Abstract
Background
Recent research has implicated that mutations in the neurexin-1 (NRXN1) gene on chromosome 2p16.3 might play a role in schizophrenia, autism, and nicotine dependence. In order to explore the association of NRXN1 polymorphisms with schizophrenia, we made a case-control association study in Chinese Han population.
Methods
We examined six tag single nucleotide polymorphisms (SNPs) spanning 116.7 kb of NRXN1 in 768 schizophrenic patients and 738 healthy control subjects. The association of NRXN1 polymorphisms with schizophrenia and the age-at-onset of this disease were explored.
Results
Our results showed that four SNPs of NRXN1 gene were significantly associated with schizophrenia (rs10490168: G > A, p = 0.017; rs2024513: A > G, p = 0.006; rs13382584: T > C, p = 0.009; and rs1558852: G > A, p = 0.031). Furthermore, the association of SNP rs2024513 with schizophrenia remained significance after the Bonferroni correction. Haplotypes consisting of above six SNPs also showed significantly associated with schizophrenia (global chi-square = 14.725, p = 0.022). A protective haplotype AGTGCA remained associated with schizophrenia, even after 10,000 permutation tests (empirical p-value = 0.043). However, we did not find any association with age-at-onset of schizophrenia with NRXN1 polymorphisms.
Conclusions
Our findings suggest that NRXN1 might represent a major susceptibility gene for schizophrenia in Chinese Han population.
Keywords: Schizophrenia, Neurexin-1 (NRXN1), Haplotype, age-at-onset
Background
Schizophrenia is a common psychiatric disorder characterized by profound disturbances of thinking, emotion, and social functioning. It affects approximately 1% of the worldwide population and accounts for about 2.5% of healthcare costs [1]. Aberrant synaptic connectivity is a prominent feature of schizophrenia's neuropathology [2]. Recently, several groups have identified schizophrenic patients who showed microdeletions disrupting the promoter and N-terminal encoding exons of neurexin 1 (NRXN1) alpha [3-5]. Kirov et al. have found a deletion disrupting the NRXN1 gene at 2p16.3 in a mother and two affected siblings from one family, and identical twins concordant for child-onset schizophrenia, respectively [3-5]. Furthermore, copy number variations (CNVs) in NRXN1 gene have also previously been implicated in autism, mental retardation and nicotine dependence [6-11].
Neurexins, including NRXN1, NRXN2, NRXN3, are cell-surface receptors that bind neuroligins (NLGNs) and form a Ca2+-dependent neurexin/neuroligin complex at synapses in the central nervous system [12,13]. The NRXN - NLGN complex plays an important role during the GABAergic and glutamatergic neurotransmission [12]. There is prevailing evidence implicating both neurexins and neuroligins as primary factors in neuropsychiatric disorders [3-11].
All these indicate that NRXN1 might be a strong candidate gene for schizophrenia [3-6]. Resequencing has been applied to identify the rare mutations of this gene, but the linkage and association studies of NRXN1 polymorphisms in Chinese Han population remained still insufficient. In this study, we attempted to investigate the association between the NRXN1 gene polymorphisms and schizophrenia and age-at-onset of this disease in 768 patients with schizophrenia and 738 healthy control subjects of Chinese Han population.
Methods
Our study sample consisted of 768 patients with schizophrenia (360 men and 408 women; mean age: 33.5 ± 8.7 years) and 738 healthy controls (358 men and 380 women; mean age: 32.2 ± 6.4 years). All of the participants were Chinese Han descendants. The patients were recruited from the Institute of Mental Health, Peking University, China. The consensus diagnoses were made by at least two experienced psychiatrists according to the Diagnosis and statistic manual of mental disorders, 4th edition (DSM-IV) criteria for schizophrenia. The age of clinical symptoms onset (12~53 years, mean 23.8 ± 7.8 years) was retrospectively estimated as the time of emergence of any schizophrenic symptoms according to the DSM-IV. None had severe medical complications. Healthy controls were recruited from communities with simple non-structured interview performed by psychiatrists, who excluded individuals with history of mental health and neurological diseases. The control subjects live in the same area with patients, and were group-matched to patients by gender, age, and ethnicity. The objectives and procedures of the study were explained to all participants and written informed consent was obtained. Research ethics committee approval was obtained from the Ethical Committee of Institute of Mental Health, Peking University.
