg7f/f;Alb-Cre mouse. Bar, 10 µm. (F) Phospho-histone H2A.X-positive cells in nontumor and tumor regions of 12-mo-old mice of the indicated genotypes. Arrows, cells positive for Phospho-histone H2A.X. Bar, 100 µm.
Autophagy deficiency in hepatocytes is accompanied by mitochondria dysfunction and genomic instability
Electron microscopy showed that the phenotype of nontumor regions of Atg7-deficient liver of 12-mo-old mice was basically similar to that previously observed in the 3-mo-old Atg7-deficient liver (Komatsu et al., 2005). Specifically, abundance of smooth ER, presence of numerous deformed mitochondria and peroxisomes, and appearance of concentric membranous structures and aggregates were noted (Fig. 1 D, a and b). Many peroxisomes were as big as the mitochondria and often surrounded by rough ER-derived double-membrane structures (Fig. 1 D, b). Although the tumor regions showed similar changes, the sections contained larger proportions of deformed mitochondria and scarce concentric membranous structures (Fig. 1 D, c and d).
To determine whether the deformed mitochondria observed in Atg7-deficient hepatocytes and tumor cells are functionally intact, we examined the activity of cytochrome c oxidase (COX) enzyme by histochemistry (Inoue et al., 2000). As shown in Fig. 1 E, the activity was markedly reduced by loss of Atg7 for 4 mo, and the reduction worsened with age. Intriguingly, the hepatocellular adenoma derived from Atg7-deficient livers also exhibited reduced COX activity (Fig. 1 E). These results indicate reduced mitochondrial activity in Atg7-deficient hepatocytes and adenoma cells. We also often recognized cells with nuclei positive for phospho-histone H2A.X, representing genomic instability, in both the nontumor and tumor regions of Atg7-deficient livers, whereas such cells were not detected in the control tissues at all (Fig. 1 F). Taken together, it is concluded that autophagy deficiency in mouse livers causes tumorigenesis associated with chronic inflammation, blockade of mitochondrial activity, and genome instability.
Activation of Nrf2 in Atg7-deficient tumors
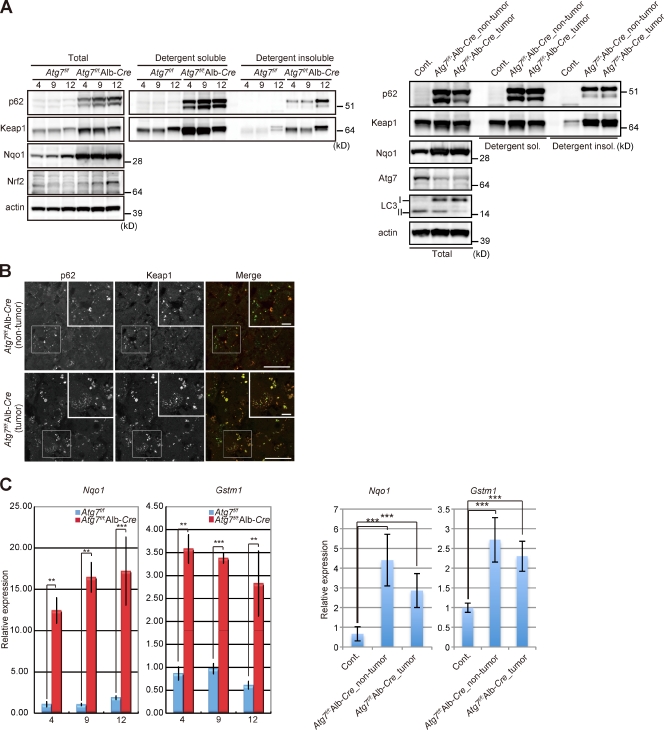
The next series of experiments served to investigate the chronologic changes in the Nrf2–Keap1 pathway in Atg7-deficient livers. Immunoblot analysis showed marked accumulation of p62 in livers of 4-mo-old Atg7f/f;Alb-Cre mice, and the presence of both the soluble and insoluble fractions of p62 (Fig. 2 A). A p62-splicing variant product that migrates faster was also accumulated in both fractions. The quantity of insoluble p62 increased with age, and Keap1 was also fractionated into the insoluble fraction in proportion to the amount of insoluble p62 (Fig. 2 A). Interestingly, accumulation of p62 was also noted in Atg7-deficient tumor regions as well as in the nontumor regions, and significant amounts of p62 and Keap1 were recovered in the insoluble fraction (Fig. 2 A), implying sequestration of Keap1 into p62 aggregate structures. Indeed, double immunofluorescence microscopy showed that most of the p62-containing aggregates in Atg7-deficient nontumor and tumor cells were also positive for Keap1 (Fig. 2 B). We confirmed not only the induction of gene expression of Nrf2 targets such as NAD(P)H dehydrogenase quinone 1 (Nqo1) and glutathione S-transferase mu1 (Gstm1; Fig. 2 C), but also increased quantity of Nrf2 and Nqo1 proteins in Atg7-deficient livers and tumor region (Fig. 2 A). Taken together, these results indicate persistent activation of Nrf2 in Atg7-deficient tumor regions as well as nontumor regions due to accumulation of p62.
