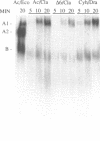
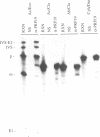
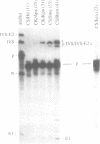
Abstract
Assembly of the spliceosome is a step-wise process and involves sequential binding of snRNAs to the pre-mRNA to form pre-splicing complex A2-1. Subsequent dissociation of U4 from the spliceosome is accompanied by formation of complex A1 (Genes Dev. 1, 1014-1027, 1987). We show that the 3' region of the intron sequence is not required for efficient assembly of the yeast spliceosome. Truncated precursor mRNA retaining only four or five nucleotides 3' to the TACTAAC box formed pre-splicing complex A1, kinetically the last pre-mRNA containing splicing complex identified. The subsequent cleavage--ligation reaction requires at least 23 nucleotides on the 3' side of the TACTAAC box in a sequence-independent manner. Immunoprecipitation with anti-PRP19 antibody showed that association of PRP19 with the spliceosome was also independent of the 3' region of the intron.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bindereif A., Green M. R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987 Aug;6(8):2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellini A., Parker R., McMahon J., Guthrie C., Rossi J. Activation of a cryptic TACTAAC box in the Saccharomyces cerevisiae actin intron. Mol Cell Biol. 1986 May;6(5):1571–1578. doi: 10.1128/mcb.6.5.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Fractionation and characterization of a yeast mRNA splicing extract. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2387–2391. doi: 10.1073/pnas.83.8.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Newman A. N., Lin R. J., McFarland G. D., Abelson J. N. Preparation and fractionation of yeast splicing extract. Methods Enzymol. 1990;181:89–96. doi: 10.1016/0076-6879(90)81114-a. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Tarn W. Y., Tsao T. Y., Abelson J. PRP19: a novel spliceosomal component. Mol Cell Biol. 1993 Mar;13(3):1876–1882. doi: 10.1128/mcb.13.3.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. D., Grabowski P. J., Sharp P. A., Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986 Mar 28;231(4745):1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., McPheeters D. S., Abelson J. In vitro assembly of yeast U6 snRNP: a functional assay. Genes Dev. 1989 Dec;3(12B):2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- Fouser L. A., Friesen J. D. Effects on mRNA splicing of mutations in the 3' region of the Saccharomyces cerevisiae actin intron. Mol Cell Biol. 1987 Jan;7(1):225–230. doi: 10.1128/mcb.7.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouser L. A., Friesen J. D. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell. 1986 Apr 11;45(1):81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Garber R. L., Kuroiwa A., Gehring W. J. Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila. EMBO J. 1983;2(11):2027–2036. doi: 10.1002/j.1460-2075.1983.tb01696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Blanco M. A., Jamison S. F., Sharp P. A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989 Dec;3(12A):1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Smith J., Claude A., Lin R. J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992 Jun;11(6):2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Lin R. J., Lustig A. J., Abelson J. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1987 Mar;1(1):7–18. doi: 10.1101/gad.1.1.7. [DOI] [PubMed] [Google Scholar]
- Lin R. J., Newman A. J., Cheng S. C., Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985 Nov 25;260(27):14780–14792. [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992 Nov 27;71(5):803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Lin R. J., Cheng S. C., Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985 Aug;42(1):335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rymond B. C., Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. 1986 Nov 27-Dec 3Nature. 324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Reich C. I., VanHoy R. W., Porter G. L., Wise J. A. Mutations at the 3' splice site can be suppressed by compensatory base changes in U1 snRNA in fission yeast. Cell. 1992 Jun 26;69(7):1159–1169. doi: 10.1016/0092-8674(92)90637-r. [DOI] [PubMed] [Google Scholar]
- Rodriguez J. R., Pikielny C. W., Rosbash M. In vivo characterization of yeast mRNA processing intermediates. Cell. 1984 Dec;39(3 Pt 2):603–610. doi: 10.1016/0092-8674(84)90467-7. [DOI] [PubMed] [Google Scholar]
- Rosbash M., Séraphin B. Who's on first? The U1 snRNP-5' splice site interaction and splicing. Trends Biochem Sci. 1991 May;16(5):187–190. doi: 10.1016/0968-0004(91)90073-5. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991 Mar;7(3):79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Specific and stable intron-factor interactions are established early during in vitro pre-mRNA splicing. Cell. 1985 Nov;43(1):131–142. doi: 10.1016/0092-8674(85)90018-2. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Rosbash M. Cleavage of 5' splice site and lariat formation are independent of 3' splice site in yeast mRNA splicing. Nature. 1985 Oct 24;317(6039):735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Torrey D. D., Rosbash M. A novel role for the 3' region of introns in pre-mRNA splicing of Saccharomyces cerevisiae. Genes Dev. 1987 May;1(3):238–246. doi: 10.1101/gad.1.3.238. [DOI] [PubMed] [Google Scholar]
- Sawa H., Abelson J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5' splice site during the splicing reaction in yeast. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H., Shimura Y. Association of U6 snRNA with the 5'-splice site region of pre-mRNA in the spliceosome. Genes Dev. 1992 Feb;6(2):244–254. doi: 10.1101/gad.6.2.244. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., Hazelrigg T. I., Polisky B. A., Pirrotta V., Scalenghe F., Kaufman T. C. The molecular organization of the Antennapedia locus of Drosophila. Cell. 1983 Dec;35(3 Pt 2):763–776. doi: 10.1016/0092-8674(83)90109-5. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Sontheimer E. J., Steitz J. A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993 Dec 24;262(5142):1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kandels-Lewis S. 3' splice site recognition in S. cerevisiae does not require base pairing with U1 snRNA. Cell. 1993 May 21;73(4):803–812. doi: 10.1016/0092-8674(93)90258-r. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W. Y., Lee K. R., Cheng S. C. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol Cell Biol. 1993 Mar;13(3):1883–1891. doi: 10.1128/mcb.13.3.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W. Y., Lee K. R., Cheng S. C. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10821–10825. doi: 10.1073/pnas.90.22.10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U., Parker R., Tamm J., Iimura Y., Rossi J., Abelson J., Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986 Jul;5(7):1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992 Sep 25;257(5078):1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Whittaker E., Lossky M., Beggs J. D. Affinity purification of spliceosomes reveals that the precursor RNA processing protein PRP8, a protein in the U5 small nuclear ribonucleoprotein particle, is a component of yeast spliceosomes. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2216–2219. doi: 10.1073/pnas.87.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean S. L., Lin R. J. U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. Mol Cell Biol. 1991 Nov;11(11):5571–5577. doi: 10.1128/mcb.11.11.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]