Peripheral blood samples were collected from all subjects. Genomic DNA was extracted using the Qiagen QIAamp DNA Mini Kit. Considering Kirov et al.'s report that micordeletions spanning the promoter and the first exon of the NRXN1 gene might increase the risk of developing schizophrenia, we selected six tag single nucleotide polymorphisms (tagSNPs) locating the promoter and the first exon of this gene, according to the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP) [3-5]. All of the six tagSNPs were genotyped with the TaqMan genotyping methods according to the manufacturer's protocol, where allelic discrimination and analysis were performed using an ABI Prism 7300 Sequence Detection System (Applied Biosystems, CA). The information of ABI Taqman assays and context sequences of six SNPs are given in Table 1. The standard TaqMan® SNP genotyping assay protocol was performed for polymerase chain reactions (PCRs), which contained 10 ng of DNA, 5 μL of 2 ×TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA), 0.5 μL of 20 × SNP Genotyping Assay Mix (Applied Biosystems, Foster City, CA), and 4.5 μL of water, for a total volume of 10 μL. The conditions of PCR amplification were as follows: 1 enzyme activation step at 95.0°C for 10 min, and 40 alternating cycles of denaturation at 92.0° C for 15 s and re-annealing and extension at 60.0°C for 60 s. To evaluate the quality of genotyping, 5% of the samples were randomly selected and re-genotyped, and the genotyping consistency rate was more than 98%.
Table 1.
Information of TaqMan assays and context sequences of 6 SNPs examinaed
| ID | SNPs | Position | Location | Context Sequence ([VIC/FAM]) |
|---|---|---|---|---|
| 1 | rs10490168 | 50988978 | intron | TGAATTGATAAAACAGTAACACTGA[C/T]AAAAATACATACACACATTCACACAC |
| 2 | rs2024513 | 51005523 | intron | GAAGTGTTCTTTCTTAGATACATGA[A/G]GTCTTGGTAACCTTAATGGCTATTT |
| 3 | rs10174398 | 51049105 | intron | ACCAGTCCACCTCTATCTGTACCCA[C/T]GCTCTCTCCATATAATGTTCTTCTC |
| 4 | rs10195460 | 51082210 | intron | CTAAAGGGAGATCAATTAAGCATAC[A/G]TTTTTCCACCTTTTAAAATAATACT |
| 5 | rs13382584 | 51100798 | intron | TCTTTACAAATGTAACCACCACCCA[C/T]ATCATGCCCAATGCTCCATTGTTTT |
| 6 | rs1558852 | 51105641 | intron | TTAATACATGATCTGTATTGGGAGA[A/G]TCATCCATTCTCAATTAATTATTCA |
The statistic power of our sample size was calculated by the genetic power calculator (GPC, http://pngu.mgh.harvard.edu/~purcell/gpc/cc2.html) [14]. Deviation of the genotype counts from the Hardy-Weinberg equilibrium was tested using a chi-square goodness-of-fit test. The pairwise linkage disequilibrium (LD) analysis was applied to detect the inter-marker relationship, using D'-values. The case-control association analysis was performed by the Haploview version 4.1 (http://www.broad.mit.edu/mpg/haploview/), a powerful software platform for computing single locus and multi-marker haplotype association tests [15]. Bonferroni corrections for multiple tests were carried out to exclude type I errors. For the haplotype analyses, 10,000 times of permutation tests were performed to control false positive results. The LD plot of 6 SNPs at the 5'UTR of NRXN1 gene and its regional plots across this gene were constructed by using the Haploview v4.1 software in 738 healthy control subjects. To indicate the relative position of above 6 SNPs and the other SNPs across the NRXN1 gene download from the HapMap stage II dataset, we then further constructed the regional LD plots using the LocusZoom (http://csg.sph.umich.edu/locuszoom/) [16]. Associations between age-at-onset and different genotype carriers were determined by one-way ANOVA tests with the SPSS 13.0. Considering that the mean age of control group was younger than that of case group, we implemented the logistic regression analysis, with diagnosis as dependent variable, and 6 tagSNPs and age at enrolled of subjects as independent variables. Results were considered significant at two-tailed P < 0.05.