Figure 2.
Activation of Nrf2 in Atg7-deficient hepatocellular adenoma. (A) Immunoblot analysis. Total, soluble, and insoluble fractions were subjected to SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. Note that the decrease in conversion of LC3-I to LC3-II, which signifies defective autophagosome formation, was confirmed in both the nontumor and tumor regions of the liver of Atg7-deficient mice (Atg7f/f;Alb-Cre). Data were obtained from three independent experiments. (B) Immunofluorescence analysis. Liver nontumor or tumor sections from 12-mo-old mice of the indicated genotypes were immunostained with anti-p62 and anti-Keap1 antibodies. The right columns show the merged images of Keap1 (green) and p62 (red). Each inset is a magnified image of the boxed region. Bars, 20 µm. (C) qRT-PCR analysis. Total RNAs were prepared from liver or tumor shown in A. Values were normalized to the amount of mRNA in the Atg7f/f liver of 4-mo-old (left) and 12-mo-old mice (right). Data are means ± SD of three experiments. **, P < 0.01; ***, P < 0.001.
Activation of Nrf2 in human hepatocellular carcinoma
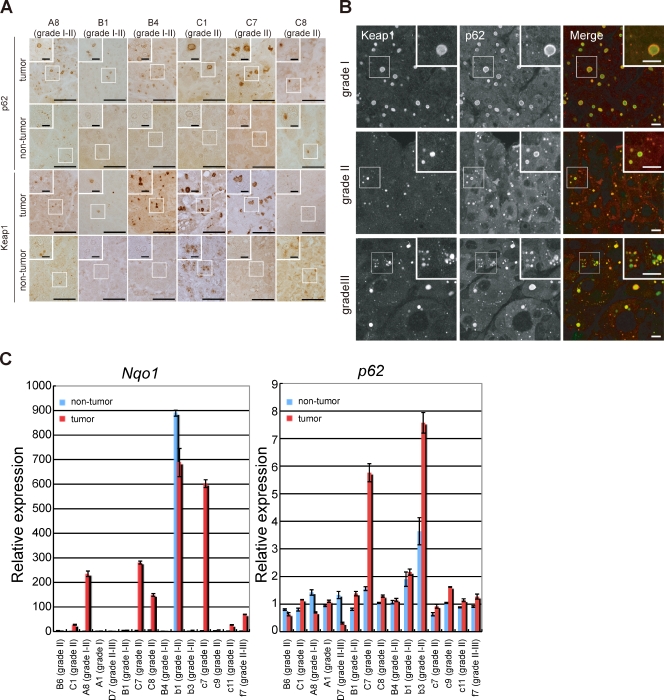
To investigate whether similar events in Atg7-deficient conditions occur in human liver disorders, including hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC), we conducted immunohistochemistry for human liver disease tissue arrays using anti-p62 and anti-Keap1 antibodies. As reported previously (Zatloukal et al., 2002), immunohistochemical analysis with anti-p62 antibody showed abundance of p62 and p62-positive aggregates in several cases of human HCC (Fig. 3 A). Aggregates positive for Keap1 were also found in the HCC cases that showed p62 aggregates (Fig. 3 A). As expected, double immunofluorescence microscopy demonstrated extensive colocalization of p62 and Keap1 in numerous aggregates in HCC regardless of the pathological grade (Fig. 3 B). Quantitative analysis showed that 25.5% (26 of 102 patients) and 21.3% (13 of 61 patients) of HCC and adjacent tissue of HCC contained p62- and Keap1-positive aggregates, respectively (Table I), albeit the aggregates in the adjacent tissue of HCC tended to be smaller in size and number (Fig. 3 A). There was no significant correlation between the presence of p62 and Keap1 double-positive aggregates and sex, age of tumor onset, or pathological grade (Table S2). These aggregates were also detected in tissues of patients with hepatitis, liver cirrhosis, and non-HCC tumors, though at lower incidence compared with those in HCC and HCC-adjacent tissues (Table I).
Figure 3.
Activation of Nrf2 in human HCC containing p62- and Keap1-positive aggregates. (A) Immunohistochemical images. Paraffin sections of a liver cancer tissue array were stained with anti-p62 or anti-Keap1 antibody. 6 out of 26 HCC cases containing the typical inclusions are shown. Case codes with pathological grade in parenthesis are indicated at the top. Higher magnification views of the boxed regions are shown in the insets. Bars: (panels) 50 µm; (insets) 10 µm. (B) Double immunofluorescence microscopy. Three cases of HCC containing typical inclusions were immunostained with anti-Keap1 and anti-p62 antibodies. The merged images of Keap1 (green) and p62 (red) are shown on the right. Pathological grades are indicated on the left. Bars, 10 µm. (C) qRT-PCR analyses. Total RNAs were prepared from both nontumor and tumor regions of 15 HCCs. Values were normalized to the amount of mRNA in the nontumor region of B6 where p62- and Keap1-positive aggregates were not recognized. Data are means ± SD of three experiments.
Table I.
Presence of Keap1 and p62 double-positive aggregates in human liver diseases.
| Pathological diagnosis | Total number | p62 and Keap1 double-positive lesions | Percentage |
| Normal | 12 | 0 | 0.0 |
| Cellular hypertrophy | 2 | 0 | 0.0 |
| Hepatitis | 13 | 1 | 7.7 |
| Cirrhosis | 40 | 3 | 7.5 |
| Hepatocellular carcinoma | 102 | 26 | 25.5 |
| Tissue adjacent to hepatocellular carcinoma | 61 | 13 | 21.3 |
| Non-HCC tumor | 14 | 1 | 7.1 |
| Clear cell carcinoma | 5 | 0 | – |
| Bile duct carcinoma | 4 | 1 | – |
| Metastasis adenoma | 3 | 0 | – |
| Sarcoma | 1 | 0 | – |
| Hemangioma | 1 | 0 | – |
Is Nrf2 activated in HCC similar to autophagy-deficient tumors? To answer this question, quantitative real-time PCR (qRT-PCR) was conducted with mRNAs prepared from nontumor and tumor regions of HCC patients. The results showed marked induction of Nqo1, an Nrf2 target, in 8 out of 15 HCCs positive for p62 and Keap1 aggregates (Fig. 3 C). These results imply hyperactivation of Nrf2 associated with p62 accumulation in certain types of human HCC.
Nrf2 activation by p62 contributes to tumor growth
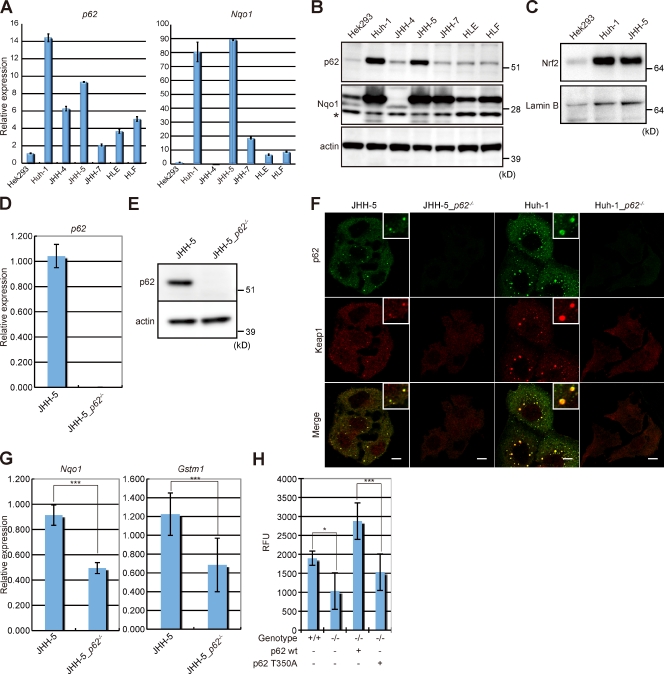
To further clarify the pathological significance of Nrf2 activation in human HCC, we used human HCC cell lines. Among the six human HCC cell lines examined, Huh-1 and JHH-5 cell lines expressed p62 and Nqo1 mRNAs and proteins at relatively high levels compared with an immortalized human embryonic kidney cell line, HEK293 (Fig. 4, A and B). Large amounts of nuclear Nrf2 protein were also found in those two cell lines (Fig. 4 C). Next, p62 knockout clones were generated from these two cell lines. The p62 in both cell lines was successfully deleted using a zinc finger nuclease system comprised of a class of engineered DNA-binding proteins that facilitate targeted editing of the genome by creating double-strand breaks in DNA at specified locations (Meng et al., 2008; Santiago et al., 2008). Disruption of p62 was confirmed by the absence of both p62 transcript and the protein in p62-deficient cell lines (JHH-5_p62−/− and Huh-1_p62−/−; Fig. 4, D and E; and unpublished data). Importantly, although p62- and Keap1-positive aggregates were frequently detected in the cytoplasm of both JHH-5 and Huh-1 cell lines, such aggregates were completely dispersed by loss of p62 (Fig. 4 F), indicating that like a case in autophagy-deficient tissues (Komatsu et al., 2007), p62 is responsible for formation of the aggregates.