Results
Six SNPs in the NRXN1 gene were genotyped in 768 schizophrenia patients and 738 healthy control subjects of Chinese Han population. The size of our sample was sufficient to detect a significant difference with a power of more than 90% assuming an odds ratio (OR) values of AA as 1.7 with a minor allele frequency of 0.1. The genotype distributions of the six SNPs for patients and controls were in Hardy-Weinberg equilibrium (Table 2). Allele and genotype frequencies of the six SNPs between patients and controls are shown in Table 2. Significant differences were found in allele frequencies between patients and controls at four SNPs, rs10490168 (G > A, p = 0.017), rs2024513 (A > G, p = 0.006), rs13382584 (T > C, p = 0.009), and rs1558852 (G > A, p = 0.031). However, only the difference in the allele frequencies of rs2024513 remained significance after the Bonferroni correction. For control the confounding effects of age of subjects, we used the logistic regression to identify whether the age influence the marker-diagnosis effects. However, the age of subjects when they were enrolled in the current study did not showed any significant confounding effects on maker-diagnosis association (p = 0.058).
Table 2.
Comparison of genotype and allele frequencies of six SNPs at the NRXN1 gene between schizophrenic patients and healthy control subjects
| Makers | Genotype N (Freq.) | Chi-square (df = 2) | p- value | HWEP | Allele N (Freq.) | Chi-square (df = 1) | p- value | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10490168 | AA | AG | GG | A | G | ||||||
| Cases | 10(0.013) | 173(0.225) | 585(0.762) | 6.049 | 0.048 | 0.485 | 193(0.126) | 1343(0.874) | 5.677 | 0.017 | 0.78(0.63-0.96) |
| Controls | 13(0.018) | 204(0.276) | 521(0.706) | 0.168 | 230(0.156) | 1246(0.844) | |||||
| rs2024513 | AA | AG | GG | A | G | ||||||
| Cases | 544(0.708) | 206(0.268) | 18(0.023) | 7.521 | 0.023 | 0.772 | 1294(0.842) | 242(0.158) | 7.327 | 0.006 | 1.30(1.07-1.56) |
| Controls | 480(0.650) | 228(0.309) | 30(0.041) | 0.655 | 1188(0.805) | 288(0.195) | |||||
| rs10174398 | CC | CT | TT | C | T | ||||||
| Cases | 276(0.359) | 359(0.467) | 133(0.173) | 0.727 | 0.695 | 0.382 | 911(0.593) | 625(0.407) | 1.064 | 0.397 | 1.06(0.92-1.23) |
| Controls | 254(0.344) | 345(0.467) | 139(0.188) | 0.256 | 853(0.578) | 623(0.422) | |||||
| rs10195460 | AA | AG | GG | A | G | ||||||
| Cases | 61(0.079) | 318(0.414) | 389(0.507) | 1.197 | 0.549 | 0.721 | 440(0.286) | 1096(0.714) | 0.972 | 0.323 | 0.92(0.79-1.08) |
| Controls | 62(0.084) | 323(0.438) | 353(0.478) | 0.321 | 447(0.303) | 1029(0.697) | |||||
| rs13382584 | CC | CT | TT | C | T | ||||||
| Cases | 14(0.018) | 200(0.260) | 554(0.721) | 7.075 | 0.029 | 0.404 | 228(0.148) | 1308(0.852) | 6.821 | 0.009 | 0.77(0.64-0.94) |
| Controls | 23(0.031) | 225(0.305) | 489(0.664) | 0.638 | 271(0.184) | 1203(0.816) | |||||
| rs1558852 | AA | AG | GG | A | G | ||||||
| Cases | 234(0.305) | 383(0.499) | 151(0.197) | 4.666 | 0.097 | 0.799 | 851(0.554) | 685(0.446) | 4.627 | 0.031 | 0.85(0.74-0.99) |
| Controls | 259(0.351) | 357(0.484) | 122(0.165) | 0.956 | 875(0.593) | 601(0.407) | |||||
Freq., frequency; HWEP: the p-value for Hardy-Weinberg equilibrium tests; OR, odds ratio; CI, confidence interval.
To further analyze the haplotype structure in our sample, we computed pairwise LD for all possible combinations of the six SNPs using D' and r2 values. Figure 1 showed the LD plot of constructed by the six SNPs locating at the 5'UTR of NRXN1 and regional LD plots across the NRXN1 gene. Haplotypes consisting of above six SNPs also showed significantly associated with schizophrenia (global chi-square = 14.725, p = 0.022). A protective haplotype AGTGCA remained associated with schizophrenia, even after 10,000 permutation tests (empirical p-value = 0.043).
Figure 1.
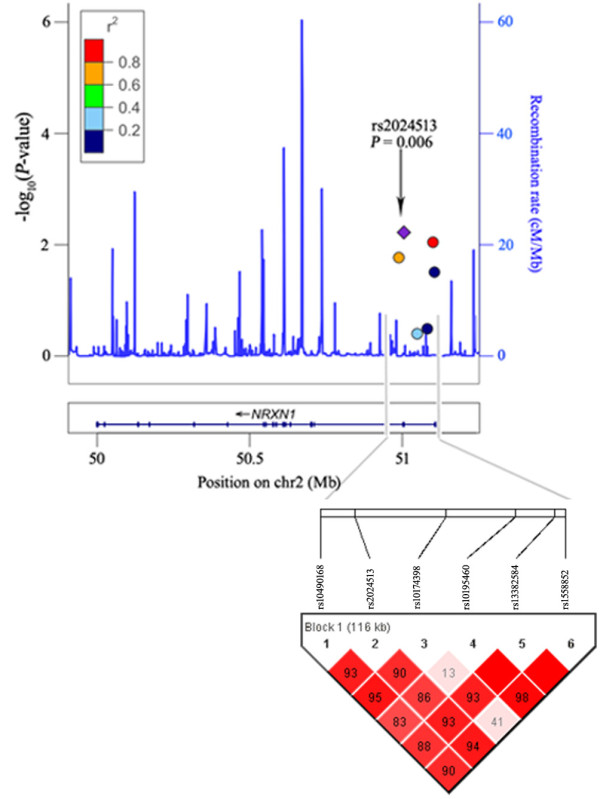
Linkage disequilibrium plot consist of 6 SNPs at the NRXN1 gene and its regional plots. Pairwise linkage disequilibrium (LD) was computed for all possible combinations of the six SNPs using D' values. Six SNPs were plotted with the -log10(P-values) in the NRXN1 genomic position (in the University of California Santa Cruz March 2006 human reference sequence, hg18). Estimated recombination rates of SNPs in NRXN1 regions (taken from HapMap Japanese in Tokyo (JPT) + Chinese Han in Beijing (CHB)) are plotted to reflect the local LD structure. The index association SNP is represented by a diamond. Color of the remaining SNPs (circles) indicates LD with the index SNP based on pairwise r2 values from HapMap data.
To explore the potential association of age-at-onset of schizophrenia with NRXN1 polymorphisms, we carried out association analysis tests using the six SNPs at NRXN1 gene. However, the results of one-way ANOVA tests revealed there was no association of age-at-onset of schizophrenia with six SNPs at NRXN1 gene (p > 0.05) (data not shown). Furthermore, we also did not find the association between NRXN1 polymorphisms and early-onset schizophrenia.
Discussion
Neurexins, including NRXN1 and other members of the neurexin family, are synaptic neuronal adhesion molecules. NRXNs could bind neuroligins (NLGNs) to form a Ca2+ - dependent NRXN - NLGN synpatic complex in the central nervous system. This NRXN - NLGN complex is required for neurotransmission at synapses and is involved in the presynaptic and postsynaptic differentiation of GABAergic and glutamatergic synapse [12,17-20]. The de novo alterations of the NRXN1 gene and their roles in the pathogenesis of autism and schizophrenia have been reported by some studies [4,21,22]. Based on these findings, we hypothesized that the NRXN1 might be a strong candidate gene for schizophrenia. However, there has been no further evidence of association between NRXN1 polymorphisms and schizophrenia in Chinese Han population. In the present study, we investigated the association of NRXN1 polymorphisms and schizophrenia in 768 cases and 738 healthy control subjects of Chinese Han population. We detected six tagSNPs of NRXN1 gene in our samples. Significant differences were found in allele frequencies between patients and controls at four SNPs (rs10490168, rs2024513, rs13382584, rs1558852). However, only the difference in the allele frequencies of rs2024513 remained significance after the stringent Bonferroni multiple-test correction. Further association analysis of haplotypes consist of six SNPs at the NRXN1 gene with schizophrenia were performed. Three haplotypes at the NRXN1 gene were found to be associated with schizophrenia. The protective haplotype AGTGCA consist of above 6 SNPs remained significantly associated with schizophrenia, even after Bonferroni correction (Table 3).
Table 3.