Figure 4.
Deletion of p62 suppressed anchorage-independent growth of human HCC cell line. (A) qRT-PCR analyses of p62 and Nqo1 in HCC cell lines. Values were normalized to the amount of mRNA in Hek293 cells. Data shown are representative of three separate experiments. (B) Immunoblot analysis. Total lysates from the indicated cell lines were subjected to SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. Asterisk indicates a nonspecific band. Data were obtained from three independent experiments. (C) Immunoblot analysis of nuclear Nrf2. Nuclear fractions were prepared from the indicated cell lines, subjected to SDS-PAGE, and analyzed by immunoblotting with antibodies against Nrf2 and lamin B (as control). Data were obtained from three independent experiments. (D) qRT-PCR analysis. Total RNAs were prepared from JHH-5 and JHH-5_p62−/− cells. Values were normalized to the amount of mRNA in JHH-5 cells. (E) Immunoblot analysis of p62. Total lysates from JHH-5 and JHH-5_p62−/− cells were subjected to SDS-PAGE and analyzed by immunoblotting with p62 and actin antibodies. (F) Double immunofluorescence microscopy. The indicated cell lines were immunostained with anti-p62 and anti-Keap1 antibodies. Bottom row: merged images of p62 (green) and Keap1 (red). Each inset is a magnified image. Bars, 10 µm. (G) qRT-PCR analyses of Nrf2 target genes under three-dimensional (3D) culture conditions. Total RNAs were prepared from JHH5 and JHH-5_p62−/− cells under 3D culture condition. Values were normalized to the amount of mRNA in the JHH-5. Data are means ± SD of three experiments. ***, P < 0.001. (H) Soft agar colony formation assay. The JHH-5, JHH-5_p62−/−, and wild-type p62- or p62 mutant (p62 T350A)–introduced JHH5_p62−/− cells were tested for their ability to grow in soft agar. The transformation was determined according to the protocol supplied by the manufacturer. RFU, relative fluorescent unit. Data are means ± SD of three experiments. *, P < 0.05; ***, P < 0.001.
Next, to mimic the physiological environment allowing cells to assemble into the three-dimensional (3D) structures similar to what is observed in vivo, the parent and p62−/− cell lines were subjected to 3D culture. Because the Huh-1 cell line did not grow in 3D culture conditions, we used the JHH-5 cell line to examine the effect of loss of p62. Quantitative analysis showed significant suppression of Nqo1 and Gstm1 expression in JHH-5 cells by loss of p62 under the 3D culture conditions (Fig. 4 G). Furthermore, the soft agar colony formation assay showed a reduced colony formation in JHH-5_p62−/− than parental JHH-5 (Fig. 4 H), indicating less anchorage-independent growth. As expected, such defect was recovered by forced expression of wild-type p62 but not p62 T350A mutant defective in Keap1 interaction (Komatsu et al., 2010) into JHH-5_p62−/− (Fig. 4 H and Fig. S1). Collectively, these results emphasize the potential role of Nrf2 activation through Keap1 inactivation (by accumulation of p62) in hepatoma development.
Conclusion
Autophagy has become a field of interest for cancer researchers since Beclin 1, which is involved in both autophagy and endocytosis (He and Levine, 2010), was reported as a tumor suppressor gene (Liang et al., 1999), and mice heterozygous for this protein developed cancers spontaneously (Qu et al., 2003; Yue et al., 2003). How does autophagy deficiency in mouse livers cause tumorigenesis? Systemically mosaic Atg5-deficient mice show accumulation of deformed mitochondria followed by oxidative stress and genome instability in the hepatocytes, culminating in hepatocellular adenoma (Takamura et al., 2011). Consistent with this mouse model, we found that loss of Atg7 in mouse livers was associated with tumorigenesis, which was probably triggered by mitochondrial dysfunction followed by genome instability (Fig. 1, D–F). In addition to these alterations, chronic inflammation was observed in liver-specific Atg7-deficient mice (Fig. 1, B and C). Thus, it is conceivable that the inflammatory changes also contribute to the tumorigenesis.