Comparison of haplotype frequencies at the NRXN1 gene between schizophrenic patients and healthy control subjects
| Haplotype | Case (Freq.) | Control (Freq.) | Chi - square | p - value | OR (95%CI) |
|---|---|---|---|---|---|
| AGTGCA | 167.76(0.109) | 207.23(0.141) | 6.770 | 0.009 | 0.75(0.60-0.93) |
| GACATA | 208.91(0.136) | 233.02(0.158) | 2.904 | 0.088 | 0.84(0.68-1.03) |
| GACGTA | 164.41(0.107) | 134.86(0.091) | 2.072 | 0.150 | 1.19(0.94-1.52) |
| GACGTG | 510.60(0.332) | 453.08(0.307) | 2.287 | 0.130 | 1.13(0.97-1.32) |
| GATATA | 214.77(0.140) | 192.85(0.131) | 0.542 | 0.461 | 1.08(0.88-1.34) |
| GATGTG | 157.26(0.102) | 135.07(0.092) | 1.018 | 0.312 | 1.13(0.89-1.44) |
| GGTGCA | 39.15(0.025) | 49.39(0.034) | 1.682 | 0.194 | 0.76(0.49-1.16) |
| Global | 14.725 | 0.022 |
Freq., frequency; OR: odds ratio; CI: confidence interval
By microarray analysis in 2,977 European patients with schizophrenia and 33,746 European controls, Rujescu et al. identified 66 deletions and 5 duplications in the NRXN1 gene: 12 deletions and 2 duplications occurred in schizophrenic patients (0.47%), compared to 49 and 3 in controls (0.15%) [5]. There was no common breakpoint, and the CNVs varied from 18 to 420 kb. No CNVs were found in the NRXN2 or NRXN3 genes. By restricting the association analysis to CNVs that disrupted exons, they identified a significant association with a high odds ratio (0.24% cases vs. 0.015% controls, p = 0.0027, OR = 8.97). Rujescu et al suggested that NRXN1 deletions affecting exons may confer risk of schizophrenia [5]. Several previous studies have suggested that the NRXN1 gene might be the susceptibility gene of schizophrenia and other neurodevelopmental disorders.
Kirov et al.'s reports CNVs in NRXN1 gene implicated with schizophrenia and other mental disorders mostly are the rare polymorphisms [3]. Several other published literatures also supported that the rare CNVs in NRXN1 gene may be associated with schizophrenia and other mental disorders. However, schizophrenia has been regarded as a complex disease, so there might be many susceptibility genes associated with this disease. To explain the association of polymorphisms with complex disorders, we have two models, the 'common disease/common variant' (CDCV) and 'common disease/rare variant' (CDRV) model [23]. The CDCV model of schizophrenia suggests that relative common (often > 5%) genetic variants in population might confer minor or modest risk (i.e., OR = 1.1-1.5). On the other hand, the CDRV model postulates that complex traits might derive from many rare mutations for individuals (usually <1%) but with relative strong effects (i.e., perhaps OR > 10). In the present study, we want to discuss that common SNPs in NRXN1 gene, other than rare CNVs, might also be associated with schizophrenia in Chinese Han population. We focused on the association of six SNPs spanning a 117.6 kb region locating in an intron near the NRXN1 5'UTR with schizophrenia. And our data of both SNPs and haplotypes showed that the NRXN1 might be a susceptibility gene of schizophrenia in Chinese Han population.
Walsh et al. reported that novel deletions and duplications of NRXN1 gene presented more frequently cases (15%) and young-onset cases (20%) with schizophrenia than those in controls (5%) [4]. In the present study, we did not do any association analyses of child-onset schizophreia with NRXN1 polymorphisms, since we have only five patients with age-at-onset less than 12 years old in our 768 patients with schizophrenia. Although we did association tests between early-onset (<18 years old) schizophrenia and NRXN1 polymorphisms, we did not find an association. In all, we did not find any association of NRXN1 polymorphisms with age-at-onset of schizophrenia. This may be partly explained by the ethnic difference across various populations. The diversity of genotype and allele distributions between two populations might be responsible for the discrepancy. Another possibility is that different genetic markers (SNPs and CNVs) might be linked with different disease status or clinical characteristics. Furthermore, the definition of age-at-onset of schizophrenia may also vary across studies. These diversities of clinical assessment also in some extent elucidated for the inconsistent findings across studies. Other clinical examinations or endophenotypes of schizophrenia need to be evaluated when further genetic association studies are implemented. The relationship between the genetic variants in the middle or 3'-end region of NRXN1 gene and schizophrenia would be verified in the future study.
Conclusions
Our case-control association study suggested that NRXN1 gene may play a role in genetic susceptibility to schizophrenia. On the other hand, the different ethnic genetic structure of NRXN1 gene might influence the association of the polymorphisms with age-at-onset. Furthermore, our results also reinforce the need for the detailed LD mapping, CNV analysis of NRXN1 in different population or other neurodevelopmental disorders. The biological effect of risk variants and haplotypes in the NRXN1 gene associated with schizophrenia, and their role in disease pathogenesis and clinical characteristics, needs to be explored.
List of abbreviations
NRXN1: neurexin-1; SNP: single nucleotide polymorphism; CNV: copy number variations; NLGN: neuroligins; DSM-IV: Diagnosis and statistic manual of mental disorders, 4th edition; PCR: polymerase chain reactions; GPC: genetic power calculator; LD: linkage disequilibrium; OR: odds ratio; ASD: autism spectrum disorder; CDCV: common disease/common variant.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DZ and WY participated in the design of the study and made final approval of the version to be published. WY and YY were involved in drafting the manuscript and data analysis. WY, YY, YZ, TL, XH, LW, and YR carried out the molecular genetic examination. WY, YY, YZ, XH, LW, and LL conducted sample selection and data management. All authors read and approved the final manuscript.
Contributor Information
Weihua Yue, Email: dryue@bjmu.edu.cn.
Yongfeng Yang, Email: yongfeng_200888@126.com.
Yanling Zhang, Email: ylzhang@bjmu.edu.cn.
Tianlan Lu, Email: tianlan_lu@126.com.
Xiaofeng Hu, Email: huxf2008ok@163.com.
Lifang Wang, Email: lifangwang@bjmu.edu.cn.
Yanyan Ruan, Email: ruanyanyan@hotmail.com.
Luxian Lv, Email: lvx928@126.com.
Dai Zhang, Email: zhangdai@bjmu.edu.cn.
Acknowledgements and Fundings
We extend our gratitude to all subjects who participated in this study. This work was supported by grants from the National Natural Science Foundation of China (30870896, 81071087), the National High Technology Research and Development Program of China (2009AA022702), the National Basic Research Program of China (2007CB512301), National Key Technology R&D Program (2007BAI17B04).
References
- Meltzer D. Perspective and the measurement of costs and benefits for cost-effectiveness analysis in schizophrenia. J Clin Psychiatry. 1999;60(Suppl 3):32–35. discussion 36-37. [PubMed] [Google Scholar]
- Stephan KE, Baldeweg T, Friston KJ. Synaptic plasticity and dysconnection in schizophrenia. Biol Psychiatry. 2006;59(10):929–939. doi: 10.1016/j.biopsych.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M. et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17(3):458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM. et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietiläinen OP, Barnes MR, Toulopoulou T. et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18(5):988–96. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A. et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409(1):10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ. et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J. et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir FR, Baross A, Delaney AD, Eydoux P, Fernandes ND, Pugh T. et al. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2007;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- Nussbaum J, Xu Q, Payne TJ, Ma JZ, Huang W, Gelernter J, Li MD. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum Mol Genet. 2008;17(11):1569–1577. doi: 10.1093/hmg/ddn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF. et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissner C, Klose M, Fairless R, Missler M. Mutational analysis of the neurexin/neuroligin complex reveals essential and regulatory components. Proc Natl Acad Sci USA. 2008;105(39):15124–15129. doi: 10.1073/pnas.0801639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Fernandez-Chacon R, Südhof TC. The making of neurexins. J Neurochem. 1998;71(4):1339–1347. doi: 10.1046/j.1471-4159.1998.71041339.x. [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudanova I, Tabuchi K, Rohlmann A, Sudhof T, Missler M. Deletion of alpha-neurexins does not cause a major impairment of axonal pathfinding or synapse formation. J Comp Neurol. 2007;502(2):261–274. doi: 10.1002/cne.21305. [DOI] [PubMed] [Google Scholar]
- Kang Y, Zhang X, Dobie F, Wu H, Craig A. Induction of GABAergic post-synaptic differentiation by alpha-neurexins. J Biol Chem. 2008;283(4):2323–2334. doi: 10.1074/jbc.M703957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lise M, El-Husseini A. The neuroligin and neurexin families: from structure to function at the synapse. Cell Mol Life Sci. 2006;63(16):1833–1849. doi: 10.1007/s00018-006-6061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer R, Gottmann K, Sudhof T. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423(6943):939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Kirov G, Rujescu D, Ingason A, Collier DA, O'Donovan MC, Owen MJ. Neurexin 1 (NRXN1) deletions in schizophrenia. Schizophr Bull. 2009;35(5):851–854. doi: 10.1093/schbul/sbp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y. et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82(1):199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Steering Committee. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14(1):10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]