Growing lines of evidence indicate that p62 is a scaffold protein for cell survival and death-signaling pathways such as the NF-κB pathway (Sanz et al., 1999; Moscat et al., 2007; Moscat and Diaz-Meco, 2009), Wnt signaling (Gao et al., 2010), and apoptosis (Jin et al., 2009). Accordingly, it is plausible that excess p62 leads to dysregulated activation of these signaling pathways. In agreement with this hypothesis, p62 has been identified as a component of inclusion bodies found in various human diseases including HCC and malignant glioma (Zatloukal et al., 2002). Furthermore, the loss of p62 suppressed Ras-induced lung adenocarcinoma (Duran et al., 2008). Moreover, dysregulation of NF-κB signal in autophagy-incompetent cells was due, at least in part, to accumulation of p62, which subsequently enhanced tumorigenesis (Mathew et al., 2009). Those lines of evidence suggest the involvement of aberrant signals related to p62 accumulation in tumorigenesis. We also propose here that persistent activation of Nrf2 through excess accumulation of p62 also contributes to tumor development, as described in the following paragraph.
It has been considered over a long time that marked induction of detoxification enzymes in tumor cells is driven by constant activation of the normal stress response. Recently, somatic mutations in Keap1 and Nrf2 were identified in lung, gall bladder, and head and neck tumors (Padmanabhan et al., 2006; Singh et al., 2006; Shibata et al., 2008a,b). These mutations lead to constitutive activation of Nrf2, followed by induction of Nrf2 target genes, such as cytoprotective enzymes, antioxidant proteins, and multi-drug efflux pumps, thus conferring resistance of tumor cells to oxidative stress and anticancer agents (Shibata et al., 2008b; Hayes and McMahon, 2009). In this regard, the present study showed the presence of p62- and Keap1-positive aggregates and induction of Nrf2 targets in HCC (Fig. 3). We also noted that cells in areas adjacent to the tumor contained similar aggregates, at an incidence (21.3%) comparable to that of tumor region (25.5%). Both rates were more than threefold that of hepatitis, liver cirrhosis, and non-HCC tumors (Table I). This implies that accumulation and/or aggregation of p62 and Keap1 could predispose to hepatocarcinogenesis. Importantly, the marked induction of Nqo1 in tumor regions with massive accumulation of p62 was not associated with increased p62 mRNA expression (Fig. 3 C), suggesting that the excess p62 was likely related to the suppression of autophagy. Considering cancers with somatic mutation of Nrf2 or Keap1, some types of HCC with excess levels of p62 would be resistant to oxidative stress and anticancer agents through Nrf2 activation without somatic mutations in Nrf2 or Keap1. Actually, loss of p62 in human HCC cell line JHH-5 exhibited less anchorage-independent growth than parental cells, and such defect was recovered by forced expression of p62 but not the mutant defective in Keap1 interaction (Fig. 4 H). These results suggest that continuous activation of Nrf2 through excess accumulation of p62 directly contributes to tumor development. Thus, a better understanding of the cellular fluctuations associated with p62 accumulation and/or failure of autophagy could provide the basis for the development of new therapies for HCC.
Materials and methods
Mice
The Atg7f/f;Alb-Cre mice described previously by our group (Komatsu et al., 2007) were used in this study. Mice were housed in specific pathogen-free facilities and the experimental protocol was approved by the Ethics Review Committee for Animal Experimentation of the Tokyo Metropolitan Institute of Medical Science and Juntendo University.
Cell culture and gene targeting
The media and reagents for cell culture were purchased from Life Technologies. HCC cell lines were provided by Health Science Research Resources Bank (Osaka, Japan). Huh-1 and JHH-5 cells were grown in William’s E medium containing 10% fetal bovine serum (FBS), 5 U/ml penicillin, and 50 µg/ml streptomycin. The 3D structures were prepared using the AlgiMatrix 3D culture system (Invitrogen). Cells were resuspended, inoculated at 106 cells/well, and then cultured for 7 d. The culture medium was changed every other day. The gene targeting for p62 of Huh-1 and JHH-5 cells was conducted using the CompoZr Zinc Finger Nuclease (Sigma-Aldrich). Wild-type p62 and p62 T350A mutant were subcloned into retrovirus vector, pMXs-puro (Kitamura et al., 2003). As described recently (Ichimura et al., 2008), each p62 and p62 T350A vector was introduced into established JHH-5_p62−/− cells.
Immunological analysis
Livers were homogenized in 0.25 M sucrose, 10 mM Hepes, pH 7.4, and 1 mM DTT. The resultant homogenates were fractionated into 0.5% Triton X-100 soluble and insoluble fractions. The samples were separated by the NuPage system (Invitrogen) with 12% Bis-Tris gels and MOPS-SDS buffer and transferred to a polyvinylidene difluoride membrane (Komatsu et al., 2010). The antibodies for p62 (GP62, Progen Biotechnik), Keap1 (Proteintech Group, Inc.), Nqo1 (Abcam), Nrf2 (H-300; Santa Cruz Biotechnology, Inc.), actin (MAB1501R; Millipore), and Lamin B (M-20; Santa Cruz Biotechnology, Inc.) were purchased from the indicated suppliers. The rabbit polyclonal antibodies against Atg7 (Tanida et al., 1999) and LC3 (Komatsu et al., 2005) have been described previously.
Human tissue samples
Glass slides with tissue array of various liver diseases were purchased from Shanghai Outdo Biotech Co.; Biochain Institute, Inc.; and Biomax, Inc. Frozen tissues and the corresponding mRNAs were purchased from Shanghai Outdo Biotech Co. This part of the study was approved by the institutional review board of the Tokyo Metropolitan Institute of Medical Science.
Immunofluorescence microscopy
Cells grown on coverslips were fixed in 4% paraformaldehyde in 0.1 M phosphate buffer (PB), pH 7.4, for 15 min, permeabilized with 50 µg/ml digitonin in PB for 5 min, blocked with 0.1% (vol/vol) gelatin (#G-9391; Sigma-Aldrich) in PB for 30 min, and then incubated with primary antibodies for 1 h. After washing, cells were incubated with Alexa Fluor 488–conjugated goat anti–guinea pig and Alexa Fluor 647–conjugated goat anti–rabbit IgG secondary antibodies (Invitrogen) for 30 min. Cells were imaged using a laser-scanning microscope (LSM510 META; Carl Zeiss, Inc.) with a Plan Apochromat 63x NA 1.4 oil differential interference contrast objective lens. Image contrast and brightness were adjusted using Photoshop CS4 (Adobe).
Histological examination
Fixation and embedding procedures for immunohistochemistry were described previously (Waguri and Komatsu, 2009). In brief, mouse livers were quickly excised, cut into small pieces, and then fixed by immersing in 4% paraformaldehyde/4% sucrose in PB. After rinsing, they were embedded in paraffin for H&E and Keap1 and p62 stainings, or in OCT compound for the staining of phospho-histone H2A.X. As antigen-retrieval procedures, paraffin sections were autoclaved for 10 min at 121°C in buffer at pH 9.0 and 6.0 (Nichirei) for Keap1 and p62 stainings, respectively, whereas cryosections were treated with pepsin solution (Nichirei) for 10 min at 37°C for the staining of phospho-histone H2A.X. Sections were blocked and incubated for 2–3 d at 4°C with the following primary antibodies: guinea pig polyclonal antibody against p62 (Progen), rabbit polyclonal antibody against Keap1 (Proteintech Group), and rabbit monoclonal antibody against phospho-histone H2A.X (Cell Signaling Technology). They were then incubated for 1 h with HRP-conjugated donkey anti–guinea pig IgG (Millipore) or goat anti–rabbit IgG (Histofine simple stain mouse MAX-PO[R]; Nichirei) followed by the diaminobenzidine reaction. Double immunofluorescence for p62 and Keap1 was performed using Alexa 549–conjugated goat anti–guinea pig IgG (Invitrogen) and Dylight 488–conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.), respectively. Cytochrome c oxidase (COX) staining was performed as reported previously (Seligman et al., 1968; Inoue et al., 2000). In brief, air-dried cryosections (10 µm thick) were incubated in reaction medium: 5 mg diaminobenzidine tetrahydrochloride (DAB), 9 ml sodium phosphate buffer (0.05 M, pH 7.4), 1 ml catalase (20 µg/ml), 10 mg cytochrome c, and 750 mg sucrose. COX activity was visualized as a brown DAB reaction product.
Images were acquired with a microscope (BX51; Olympus) equipped with a 60x objective lens (PlanApoN, NA 1.42, oil; Olympus) and a cooled CCD camera system (DP-71; Olympus), or a laser-scanning confocal microscope (FV1000; Olympus) with a 60x objective lens (UPlanSApo, NA 1.35, oil; Olympus). After image acquisition, contrast and brightness were adjusted using Photoshop CS4 (Adobe).
Electron microscopy
For conventional electron microscopy, small pieces of the liver were fixed by immersing in 2% paraformaldehyde/2% glutaraldehyde in PB, post-fixed with 1% OsO4, and embedded in Epon 812 (Waguri and Komatsu, 2009). After sectioning and staining with uranyl acetate and lead citrate, they were observed under an electron microscope (JEM 1200EX; JEOL).
Quantitative real-time PCR (qRT-PCR)
Using a Transcriptor First Strand cDNA Synthesis kit (Roche), the cDNA was synthesized from 1 µg of total RNA. Quantitative PCR was performed using LightCycler 480 Probes Master (Roche) in a LightCycler 480 (Roche). Signals were normalized to β-glucuronidase (GUS) for mouse samples and to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for human samples. The sequences of the primers used for mouse samples were: Nqo1F, 5′-AGCGTTCGGTATTACGATCC-3′; Nqo1R, 5′-AGTACAATCAGGGCTCTTCTCG-3′; Gstm1F, 5′-CTACCTTGCCCGAAAGCAC-3′; Gstm1R, 5′-ATGTCTGCACGGATCCTCTC-3′; GusF, 5′-CTCTGGTGGCCTTACCTGA-3′; and GusR, 5′-CTCAGTTGTTGTCACCTTCACC-3′. The sequences of the primers used for human samples were: Nqo1F, 5′-AGCGTTCGGTATTACGATCC-3′; Nqo1R, 5′-AGTACAATCAGGGCTCTTCTCG-3′; Gstm1F, 5′-TGCCCATGATACTGGGGTA-3′; Gstm1R, 5′-GCCACTGGCTTCTGTCATAAT-3′; p62F, 5′-CAGAGAAGCCCATGGACAG-3′; p62R, 5′-AGCTGCCTTGTACCCACATC-3′; GapdhF, 5′-CTCTGGTGGCCTTACCTGA-3′; and GapdhR, 5′-CTCAGTTGTTGTCACCTTCACC-3′.
Transformation in soft agar
Anchorage-independent transformation assays were performed using a CytoSelect 96-well cell transformation assay kit (Cell Biolabs, Inc.). In brief, cells were plated in soft agar in a 96-well plate at 5,000 cells/well and cultured for 5 d. The transformation was determined according to the protocol provided by the manufacturer.
Statistical analysis
Values are mean ± SD, including those displayed in the graphs. Statistical analysis was performed using the unpaired t test (Welch test). Correlation analyses between p62/Keap1 immunoreactivity and sex and age were performed using the Mann-Whitney test, whereas those among immunoreactivity and histological grades were performed using the Kruskal-Wallis test. A P-value less than 0.05 denoted the presence of a statistical significance.
Online supplemental material
Fig. S1 shows inactivity of the p62 T350A mutant in a Keap1 interaction. Table S1 lists frequency of tumorigenesis in Atg7-deficient livers. Table S2 lists proportions of p62 and Keap1 double-positive cases in human liver diseases. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201102031/DC1.
Acknowledgments
We thank A. Yamada, K. Kanno, and A. Yabashi (Fukushima Medical University School of Medicine) and T. Mita (Tokyo Metropolitan Institute of Medical Science) for their help in histological studies.
This work was supported by Grants-in-Aid for Specially Promoted Research (to K. Tanaka) from the Ministry of Education, Culture, Sports, Science and Technology, Japan; Funding Program for Next Generation World-Leading Researchers (to M. Komatsu); and the Global Research Laboratory Grant of the National Research Foundation of Korea (to M.-S. Lee and M. Komatsu).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Gstm1
- glutathione S-transferase mu1
- HCC
- hepatocellular carcinoma
- Keap1
- kelch-like ECH-associated protein 1
- Nqo1
- NAD(P)H dehydrogenase quinone 1
- Nrf2
- nuclear factor erythroid 2–related factor 2
References
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copple I.M., Lister A., Obeng A.D., Kitteringham N.R., Jenkins R.E., Layfield R., Foster B.J., Goldring C.E., Park B.K. 2010. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J. Biol. Chem. 285:16782–16788 10.1074/jbc.M109.096545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A., Linares J.F., Galvez A.S., Wikenheiser K., Flores J.M., Diaz-Meco M.T., Moscat J. 2008. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 13:343–354 10.1016/j.ccr.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Gao C., Cao W., Bao L., Zuo W., Xie G., Cai T., Fu W., Zhang J., Wu W., Zhang X., Chen Y.G. 2010. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol. 12:781–790 10.1038/ncb2082 [DOI] [PubMed] [Google Scholar]
- Hayes J.D., McMahon M. 2009. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 34:176–188 10.1016/j.tibs.2008.12.008 [DOI] [PubMed] [Google Scholar]
- He C., Levine B. 2010. The Beclin 1 interactome. Curr. Opin. Cell Biol. 22:140–149 10.1016/j.ceb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. 2008. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283:22847–22857 10.1074/jbc.M802182200 [DOI] [PubMed] [Google Scholar]
- Inoue K., Nakada K., Ogura A., Isobe K., Goto Y., Nonaka I., Hayashi J.I. 2000. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat. Genet. 26:176–181 10.1038/82826 [DOI] [PubMed] [Google Scholar]
- Itakura E., Mizushima N. 2011. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 192:17–27 10.1083/jcb.201009067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Lamark T., Sjøttem E., Larsen K.B., Awuh J.A., Øvervatn A., McMahon M., Hayes J.D., Johansen T. 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285:22576–22591 10.1074/jbc.M110.118976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Li Y., Pitti R., Lawrence D., Pham V.C., Lill J.R., Ashkenazi A. 2009. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 137:721–735 10.1016/j.cell.2009.03.015 [DOI] [PubMed] [Google Scholar]
- Johansen T., Lamark T. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 7:1–18 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., McEwan D.G., Novak I., Dikic I. 2009. A role for ubiquitin in selective autophagy. Mol. Cell. 34:259–269 10.1016/j.molcel.2009.04.026 [DOI] [PubMed] [Google Scholar]
- Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31:1007–1014 [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 131:1149–1163 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12:213–223 [DOI] [PubMed] [Google Scholar]
- Kraft C., Peter M., Hofmann K. 2010. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 12:836–841 10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- Lau A., Wang X.J., Zhao F., Villeneuve N.F., Wu T., Jiang T., Sun Z., White E., Zhang D.D. 2010. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell. Biol. 30:3275–3285 10.1128/MCB.00248-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell. 132:27–42 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 402:672–676 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- Mathew R., Karp C.M., Beaudoin B., Vuong N., Chen G., Chen H.Y., Bray K., Reddy A., Bhanot G., Gelinas C., et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 137:1062–1075 10.1016/j.cell.2009.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Noyes M.B., Zhu L.J., Lawson N.D., Wolfe S.A. 2008. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 26:695–701 10.1038/nbt1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco M.T. 2009. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 137:1001–1004 10.1016/j.cell.2009.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco M.T., Wooten M.W. 2007. Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 32:95–100 10.1016/j.tibs.2006.12.002 [DOI] [PubMed] [Google Scholar]
- Motohashi H., Yamamoto M. 2004. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10:549–557 10.1016/j.molmed.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Nezis I.P., Simonsen A., Sagona A.P., Finley K., Gaumer S., Contamine D., Rusten T.E., Stenmark H., Brech A. 2008. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 180:1065–1071 10.1083/jcb.200711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan B., Tong K.I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Kang M.I., Kobayashi A., Yokoyama S., Yamamoto M. 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 21:689–700 10.1016/j.molcel.2006.01.013 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., et al. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112:1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J.D., White E. 2010. Autophagy and metabolism. Science. 330:1344–1348 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B.E., Kaiser S.E., Shaler T.A., Ng A.C., Hara T., Hipp M.S., Lage K., Xavier R.J., Ryu K.Y., Taguchi K., et al. 2010. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 191:537–552 10.1083/jcb.201005012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C. 2006. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 443:780–786 10.1038/nature05291 [DOI] [PubMed] [Google Scholar]
- Santiago Y., Chan E., Liu P.Q., Orlando S., Zhang L., Urnov F.D., Holmes M.C., Guschin D., Waite A., Miller J.C., et al. 2008. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA. 105:5809–5814 10.1073/pnas.0800940105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Sanchez P., Lallena M.J., Diaz-Meco M.T., Moscat J. 1999. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J. 18:3044–3053 10.1093/emboj/18.11.3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman A.M., Karnovsky M.J., Wasserkrug H.L., Hanker J.S. 1968. Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB). J. Cell Biol. 38:1–14 10.1083/jcb.38.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Kokubu A., Gotoh M., Ojima H., Ohta T., Yamamoto M., Hirohashi S. 2008a. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 135:1358–1368: 1368: e1–e4 10.1053/j.gastro.2008.06.082 [DOI] [PubMed] [Google Scholar]
- Shibata T., Ohta T., Tong K.I., Kokubu A., Odogawa R., Tsuta K., Asamura H., Yamamoto M., Hirohashi S. 2008b. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 105:13568–13573 10.1073/pnas.0806268105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Misra V., Thimmulappa R.K., Lee H., Ames S., Hoque M.O., Herman J.G., Baylin S.B., Sidransky D., Gabrielson E., et al. 2006. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 3:e420 10.1371/journal.pmed.0030420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. 2011. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., Kominami E. 1999. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 10:1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve N.F., Lau A., Zhang D.D. 2010. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid. Redox Signal. 13:1699–1712 10.1089/ars.2010.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waguri S., Komatsu M. 2009. Biochemical and morphological detection of inclusion bodies in autophagy-deficient mice. Methods Enzymol. 453:181–196 10.1016/S0076-6879(08)04009-3 [DOI] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A.J., Heintz N. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 100:15077–15082 10.1073/pnas.2436255100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatloukal K., Stumptner C., Fuchsbichler A., Heid H., Schnoelzer M., Kenner L., Kleinert R., Prinz M., Aguzzi A., Denk H. 2002. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 160:255–263 10.1016/S0002-9440(10)64369-6 [DOI] [PMC free article] [PubMed] [Google Scholar]